Abstract
GDNF (glial cell-line-derived neurotrophic factor), and the closely related cytokines artemin and neurturin, bind strongly to heparin. Deletion of a basic amino-acid-rich sequence of 16 residues N-terminal to the first cysteine of the transforming growth factor β domain of GDNF results in a marked reduction in heparin binding, whereas removal of a neighbouring sequence, and replacement of pairs of other basic residues with alanine had no effect. The heparin-binding sequence is quite distinct from the binding site for the high affinity GDNF polypeptide receptor, GFRα1 (GDNF family receptor α1), and heparin-bound GDNF is able to bind GFRα1 simultaneously. The heparin-binding sequence of GDNF is dispensable both for GFRα1 binding, and for activity for in vitro neurite outgrowth assay. Surprisingly, the observed inhibition of GDNF bioactivity with the wild-type protein in this assay was still found with the deletion mutant lacking the heparin-binding sequence. Heparin neither inhibits nor potentiates GDNF–GFRα1 interaction, and the extracellular domain of GFRα1 does not bind to heparin itself, precluding heparin cross-bridging of cytokine and receptor polypeptides. The role of heparin and heparan sulfate in GDNF signalling remains unclear, but the present study indicates that it does not occur in the first step of the pathway, namely GDNF–GFRα1 engagement.
Keywords: artemin, GDNF family receptor α1 (GFRα1), glial cell-line-derived neurotrophic factor (GDNF), heparan sulfate, heparin, PC12 cells
Abbreviations: ART, artemin; BMP-2, bone morphogenetic protein 2; DMEM, Dulbecco's modified Eagle's medium; FGF-2, fibroblast growth factor-2; GAG, glycosaminoglycan; GDNF, glial cell-line-derived neurotrophic factor; GFRα, GDNF family receptor α; GFL, GDNF family ligand; HS, heparan sulfate; NTN, neurturin; 2-OST, 2-O-sulfotransferase PSP, persephin; rhGDNF, recombinant human GDNF; TBS/T, Tris-buffered saline containing 0.05% Tween 20, TGF-β, transforming growth factor β
INTRODUCTION
GDNF (glial cell-line-derived neurotrophic factor), a member of the TGF-β (transforming growth factor β) cytokine superfamily, is a homodimeric cytokine of disulfide-bridged N-glycosylated subunits, each 18–22 kDa. It is the prototypic member of a subfamily of four neurotrophic factors, the GFLs (GDNF family ligands), which also comprises ART (artemin), NTN (neurturin) and PSP (persephin) [1]. The GFLs exhibit high sequence similarity and, in their canonical signalling pathway, share the cell surface tyrosine kinase Ret as the transmembrane signalling component of their receptor complexes [1]. The receptor complexes also contain a high affinity GPI (glycosylphosphatidylinositol)-linked subunit, termed a GFRα (GDNF family receptor α), with one GFRα for each GFL, which is GFRα1 in the case of GDNF. Although limited cross-talk between some GFLs and GFRαs has been reported [1], this may have little functional significance [2]. GDNF–GFRα1 interaction leads to engagement and activation of Ret [1]. Beyond this canonical GFRα–Ret signalling mechanism, there is evidence for other pathways, such as Ret-independent signalling [3–5]. In particular, NCAM (neural cell-adhesion molecule) has been proposed as a signalling co-receptor with GFRα1 for GDNF [6]. However, the physiological significance of this latter pathway may be limited [7].
A considerable body of evidence shows that GDNF is potently neuroprotective both in vivo and in vitro. Since GDNF promotes, in particular, the differentiation and survival of dopaminergic neurones, there has been much interest in the prospect of employing GDNF as a therapeutic agent in Parkinson's disease. Indeed two small-scale trials of infusion of GDNF into the putamen of patients with established Parkinsonism have resulted in sustained improvements in symptoms, in the absence of serious side effects [8,9]. These findings have yet to be replicated in a large-scale clinical trial.
Homozygous GDNF gene deletion is perinatally lethal and reveals an essential non-redundant role for GDNF in the enteric nervous system. There are also selective neuronal deficits in the central nervous system [10–12]. However, the most striking feature of the GDNF−/− phenotype is the complete absence of kidneys. Kidney defects including unilateral or bilateral kidney agenesis are also observed in a proportion of GDNF+/− heterozygous mice. The GDNF−/− phenotype is essentially recapitulated by the targeted deletion of GFRα1 [13,14] or Ret [15]. This genetic evidence, together with the analysis of GDNF and receptor expression in the embryonic tissues which give rise to the mature kidney, demonstrates that a critical early step in kidney organogenesis is the molecular encounter between GDNF, expressed in the metanephric blastema, and GFRα1 and Ret expressed on the tip of the invading ureteric bud [16,17].
In common with a number of diverse cytokines, GDNF binds strongly to the highly sulfated polysaccharide heparin [18]. Although heparin is confined to mast cell granules, the structurally related GAG (glycosaminoglycan) HS (heparan sulfate) is widely distributed on cell surfaces and in the extracellular matrix. Heparin and HS are long unbranched polysaccharides, which, owing to several enzyme modifications during biosynthesis including the variable introduction of sulfates with N-, 2-O-, 3-O- and 6-O- linkages, have considerable sequence diversity [19]. The enzymology of heparin and HS biosynthesis is complex because of the large number of enzyme-catalysed steps involved, with, in general, each of these enzymes being encoded by multiple gene loci. An exception here is HS 2-OST (2-O-sulfotransferase), which is only encoded at a single locus [19]. The homozygous targeting of this locus during a screen for genes important in development resulted in a complete absence of kidneys along with minor skeletal defects [20]. Given the considerable number of growth factors and other proteins that interact with HS and the importance of sulfate groups including 2-O-sulfates in these interactions, it is surprising that the developmental abnormalities in HS 2-OST−/− mice are rather selective. The resemblance of this phenotype to those of GDNF, GFRα1 and Ret gene deletion raises the hypothesis that loss of GDNF signalling is a primary underlying defect. This hypothesis is supported by the transitorily high expression of HS 2-OST in the metanephric blastema at the time of ureteric bud invasion [20]. We therefore characterized the specificity of the interaction between GDNF and heparin/HS and showed that it demonstrates an unusually high dependence on the presence of 2-O-sulfates [21]. This provides biochemical support for the view that the critical developmental activity of GDNF has an essential requirement for 2-O-sulfated HS. HS GAG is also seen to be essential for GDNF signalling in vitro. Barnett et al. [22] have shown that pre-treatment of both dorsal root ganglia cultures and PC12 cells with chlorate, a sulfation inhibitor, renders them insensitive to GDNF-induced neurite outgrowth. Moreover the GDNF-induced motility of Madin–Darby canine kidney cells, transfected to express Ret and GFRα1, was inhibited not only by chlorate pre-treatment, but also by digestion with heparinase III. This enzyme also abolished GDNF-induced Ret phosphorylation in these cells [23]. Low concentrations of exogenous heparin block the neurite outgrowth induced in PC12 cells by the combination of GDNF and recombinant soluble GFRα1-Fc chimaeric protein [23]. However, exogenous heparin was shown elsewhere to potentiate the activity of GDNF in the induction of tyrosine hydroxylase gene expression in neuroblastoma cells [24]. Thus the role of heparin/HS in GDNF signalling remains unclear.
Given the developmental importance of GDNF and its significant therapeutic potential in a major neurodegenerative disease, we sought a better understanding of the molecular basis of the interaction between this cytokine and heparin-related GAG, and of how this interaction affects GDNF signalling. To this end, we have investigated the localization of the heparin-binding site on GDNF by mutagenesis directed against clusters of basic amino acids. Subsequently we have employed a GDNF mutant with markedly reduced heparin-binding activity to investigate the role of heparin in GDNF–GFRα1 interactions.
