Abstract
Several rare and novel NNRTI [non-nucleoside reverse transcriptase (RT) inhibitor] resistance mutations were recently detected at codons 132 and 135 in RTs from clinical isolates using the yeast-based chimaeric TyHRT (Ty1/HIV-1 RT) phenotypic assay. Ile132 and Ile135 form part of the β7–β8 loop of HIV-1 RT (residues 132–140). To elucidate the contribution of these residues in RT structure–function and drug resistance, we constructed twelve recombinant enzymes harbouring mutations at codons 132 and 135–140. Several of the mutant enzymes exhibited reduced DNA polymerase activities. Using the yeast two-hybrid assay for HIV-1 RT dimerization we show that in some instances this decrease in enzyme activity could be attributed to the mutations, in the context of the 51 kDa subunit of HIV-1 RT, disrupting the subunit–subunit interactions of the enzyme. Drug resistance analyses using purified RT, the TyHRT assay and antiviral assays demonstrated that the I132M mutation conferred high-level resistance (>10-fold) to nevirapine and delavirdine and low-level resistance (∼2–3-fold) to efavirenz. The I135A and I135M mutations also conferred low level NNRTI resistance (∼2-fold). Subunit selective mutagenesis studies again demonstrated that resistance was conferred via the p51 subunit of HIV-1 RT. Taken together, our results highlight a specific role of residues 132 and 135 in NNRTI resistance and a general role for residues in the β7–β8 loop in the stability of HIV-1 RT.
Keywords: chimaeric Ty1/HIV-1 reverse transcriptase phenotypic assay, HIV-1, non-nucleoside reverse transcriptase inhibitor, resistance, reverse transcriptase
Abbreviations: β-gal, β-galactosidase; p66, 66 kDa subunit of reverse transcriptase; p51, 51 kDa subunit of reverse transcriptase; RLU, relative light unit; RT, reverse transcriptase; TyHRT, Ty1/HIV-1 RT; NNRTI, non-nucleoside RT inhibitor; NNRTI-BP, NNRTI-binding pocket; NRTI, nucleoside RT inhibitor; WT, wild-type
INTRODUCTION
HIV-1 RT (reverse transcriptase) is a multifunctional enzyme that catalyses the conversion of the viral single-stranded RNA into double-stranded DNA. Due to its essential role in HIV-1 replication, RT is a major target for the development of antiretroviral agents [1]. To date, two therapeutic classes of RT inhibitors have been identified: the NRTIs (nucleoside RT inhibitors) and the NNRTIs (non-nucleoside RT inhibitors). NRTIs bind at the active site of RT and act as competitive, chain-terminating inhibitors of DNA polymerization [2]. By contrast, the NNRTIs bind to a non-active site pocket in HIV-1 RT [termed the NNRTI-BP (NNRTI-binding pocket)] and act as allosteric inhibitors of DNA polymerization [3]. Although combinatorial therapies that contain two or more RT inhibitors have profoundly reduced morbidity and mortality associated with AIDS, their long-term efficacy is limited by the selection of drug-resistant variants of HIV-1.
The detection and characterization of drug-resistant variants is important for the effective management of HIV-1 infected individuals. The TyHRT (Ty1/HIV-1 RT) assay, which uses hybrid yeast Ty1/HIV-1 RT retroelements, was previously developed to provide a simple phenotypic assay for HIV-1 RT activity [4]. Expression of TyHRT elements carrying a reverse transcription indicator gene generates HIV-1 RT-mediated events at a high frequency and the activity of HIV-1 RT variants can be differentiated and characterized over a 10000-fold range. Since HIV-1 RT activity is inhibited by NNRTI in yeast [5], this assay can be used to determine the inhibitor susceptibility of individual RTs derived from laboratory clones or from clinical sample isolates [6] and can be used to detect drug resistant viral variants present at frequencies of less than 1% of the virus population [6,7]. In this regard, we recently detected several rare and novel NNRTI resistance mutations at codons 132 and 135, in particular I132M and I135M, in HIV-1 RT [8,8a]. However, the genetic background of the clinical RT isolates in these studies varied from canonical WT (wild-type) strains and contained multiple polymorphisms that may also have affected RT activity and inhibitor susceptibility. Accordingly, the primary objective of the present study was to elucidate the role of these residues in RT structure–function and drug resistance in a defined RT background.
MATERIALS AND METHODS
Materials
TyHRT reverse transcription assays were carried out in Saccharomyces cerevisiae strain GRY 1990(MATα ura3–167 trp1:pgk-lacZ spt3–101 his3▵200). Yeast two-hybrid analyses were carried out using the S. cerevisiae strain CTY10–5d (MATα ade2 trp1–901 leu2-lacZ his3–200 gal4−gal80− URA3:lexA-lacZ). Efavirenz and nevirapine were obtained from the NIH (National Institutes of Health) AIDS Research and Reference Reagent Program. Delavirdine was purchased from BIOMOL. All