EXPERIMENTAL
Materials
Recombinant human and rat GDNFs, human ART, human NTN, and human GFRα1–Fc chimaeric protein, together with the affinity-purified goat polyclonal antibodies with respective specificities were purchased from R&D Systems Europe (Abingdon, Oxon, U.K.). Affinity purified rabbit anti-GDNF antibody (sc-328) was from Santa Cruz Biotechnology. Alkaline phosphatase-conjugated rabbit anti-goat IgG, anti-FLAG M2 monoclonal antibody, collagen, poly-D-lysine (70–150 kDa; catalogue number P0899), porcine intestinal mucosal heparin (sodium salt, grade I-A) and BSA (96% electrophoresis grade), used for immunoassay blocking, were obtained from Sigma–Aldrich. Affinity-purified anti-FLAG rabbit polyclonal antibody was purchased form Cayman Chemical Co. (Ann Arbor, MI, U.S.A.). Hi-Trap Heparin HP columns (1ml), PD-10 desalting columns and Q-Sepharose were obtained from GE Amersham Biosciences. SP-Trisacryl was obtained from Sigma–Aldrich. NUNC Maxisorb 96-well ELISA plates were obtained from Life Technologies. Enhanced chemiluminescence reagents (Super Signal West Pico, Pierce) were purchased from Perbio Science U.K.
Heparin-binding ELISA
A covalent heparin–BSA complex was prepared by reducing end-coupling as described fully elsewhere [21,25], as was mock-conjugated BSA, which was treated similarly with coupling agent, but in the absence of heparin. ELISA wells were coated with either 10 ng of heparin–BSA complex or the same amount of mock-conjugated BSA in 100μl of 50 mM Tris/HCl (pH 7.4) containing 12.7 mM EDTA. After overnight incubation at 4 °C, the wells were washed three times with PBS, and blocked with 200μl of blocking buffer 1 [PBS containing 2% (w/v) BSA] per well for 1 h at room temperature (17 °C) on a rotating platform. After washing three times in PBS, the samples were incubated for 2 h with GFL diluted in blocking buffer 1, and then washed three times with PBS containing 0.05% Tween 20. The respective anti-GFL antibodies, diluted 1:200 in blocking buffer 1, were added for 1 h. Following washes with PBS containing 0.05% Tween 20, alkaline phosphatase-coupled second antibody diluted 1:1000 in blocking buffer 1 was added for 30 min. After three washes with PBS containing 0.05% Tween 20, and two washes with PBS alone, the wells were incubated with 100μl of p-nitrophenol solution per well at 37 °C in a shaking microtitre plate incubator. Absorbance at 405 nm was read in an Emax plate reader (Molecular Devices, Sunnyvale, CA, U.S.A.), typically after 30 min. Uncoated, but blocked, wells containing substrate solution only were used as blanks. In a competitive variant of the ELISA, GFLs were pre-incubated with increasing concentrations of soluble heparin in PBS for 30 min at room temperature, prior to addition to the coated ELISA wells.
Cloning and expression of GDNF
A plasmid, kindly provided by Dr Carlos Ibanez (Karolinska Institute, Stockholm, Sweden), encoding the full-length sequence for wild-type rat GDNF precursor protein was digested with BamHI, and a 600 bp product containing the coding sequence was extracted from a 1% (w/v) agarose gel. The pFastBac1 vector (Invitrogen) was also digested with BamHI, and dephosphorylated with 1 unit of calf intestinal alkaline phosphatase (Promega) in restriction enzyme buffer. Treated vector (100 ng) was ligated with either 20 or 40 ng of coding DNA using T4 DNA ligase (Fermentas, Helena Biosciences, Sunderland, U.K.), and transformed into DH5α1 Escherichia coli cells (Invitrogen). Plasmid DNA, isolated from resulting colonies, was digested with BamHI to confirm insertion, and clones were further selected for appropriate orientation of the insert according to the presence of a 300 bp product after PstI digestion. The insert sequence was verified by nucleotide sequencing of a PCR product generated using the following primers: forward primer, 5′-TCCGGATTATTCATACCGTCC-3′ (corresponding to a sequence on the 5′ side of the multiple cloning site); and, a reverse primer, 5′-ATGATCCTCTAGTACTTCTCG-3′ (located downstream of the multiple cloning site). A selected clone was then transformed into DH10Bac E. coli cells (Invitrogen) according to the supplier's protocol. A single transformed colony was re-cloned and grown as a 200 ml liquid culture in Luria–Bertani medium containing selection antibiotics. The bacmid virus DNA was then isolated and stored at −20 °C. Sf9 insect cells (Invitrogen) were then transfected with the viral DNA, following the supplier's protocols, to produce a P1 virus, which was used to infect Sf9 cells to express the protein.
Site-directed mutagenesis of GDNF
Two double-point mutations, K81A/K84A and R88A/R90A, and two partial deletions in the N-terminus, NΔ1 and NΔ2, were generated by overlap extension PCR. The mutagenesis primers used were as follows: K81A/K84A forward, 5′-ATGTACGACGCAATACTAGCAAATCTGTCTCGAAGT-3′; K81A/K84A reverse, 5′-ACTTCGAGACAGATTTGCTAGTATTGCGTCGTACAT-3′; R88A/R90A forward, 5′-CTGTCTGCAAGTGCAAGGCTAACAAGTGAC-3′; R88A/R90A reverse, 5′-GTCACTTGTTAGCCTTGCACTTGCAGACAG-3′; NΔ1 forward, 5′-AAAGGTCACCAGATGAGAATTCCA-3′; NΔ1 reverse, 5′-CTGGAATTCTCATCTGGTGACCTT-3′; NΔ2 forward, 5′-GCTGCCAGCCCAGGGTGCGTCTTAA-3′; NΔ2 reverse, 5′-TTAAGACGCACCCTGGGCTGGCAG-3′.
In the case of the alanine substitutions, the mismatched codons are underlined. For each mutein, two PCRs were performed using the proof-reading polymerase Platinum Pfx (Invitrogen) and 1 ng of GDNF wild-type DNA as template, employing one of the mutagenesis primers with either the forward or reverse multiple cloning site primer as appropriate, with an initial cycle of 3 min at 92 °C and 5 min at 55 °C, followed by 26 cycles of 30 s at 90 °C, 45 s at 55 °C and 90 s at 72 °C. The resulting amplified fragment was recovered from an agarose gel and purified. Paired fragments were then annealed in a second round of PCR using 10 ng of each fragment and forward and reverse multiple cloning site primers. The product containing the full-length mutated GDNF cDNA was resolved on an agarose gel, purified, digested with BamHI and cloned in to pFastBac1 vector (Invitrogen).
Cloning and expression of soluble GFRα–FLAG
A plasmid encoding the extracellular domain of human GFRα1, splice variant 2, fused at Ile335 to a FLAG tag sequence was digested with XhoI and BamHI, and the 1400 bp fragment containing the GFRα1 cDNA was extracted from a 1% (w/v) agarose gel. The vector pFastBac1 was digested with the same endonuclease pair and dephosphorylated. The fragment was cloned into the vector as described above and the insertion was verified by sequencing. Expression of the GFRα1 protein in Sf9 cells was then carried out as described for GDNF.
Ion-exchange chromatography
Conditioned supernatants from cultures expressing GDNF and variants thereof were dialysed against 25mM sodium phosphate buffer (pH 7.6) containing 5mM sodium EDTA. Dialysed supernatants, 50–100 ml, were then applied at a flow rate of 5 ml/h to SP-Trisacryl columns (2.5 ml bed vols) equilibrated in the same buffer. After a 5-bed vol. wash in buffer, a linear gradient (0–1.5 M) of NaCl was applied. Fractions were assayed by heparin-binding ELISA and Western blotting, and active fractions were pooled, dialysed against PBS and frozen.
Partial purification and concentration of GFRα1–FLAG was achieved by dialysing conditioned supernatants (150–200ml) against 20 mM Tris/HCl buffer (pH 8.5) containing 1mM EDTA, and applied on to a 2 ml Q-Sepharose Fast Flow column, at a flow rate of 5 ml/h and eluted by a linear NaCl gradient. Fractions were analysed by anti-FLAG captured ELISA, and positive fractions, which were eluted by approx. 0.4 M NaCl, were pooled and desalted into PBS on PD-10 columns.
Immunoassays of GDNF
Immunoblotting of GDNF was carried out by SDS/PAGE on 15% (w/v) gels. Gels were transblotted on to nitrocellulose membranes for 1 h at 4 °C. After three 5 min washes in TBS/T (Trisbuffered saline containing 0.05% Tween 20; 50 mM Tris/HCl, pH 7.6, and 0.9% NaCl containing 0.05% Tween 20), membranes were blocked in blocking buffer 2 [TBS/T containing 2% (w/v) dried skimmed milk powder]. Membranes were then incubated overnight at 4 °C with rabbit anti-GDNF antibody diluted 1:500 in blocking buffer 2. After washing as above in TBS/T, membranes were incubated for 1 h with affinity purified alkaline phosphatase-labelled anti-rabbit antibody (Jackson Immuno-Research Laboratories) diluted 1:2000 in blocking buffer 2. After further washes in TBS/T, the membrane was developed by enhanced chemiluminescence. Dot-blotted nitrocellulose membranes were blocked and developed by the same method.
ELISA of GDNF binding to immobilized GFRα1–FLAG was performed by coating the wells of the plate with anti-FLAG monoclonal antibody (diluted 1:500 in 30 mM sodium bicarbonate buffer, pH 8.3) by incubation overnight at 4 °C. After washing the plate three times with PBS, the wells were blocked with blocking buffer 1 for 30 min. After washing three times with PBS, the wells were incubated with 100 μl/well of partially purified GFRα1–FLAG (diluted 6.7-fold in blocking buffer 1). Plates were incubated for 2 h, washed three times in PBS containing 0.05% Tween 20, and incubated for 1 h with GDNF diluted in blocking buffer 1. After washing in PBS containing 0.05% Tween 20, the plates were incubated for 1 h with anti-GDNF (diluted 1:200 in blocking buffer 1) and washed again as described above. The samples were then incubated with alkaline phosphatase coupled anti-goat antibody, washed, and developed with p-nitrophenol phosphate as described above for the heparin-binding ELISA.
ELISA of GFRα1–GDNF complexes
Complexes formed for the incubation of partially purified GFRα1–FLAG with GDNF were assayed by a combination of the above assay protocols. Wells were coated with anti-FLAG monoclonal antibody, washed and blocked with blocking buffer 1. During this time, appropriate quantities of GFRα–FLAG and GDNF were incubated in blocking buffer 1 in the presence and absence of 2 μg/ml of heparin for 30 min at room temperature. Wells were washed three times with PBS containing 0.05% Tween and 100μl aliquots of the incubation mixture were added for 2 h. The wells were then washed, followed by incubation with rabbit anti-GDNF and alkaline phosphatase-coupled secondary antibody as described for the GDNF ELISA.
Heparin affinity chromatography
Samples of recombinant GFLs, 500 ng in the case of commercial rhGDNF (recombinant human GDNF), and equivalent immunoreactivities, as adjudged by immunoblotting in the case of baculovirus expressed variants, dissolved in PBS were applied to HiTrap Heparin columns at a flow rate of 0.5 ml/min. After elution by 5 ml of PBS, a linear gradient of NaCl up to a final concentration of 1.5 M was applied at the same flow rate; 1 ml fractions were collected throughout.
PC12 cell bioassay
Rat PC12 pheochromocytoma cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) foetal calf serum, 2 mM L-glutamine, 5% (v/v) heat-inactivated horse serum and 0.2 mg/ml gentamicin. The wells of 96-well plates were coated with 10 μg/ml of poly-D-lysine for 2 h, followed by incubation with 5 μg/ml of collagen in PBS for a further 2 h. Cells were plated out at a density of 2×104 per well, and grown for 24 h. The medium was then aspirated and replaced with 100 μl of complete DMEM lacking foetal calf serum and containing only 1% (v/v) heat-inactivated horse serum. Where present, GFRα1 was added at 2 ng/well, and commercial rhGDNF was added at 10 ng/well. Baculovirus expressed wild-type GDNF and mutants thereof were added in comparable amounts as adjudged by Western blotting.
RESULTS
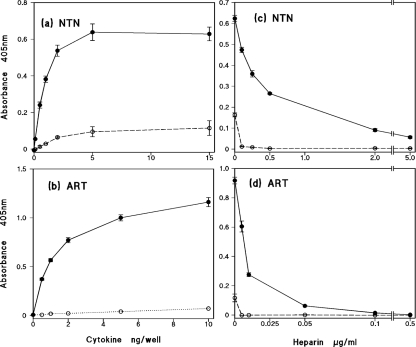
Previous work in our laboratory showed, using a heparin-binding ELISA, that rhGDNF binds strongly to heparin and highly sulfated HS [21]. In the present study, we investigated whether or not strong binding to heparin is a common feature of the GFLs. As shown in Figures 1(a) and 1(b), both NTN and ART show strong, dose-dependent binding to immobilized heparin–BSA complex, with such binding being readily detectable with 0.5 ng/well of the cytokine. By comparison, there is only low, background binding of NTN and ART in control wells coated with mock-conjugated BSA. In order to confirm that the high binding to the heparin complex indeed arises by interaction with the GAG chains, we employed pre-incubation of sub-saturating amounts of the GFLs with increasing concentrations of soluble heparin prior to addition to the wells. As shown in Figures 1(c) and 1(d), soluble heparin at low concentrations effectively competes for the cytokine, giving IC50 values of approx 0.5 and 0.1 μg/ml for NTN and ART respectively.
Figure 1. ELISA of the binding of recombinant human NTN and ART to immobilized heparin.
Dose-dependence of (a) NTN and (b) ART binding. Competitive binding after pre-incubation with increasing concentrations of soluble heparin of 2 ng/well NTN (c) and 8 ng/well ART (d). The solid lines and closed symbols represent wells coated with heparin–BSA complex; the open symbols and dotted lines indicate control wells coated with mock-conjugated BSA. The results shown are means±S.E.M. for a single representative experiment, and each data point taken is from four or five replicate wells.
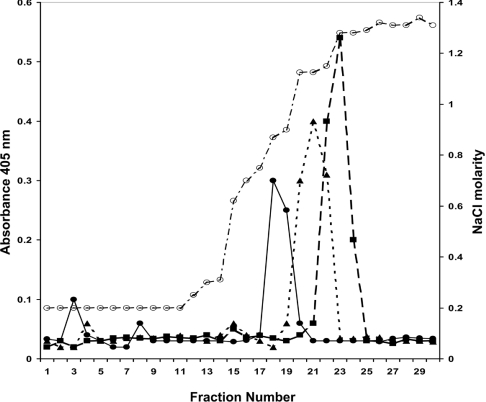
We investigated further the heparin binding properties of GDNF, NTN and ART using affinity chromatography. As shown in Figure 2, all three GFLs bind tightly to the heparin column at physiological ionic strength and pH, and elute as single peaks at high salt concentrations. These results are entirely consistent with the strong interaction of rhGDNF we found previously in the heparin ELISA [21], showing that rhGDNF is eluted from the column by a comparatively high salt concentration (0.8 M NaCl). We found that NTN and ART reproducibly showed even stronger binding to the column, with peak elution at 1.1 and 1.2 M NaCl respectively. These data confirm, by an independent experimental procedure, that these three GFLs bind strongly to heparin.
Figure 2. Heparin affinity chromatography of rat GDNF, human ART and human NTN.
Commercial recombinant GFLs (500 ng) dissolved in PBS were separately applied to a HiTrap Heparin column. Eluate fractions (1 ml) were assayed for GFL by heparin-binding ELISA. Salt concentrations (open circles, dotted and dashed line) were determined by conductivity measurements as shown on the right-hand axis, by standardizing with a range of solutions of known concentration dissolved in PBS. Fractions were assayed for GDNF (closed circles and solid line), NTN (closed triangles and dotted line), and ART (closed squares and dashed line) by heparin-binding ELISA (left-hand axis).
Since the primary sequences of all four GFLs show a high degree of similarity, they should shed light on the location of the heparin-binding sites. We first considered a cluster of basic residues on the exposed face of the α-helix of GDNF. As has been previously noted, these form a prominent positively charged patch on the surface of GDNF [26]. Interestingly, NTN has a considerable cluster of basic charges in an equivalent region of its sequence (Figure 3). We accordingly prepared two double-point mutants of rat GDNF, in each of which two of the basically charged residues comprising this basic patch were replaced by alanine (Figure 3). We termed these muteins K81A/K84A and R88A/R90A.
Figure 3. Alignment of the amino acid sequences of the GFLs.
The sequences of the mature, secreted forms of rat GDNF, and human ART, NTN and PSN are shown. The human GDNF sequence differs from that of the rat by seven residues. The residues comprising the α-helix of GDNF are indicated. The residues deleted in the two truncation mutations NΔ1 and NΔ2 are shown, and the residues subjected to alanine replacement are underlined. Arginine and lysine residues are highlighted in bold italics.
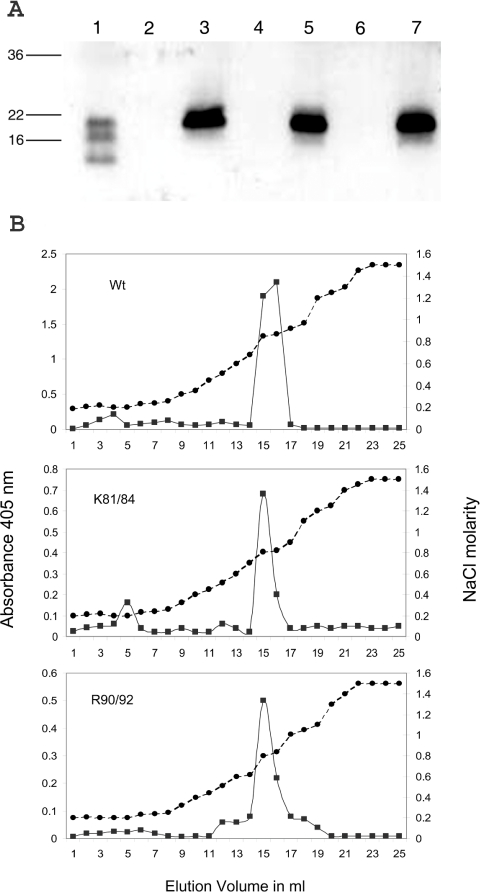
Mature wild-type rat GDNF and the K81A/K84A and R88A/R90A muteins were expressed in Sf9 insect cells. All three proteins were expressed as secreted dimers of anticipated molecular masses of approx. 40 kDa (results not shown). The GDNF immunoreactivity of the conditioned supernatants was concentrated and partially purified by ion-exchange chromatography on SP-Trisacryl. In each case, the immunoreactivity bound to the column was eluted by 0.6–1.0 M NaCl. In Figure 4(A), the immunoreactive fractions of each variant, after pooling and dialysing against PBS, gave strong bands at approx. 19 kDa on the immunoblots using reducing conditions, consistent with the anticipated size of glycosylated GDNF polypeptides. The partially purified supernatants of the three GDNF variants were analysed by heparin-binding ELISA using sample volumes adjusted to give equivalent densities on immunoblotting. All three GDNF variants showed strong binding, with dose indistinguishable response curves (results not shown) resembling the commercial rhGDNF reported previously [21]. To investigate further the possible effect of the double-point mutations on the heparin binding properties of GDNF, comparable immunoreactivities of the dialysed fractions were applied to heparin affinity columns. In Figure 4(B), in each case, a single major peak of immunoreactivity was eluted by 0.8 M NaCl. Peak fractions from each profile were subjected to immunoblotting and in each case produced single bands with mobilities indistinguishable from those shown in Figure 4(A) (results not shown). This confirms that the ELISA immunoreactivity in the column eluate fractions arose from polypeptides with the appropriate molecular masses. We therefore conclude that the heparin-binding affinity of GDNF is unaffected by these particular point substitutions.
Figure 4. Characterization of double point mutants of rat GDNF.
(A) Western blot of baculovirus-expressed mutants of GDNF. Conditioned supernatants from infected Sf9 cells were subjected to ion-exchange chromatography on SP-Trisacryl and GDNF immunoreactivity fractions were dialysed against PBS. The loadings shown were adjusted to give comparable intensities of development. Lane 1, commercial rat GDNF; lane 3, baculovirus-expressed wild-type GDNF; lane 5, K81A/K84A double point mutant; and, lane 7, R88A/R90A double point mutant. Lanes 2, 4 and 6 show similarly processed culture supernatants from mock-infected Sf9 cells, treated and grown in parallel to the cultures yielding the supernatant shown on the immediate right-hand side. (B) Heparin chromatography of wild-type (Wt) and double point mutants of GDNF. Samples of the immunoreactive SP-Trisacryl eluate fractions were dialysed against PBS and applied to 1 ml HiTrap Heparin columns. Fractions were assayed for GDNF immunoreactivity by heparin-binding ELISA (closed squares and solid line) and for conductivity (closed circles and dashed line).
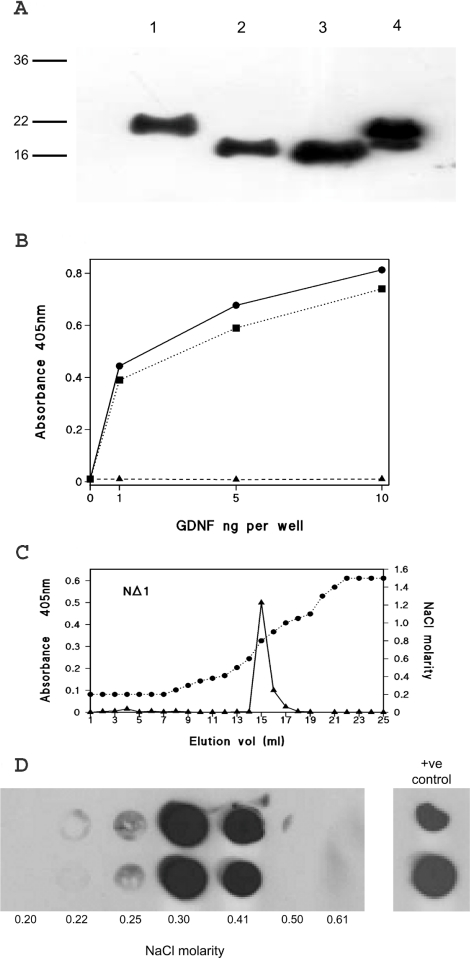
Further consideration of the sequence of GDNF revealed that the comparatively long 40 residue N-terminal sequence before the cysteine-rich domain of GDNF is rich in basic residues, containing a total of 12 arginine and lysine residues (Figure 3). Of these residues, 11 may be considered to comprise two basic-residue clusters, one of which has four arginine resides in a hexapeptide sequence, and the other has seven basic residues within a 13-residue sequence. NTN and ART have much shorter N-terminal extensions, but both of these contain multiple arginine residues (Figure 3). We therefore investigated the N-terminal region of GDNF by preparing two partial deletion mutations, NΔ1 and NΔ2, in which sequences encompassing each of the clusters were individually removed, as shown in Figure 3. The two deletion mutants were similarly produced by baculovirus expression and partially purified by SP-Trisacryl ion-exchange chromatography. By immunoblotting, the two deletion mutants were again found to be dimers (results not shown), which under reducing and denaturing conditions ran as single bands with higher electrophoretic mobilities than intact wild-type GDNF. These observations were entirely consistent with the predicted reduced molecular masses (Figure 5A). In Figure 5B, the heparin-binding ELISA revealed that the NΔ1 mutant binds to the heparin complex in a manner indistinguishable from that of intact GDNF. By contrast, an equivalent immunoreactivity of NΔ2-GDNF, as adjudged by immunoblotting, failed to bind to the heparin complex in the ELISA.
Figure 5. Characterization of partial N-terminal deleted mutants of GDNF.
(A) Western blot of baculoviral-expressed mutants of GDNF following SP-Trisacryl ion-exchange chromatography. Lane 1, wild-type rat GDNF; lane 2, NΔ1-GDNF; lane 3, NΔ2-GDNF; and, lane 4, commercial mammalian cell expressed rat GDNF. (B) Heparin-binding ELISA of wild-type rat GDNF (closed circles and solid line), NΔ1 (closed squares and dotted line), and NΔ2 (closed triangles and dashed line). Each data point is the mean of triplicate wells. (C) and (D) Heparin affinity chromatography of GDNF N-terminal deletion mutants. (C) NΔ1; immunoreactivity detected by heparin-binding ELISA. (D) NΔ2; immunoreactivity detected by dot blotting. Duplicate dot blots of fractions from the beginning of the salt gradient are shown together with the molarity determined, this region of the elution being the only one showing immunoreactivity. Duplicate applications of the input are shown on the right-hand side.
Using heparin affinity chromatography, NΔ1 bound strongly to the heparin column and was eluted by 0.8 M NaCl (Figure 5C), which was indistinguishable from the intact GDNF (Figure 4B). The presence of the NΔ1 mutant in these fractions was confirmed by dot blotting (results not shown). Since the NΔ2 mutant would be undetectable by heparin-binding ELISA (Figure 5B), its elution was monitored solely by dot blotting. This revealed that NΔ2 also binds completely to the column, but, in this case, it was eluted by 0.3 M NaCl (Figure 5d), a level only just above the physiological salt concentration (0.18 M). In the case of wild-type GDNF and the two truncation mutants, Western blotting also confirmed that immunoreactivity was confined to the ELISA or dot blotting positive fractions, and that under reducing conditions they migrated with molecular masses of approx. 19 kDa (results not shown).
Since we have demonstrated that wild-type GDNF binds strongly to heparin by two independent experimental approaches, we investigated how this interaction might affect the signalling activity of GDNF. Taking FGF-2 (fibroblast growth factor-2) as a prototypic example of a heparin-binding growth factor, both cytokine and receptor bind to a heparin chain, which cross-bridges between the two thus stabilizing their interaction [27,28]. We therefore examined whether GFRα1, the high-affinity GDNF receptor polypeptide, could also bind to heparin. Our initial studies employed a commercially obtained recombinant chimaeric protein in which the extracellular domain of GFRα1 is fused to an immunoglobulin Fc domain and a C-terminal polyhistidine tag. This protein showed strong binding to heparin in ELISA and by heparin affinity chromatography, wherein it required approx. 0.5 M NaCl for its elution (results not shown). These observations raised the prospect that heparin-like GAG chains might indeed promote GDNF–receptor interactions. Because of the considerable biological significance of this possibility we examined this further. Since this chimaeric polypeptide possesses a considerable amount of sequence not originating in the extracellular domain of the receptor, in particular an exposed polyhistidine tag, we expressed a soluble GFRα1 construct where the extracellular domain was fused to a C-terminal FLAG tag. We found that, unlike the commercial chimaera, this construct does not bind to the heparin–BSA complex in ELISA at physiological ionic strength and pH (results not shown). Moreover on heparin affinity chromatography at physiological pH and ionic strength, GFRα1 eluted, without retention, in the break-through fractions as shown in Figure 6(A).
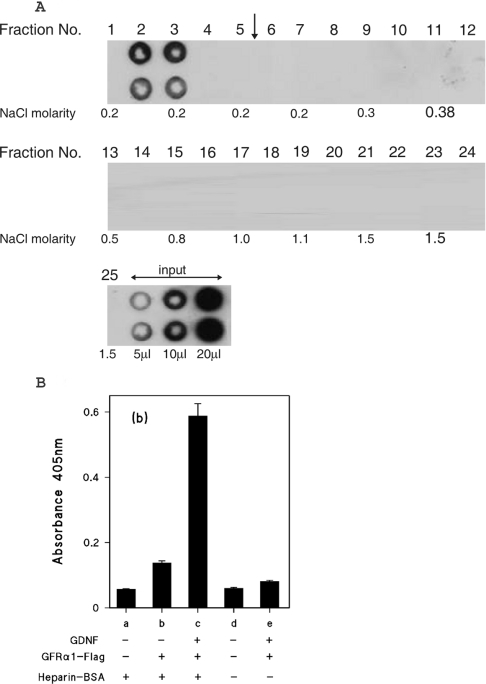
Figure 6. Heparin affinity chromatography of GFRα1–FLAG chimaeric protein.
(A) GFRα1–FLAG partially purified by ion-exchange chromatography was applied to a heparin Hi-trap column as described in the Experimental section for GDNF, except that the linear NaCl gradient was 0.18–2 M. The gradient was applied after fraction 5, as indicated by the vertical arrow. The NaCl molarities of the odd numbered fractions, as determined by conductivity measurements are shown. (B) Binding of GFRα1–FLAG to heparin-immobilized GDNF. GDNF (2.5 ng/well) was captured on wells coated with either heparin–BSA complex (columns a–c) or mock-conjugated BSA (columns d and e) as described for the heparin-binding ELISA. GFRα1–FLAG was then added to wells (columns b, c and e) at a quantity of 20 ng/well, as estimated by comparison with the GFRα1–Fc Western blotting. All wells were developed with anti-FLAG and alkaline phosphatase-labelled second antibody. The results are shown as means±S.E.M. for four replicate wells from a single representative experiment.
Since the GFRα1–FLAG protein does not itself bind to heparin, we were able to employ it in the heparin binding ELISA to investigate whether GDNF retained on heparin is still able to bind to this receptor polypeptide. As shown in Figure 6(B), heparin-complex-coated wells incubated with GDNF were able to capture GFRα1–FLAG, as revealed by strong immunoreactivity obtained after development with anti-FLAG antibody (Figure 6B, column c). By contrast there is little binding above background levels (Figure 6B, columns a and d) of GFRα1–FLAG in the absence of GDNF (Figure 6B, column b), or in wells lacking heparin (Figure 6B, column e).
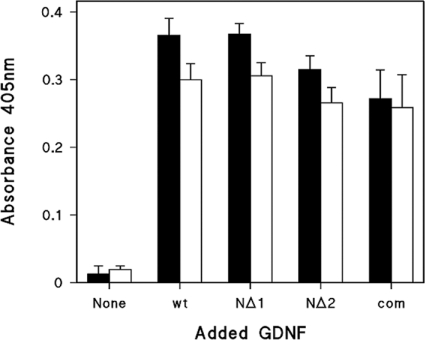
We sought to determine whether the recombinant GDNFs we had expressed were able to bind to the recombinant GFRα1–FLAG, and how heparin might affect this interaction for both the wild-type and truncated mutants. Therefore comparable quantities of the partially purified GDNF and its variants, as adjudged by Western blotting, were incubated with a fixed amount of GFRα1–FLAG prior to capture on anti-FLAG coated wells. As shown by the closed bars in Figure 7, wild-type GDNF and the NΔ1 and NΔ2 mutants all gave similar binding activities, indicating that neither deletion appreciably affects GFRα1 binding. Moreover addition of soluble heparin at 2 μg/ml to the GFRα1–FLAG and GDNF co-incubations neither substantially potentiates nor attenuates GFRα1–FLAG binding to the wild-type or truncated GDNFs (Figure 7, open bars).
Figure 7. inding of wild-type GDNF and deletion mutants NΔ1 and NΔ2 to GFRα1 in the presence and absence of heparin.
FRa1–FLAG was incubated with GDNF variants for 30 min in the absence (closed bars) or presence (open bars) of 2 μg/ml heparin, before capture on anti-FLAG coated wells and subsequent detection with anti-FLAG and secondary antibodies. The quantity of the GDNF variants used was judged by Western blotting to be comparable with 2 ng/well of commercial GDNF, a quantity giving high, but sub-maximal, absorbance. The absorbance shown was blanked against anti-FLAG coated wells with no GFRα1–FLAG or GDNF. Values are the means for triplicate wells and shown ± S.D. wt, wild-type GDNF; com; commercial recombinant rat GDNF.
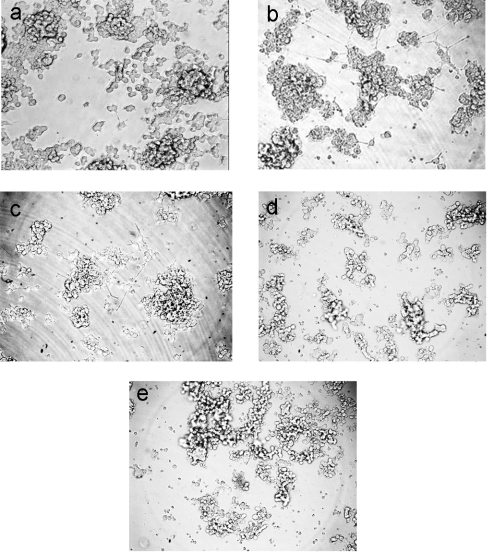
The addition of soluble GFRα1 and GDNF to PC12 cells stimulated neurite outgrowth in a minority of the cells [29]. Since Davies et al. [23] showed that low concentrations of heparin are able to inhibit such outgrowth, we compared the activity of the NΔ2 mutant with intact GDNF in this system. As shown in Figure 8(a), cells grown in the absence of GDNF, but in the presence of soluble GFRα1–Fc show a very low spontaneous rate of neurite outgrowth, and where formed, these are of modest length. The further addition of recombinant wild-type GDNF causes a marked increase in the proportion of cells producing neurite outgrowth, although this remains a minority (Figure 8b). Also more cells adopt a flattened morphology and many of the neurites formed are of considerable length. Because the cells form clumps in these cultures, the quantification of these effects is unreliable. The effects observed with the baculovirus expressed GDNF are fully consistent, in nature and extent, with those we obtained with the commercial recombinant GDNF (results not shown). The NΔ2 variant was similarly active in promoting these effects (Figure 8c). Heparin at a concentration of 1 μg/ml completely inhibited the neurite outgrowth induced by the wild-type GDNF expressed in the baculovirus system (Figure 8d), which has been reported previously for the commercial recombinant GDNF [23]. Somewhat surprisingly, the same total inhibition by heparin was also observed with the NΔ2 variant (Figure 8e), despite its considerably reduced affinity for heparin.
Figure 8. Neurite outgrowth activity in vitro of wild-type GDNF and deletion mutant NΔ2.
(a) No GDNF. (b) and (d) Wild-type GDNF. (c) and (e) NΔ2-GDNF. Incubations were performed in the absence (a–c) or presence (d and e) of 1 μg/ml heparin. GFRα1–Fc (2 ng/well) was used throughout and, where present, the baculovirus expressed GDNF variants were employed at quantities judged by Western blotting to be equivalent to 10 ng/well.
DISCUSSION
Having previously investigated the binding of GDNF to heparin and related polysaccharides, we show here by both ELISA and affinity chromatography that two other GFLs, ART and NTN, also bind heparin. Indeed higher salt concentrations are required for the elution of ART and NTN from the heparin column than for GDNF. Since the interaction between polypeptides and sulfated GAGs is largely polyionic [30], our results reveal that, within the GFLs, heparin affinity increases in the order GDNF<NTN<ART. A key finding of the present study is that the major determinant of heparin binding is located within a 16 residue sequence, which contains seven arginine and lysine residues, and is located immediately N-terminal to the TGF-β-type cysteine-knot domain. The unusually long, 40 residue N-terminal extension to the cysteine-knot domain in GDNF is not resolved in its high-resolution structure [31], which precludes structural modelling of this interaction. Thus it is not possible to suggest whether or not the two NΔ2 sequences in a GDNF dimer might combine to give a single joint site. The more N-terminal sequence deletion, NΔ1, has no effect on heparin binding. This is somewhat surprising, as this region might be more exposed and it contains a BBXB-motif, which is often advocated as a heparin-binding motif [32]. We also show that double alanine substitutions of basic residues in and around the short α-helix, a region of prominent basic charge [26], have no effect on heparin binding.
The NΔ2 mutant failed to bind in the ELISA, but binds to the heparin affinity column, despite the fact that both procedures are conducted at physiological pH and ionic strength. However, NΔ2-GDNF is eluted from the heparin column by only a marginal increase in salt concentration, demonstrating that binding affinity is very weak. The different observations from these two experimental approaches may arise from the heparin involved being from different sources, or having been subjected to different coupling methods. It might also arise from the greater degree of mechanical disturbance of binding during ELISA plate washes compared with the slow and uniform flow of eluent over the columns. Alternatively, since the heparin affinity column matrix contains a high concentration of heparin chains, which, in turn, have dense arrays of strongly anionic groups, the residual heparin binding to the affinity matrix may simply represent a non-specific weak ion-exchange interaction, bearing in mind that the NΔ2 variant, like intact GDNF, exhibits unchanged binding affinity for the sulfopropyl moiety in the ion-exchange resin used for their partial purification.
During the course of the present study, a structural study of ART has revealed that it has a high affinity for heparin [33]. These workers found that deletion of the first nine N-terminal residues, which includes two basic residues and corresponds partially to the NΔ2 sequence of GDNF, resulted in a substantial decrease in heparin binding. They further showed that a BBXB motif (Arg-Arg-Ala-Arg) close to the α-helix was also an important determinant of heparin binding of the unglycosylated ART they studied [33]. This implies the existence of a discontinuous binding site encompassing the N-terminus and the BBXB motif, but again the lack of resolution of the N-terminus in their crystallographic structure of ART precludes full insight. This central BBXB motif is absent from GDNF, indeed one of the corresponding residues in the latter is the acidic charged residue aspartate. Taking our findings together with the findings reported previously, we propose that basic residues lying immediately N-terminal to the cysteine-knot in a flexible region of the polypeptide are major determinants of heparin binding in the GFLs. However, there has been some evolutionary divergence of heparin-binding sites, such that in ART a further basic charged motif, which is central in this polypeptide, but absent from GDNF, is also involved. The weak heparin binding we observe with the NΔ2-GDNF mutant in heparin affinity chromatography means we cannot preclude the possible involvement of additional sites within GDNF. The importance of basic residue clusters immediately N-terminal to the cysteine-rich TGF-β-type domain in heparin binding is also seen in the related cytokines BMP-2 (bone morphogenetic protein 2) [34] and BMP-4 [35].
The prototypic and arguably best-understood example of a cytokine–GAG interaction is the heparin binding of FGF-2. Heparin/HS is essential for FGF-2 signalling, and high resolution structures of the FGF receptor–FGF–heparin complexes show heparin stabilizing the complex by cross-bridging the cytokine to its receptor [27,28], and in one case also cross-bridging the two FGF–FGF receptor pairs in the tetrameric assembly [27]. The issue of how applicable this model might be for other cytokine–GAG interactions remains an open question. The findings of the present study suggest that for GDNF the involvement of HS GAG in signalling does not follow this paradigm. It had been proposed that GFRα, which possesses several clusters of basic residues in its extracellular domain, might bind to heparin and HS [22,36], thereby allowing GDNF–receptor cross-bridging. Despite the fact that we found that a commercial His-tagged GFRα1–Fc chimaeric protein binds strongly to heparin, we show here that a soluble recombinant protein comprising the extracellular domain of GFRα1 fused to a C-terminal FLAG tag shows no such binding. This protein binds GDNF and is active in a standard cellular bioassay for GDNF signalling at levels comparable with the commercial chimaera (results not shown), indicating correct folding. Our results preclude the possibility that heparin/HS can cross-bridge GDNF to its high-affinity receptor protein. Therefore the GAG binding of the GFRα1–Fc chimaera must arise from either the Fc domain or polyhistidine tag to which it is fused. Since there is no record of GAG binding within the human IgG1 Fc domain involved, it is very likely that it is the latter polybasic sequence which introduces an artefactual heparin-binding site. This phenomenon has been reported previously [37], and suggests that the characterization of GAG binding of recombinant proteins bearing a polyhistidine tag is unreliable.
GDNF captured on heparin is still able to bind GFRα1–FLAG. This finding is of some physiological significance, since it suggests that GDNF immobilized on heparin-like chains does not need to be released in order to engage GFRα1, the first step in its signalling pathway. It also shows that the heparin-binding site on GDNF is physically distinct from its GFRα1-binding sites, such that one interaction does not prevent the other. Since the GFRα1-binding sites are located on the exposed tips of finger-like β-stranded loops [26], which are distant from the N-termini of the subunits, this result is consistent with our localization of the major determinant of heparin binding.
We show here that GDNF mutants with N-terminal truncations bind to GFRα1 and trigger cellular response normally. The binding of wild-type GDNF and the truncated variants to soluble GFRα1 is essentially neither inhibited nor potentiated by soluble heparin, indicating that heparin binding plays no role in this interaction. We have reported previously a modest potentiation of GDNF binding to surface-captured GFRα1–Fc [21], and we also observed this in the present study in some experiments when high concentrations of GDNF were employed. However, we cannot now exclude the possibility that under these particular conditions, multiple GDNF molecules may bind to a heparin chain, thus the receptor engagement of one cytokine molecule in such an oligomerized complex to the receptor will indirectly give rise to the binding of others. Our current conclusion is therefore that heparin plays no role on GDNF–GFRα1 interactions.
It has been shown that soluble heparin at low concentration inhibits GDNF activity in several culture systems, and that selective 2-O-desulfation of the heparin blocks this effect, whereas other modifications do not [23]. This finding fits well with a model in which GDNF activity is dependent on binding to 2-O-sulfate-rich heparin-like GAG for its physiological activity. In the present study, we show that a GDNF variant in which heparin affinity is markedly reduced still binds normally to a soluble construct of its high affinity receptor polypeptide, and that this interaction is unaffected by the presence of heparin. However, the activity of this GDNF variant in cell culture still remains sensitive to the presence of heparin. These findings indicate that the effect of heparin/HS on GDNF activity is located in a separate region of the protein from the GFRα1-binding site, and that other cell surface events in GDNF signalling require scrutiny in order to reveal the mechanism for the GAG-dependence of GDNF activity.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council. We are grateful to David McClarence for his careful reading of this manuscript.
References
- 1.Baloh R., Enomoto H., Johnson E., Milbrandt J. The GDNF family ligands and receptors: implications for neural development. Curr. Opin. Neurobiol. 2000;10:103–110. doi: 10.1016/s0959-4388(99)00048-3. [DOI] [PubMed] [Google Scholar]
- 2.Carmillo P., Dago L., Day E., Worley D., Rossomando A., Walus L., Orozco O., Buckley C., Miller S., Tse A., et al. Glial cell line-derived neurotrophic factor (GDNF) receptor α-1 (GFRα1) is highly selective for GDNF versus artemin. Biochemistry. 2005;44:2545–2554. doi: 10.1021/bi049247p. [DOI] [PubMed] [Google Scholar]
- 3.Poteryaev D., Titievsky A., Sun Y. F., Thomas-Crusells J., Lindahl M., Billaud M., Arumae U., Saarma M. GDNF triggers a novel ret-independent Src kinase family-coupled signalling via a GPI-linked GDNF receptor α1. FEBS Lett. 1999;463:63–66. doi: 10.1016/s0014-5793(99)01590-2. [DOI] [PubMed] [Google Scholar]
- 4.Trupp M., Scott R., Whittlemore S. R., Ibanez C. F. Ret-dependent and -independent mechanisms of glial cell line-derived neurotrophic factor signalling in neuronal cells. J. Biol. Chem. 1999;274:20885–20894. doi: 10.1074/jbc.274.30.20885. [DOI] [PubMed] [Google Scholar]
- 5.Popsueva A., Poteryaev D., Arighi E., Meng X., Angers-Loustau A., Kaplan D., Saarma M., Sariola H. GDNF promotes tubulogenesis of GFR1-expressing MDCK cells by Src-mediated phosphorylation of Met receptor protein kinase. J. Cell Biol. 2003;161:119–129. doi: 10.1083/jcb.200212174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paratcha G., Ledda F., Ibanez C. F. The neural cell adhesion molecule NCAM is an alternative signaling receptor for GDNF family ligands. Cell. 2003;113:867–879. doi: 10.1016/s0092-8674(03)00435-5. [DOI] [PubMed] [Google Scholar]
- 7.Enomoto H., Hughes I., Golden J., Baloh R. H., Yonemura S., Heuckeroth R. O., Johnson E. M., Milbrandt J. GFRα1 expression in cells lacking RET is dispensable for organogenesis and nerve regeneration. Neuron. 2004;44:623–636. doi: 10.1016/j.neuron.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 8.Patel N. K., Bunnage M., Plaha P., Svendsen C. N., Heywood P., Gill S. S. Intraputamenal infusion of glial cell line-derived neurotrophic factor in PD: a two-year outcome study. Ann. Neurol. 2005;57:298–302. doi: 10.1002/ana.20374. [DOI] [PubMed] [Google Scholar]
- 9.Slevin J. T., Gash D. M., Smith C. D., Gerhardt G. A., Kryscio R., Chebrolu H., Walton A., Wagner R., Young A. B. Unilateral intraputaminal glial cell line-derived neurotrophic factor in patients with Parkinson disease: response to 1 year each of treatment and withdrawal. Neurosurg. Focus. 2006;20:E1. doi: 10.3171/foc.2006.20.5.2. [DOI] [PubMed] [Google Scholar]
- 10.Moore M., Klein R. D., Farinas I., Sauer H., Armanini M., Phillips H., Reichardt L. F., Ryan A., Carver-Moore K., Rosenthal A. Renal and neuronal abnormalities in mice lacking GDNF. Nature. 1996;382:76–79. doi: 10.1038/382076a0. [DOI] [PubMed] [Google Scholar]
- 11.Pichel J. G., Shen L., Sheng H. Z., Granholm A.-C., Drago J., Grinberg A., Lee E. J., Huang S. P, Saarma M., Hoffer B. J., et al. Defects in enteric innervation and kidney development in mice lacking GDNF. Nature. 1996;382:73–76. doi: 10.1038/382073a0. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez M. P., Silos-Santiago I., Frisen J., He B., Lira S. A., Barbacid M. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature. 1996;382:70–73. doi: 10.1038/382070a0. [DOI] [PubMed] [Google Scholar]
- 13.Cacalano G., Farinas I., Wang L. C., Hagler K., Forgie A., Moore M., Armanini M., Phillips H., Ryan A. M, Reichardt L. F., et al. GFRα1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron. 1998;21:53–62. doi: 10.1016/s0896-6273(00)80514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomac A. C., Grinberg A., Huang S. P., Nosrat C., Wang Y., Borlongan C., Lin S-Z., Chiang Y.-H., Olson L., Westphal H., Hoffer B. J. Glial cell line-derived neurotrophic factor receptor α2 availability regulates glial cell line-derived neurotrophic factor signalling; evidence from mice carrying one or two mutated alleles. Neuroscience. 2000;95:1011–1023. doi: 10.1016/s0306-4522(99)00503-5. [DOI] [PubMed] [Google Scholar]
- 15.Schuchardt A., D'Agati V., Larsson-Blomberg L., Costantini F., Pachnis V. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature. 1994;367:380–383. doi: 10.1038/367380a0. [DOI] [PubMed] [Google Scholar]
- 16.Schedl A., Hastie N. Cross-talk in kidney development. Curr. Opin. Gene. Dev. 2000;10:534–549. doi: 10.1016/s0959-437x(00)00125-8. [DOI] [PubMed] [Google Scholar]
- 17.Vainio S., Lin Y. Coordinating early kidney development: lessons from gene targeting. Nat. Rev. Genetics. 2002;3:533–543. doi: 10.1038/nrg842. [DOI] [PubMed] [Google Scholar]
- 18.Lin L.-F., Zhang T. J., Collins F., Armes L. G. Purification and initial characterization of rat B49 glial line-derived neurotrophic factor. J. Neurochem. 1994;63:758–768.. doi: 10.1046/j.1471-4159.1994.63020758.x. [DOI] [PubMed] [Google Scholar]
- 19.Esko J. D., Selleck S. B. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Ann. Rev. Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 20.Bullock S. L., Fletcher J. M., Beddington R. S. P., Wilson V. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 1998;12:1894–1906. doi: 10.1101/gad.12.12.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rickard S. M., Mummery R. S., Mulloy B., Rider C. C. The binding of human glial cell-line derived neurotrophic factor (GDNF) to heparin and heparan sulfate: importance of 2-O-sulfate groups and effects on its interaction with its receptor GFRα1. Glycobiology. 2003;13:419–426. doi: 10.1093/glycob/cwg046. [DOI] [PubMed] [Google Scholar]
- 22.Barnett M. W., Fisher C. E., Perona-Wright G., Davies J. A. Signalling by glial cell line-derived neurotrophic factor (GDNF) requires heparan sulfate glycosaminoglycan. J. Cell Sci. 2002;115:4495–4503. doi: 10.1242/jcs.00114. [DOI] [PubMed] [Google Scholar]
- 23.Davies J. A., Yates E. A., Turnbull J. E. Structural determinants of heparan sulfate modulation of GDNF signalling. Growth Factors. 2003;21:109–119. doi: 10.1080/08977190310001621005. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka M., Xiao H., Kiuchi K. Heparin facilitates glial cell-line derived neurotrophic factor signal transduction. NeuroReport. 2002;13:1913–1916. doi: 10.1097/00001756-200210280-00016. [DOI] [PubMed] [Google Scholar]
- 25.Najjam S., Gibbs R. V., Gordon M. Y., Rider C. C. Characterisation of human recombinant interleukin 2 binding to heparin and heparan sulfate using an ELISA approach. Cytokine. 1997;9:1013–1022. doi: 10.1006/cyto.1997.0246. [DOI] [PubMed] [Google Scholar]
- 26.Eketjall S., Fainzilber M., Murray-Rust J., Ibanez C. F. Distinct structural elements in GDNF mediate binding to GFRα1 and the activation of the GFRα1-c-Ret receptor complex. EMBO J. 1999;18:5901–5910. doi: 10.1093/emboj/18.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelligrini L., Burke D. F., Von Delft F., Mulloy B., Blundell T. L. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 28.Schlessinger J., Plotnikov A. N., Ibrahimi O. A., Eliseenkova A. V., Yeh B. K., Yayon A., Lindhardt R. J., Mohammadi M. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 29.Treanor J., Goodman L., De Sauvage F., Stone D., Poulsen T., Beck D., Gray C., Armanini M., Pollock A., Hefti F., et al. Characterization of a multicomponent receptor for GDNF. Nature. 1996;382:80–83. doi: 10.1038/382080a0. [DOI] [PubMed] [Google Scholar]
- 30.Hileman R. E., Fromm J. R., Weiler J. M., Linhardt R. J. Glycosaminoglycan-protein interactions: definition of consensus sites in the glycosaminoglycan-binding proteins. BioEssays. 1998;20:156–167. doi: 10.1002/(SICI)1521-1878(199802)20:2<156::AID-BIES8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 31.Eigenbrot C., Gerber N. X-Ray structure of glial cell-derived neurotrophic factor at 1.9Å resolution and implications for receptor binding. Nat. Struct. Biol. 1997;4:435–438. doi: 10.1038/nsb0697-435. [DOI] [PubMed] [Google Scholar]
- 32.Cardin A. D., Weintraub H. J. Molecular modelling of protein-glycosaminoglycan interactions. Arterioscler. Thromb. Vasc. Biol. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 33.Silvian L., Jin P., Carmillo P., Boriack-Sjodin P. A., Pelletier C., Rushe M., Gong B., Sah D., Pepinsky B., Rossomando A. Artemin crystal structure reveals insights into heparan sulfate binding. Biochemistry. 2006;45:6801–6812. doi: 10.1021/bi060035x. [DOI] [PubMed] [Google Scholar]
- 34.Ruppert R., Hoffamn E., Sebald W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur. J. Biochem. 1996;237:295–303. doi: 10.1111/j.1432-1033.1996.0295n.x. [DOI] [PubMed] [Google Scholar]
- 35.Ohkawara B., Iemura S., ten Dijke P., Ueno N. Action range of BMP is defined by its N-terminal basic amino acid core. Curr. Biol. 2002;12:205–209. doi: 10.1016/s0960-9822(01)00684-4. [DOI] [PubMed] [Google Scholar]
- 36.Leppanen V.-M., Bespalov M. M., Runeberg-Roos P., Puurand U., Merits A., Saarma M., Goldman A. The structure of GFRα1 domain 3 reveals new insights into GDNF binding and RET activation. EMBO J. 2004;23:1452–1462. doi: 10.1038/sj.emboj.7600174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lacy M., Sanderson R. 6xHis promotes binding of a recombinant protein to heparan sulfate. BioTechniques. 2002;32:254–257. doi: 10.2144/02322bm04. [DOI] [PubMed] [Google Scholar]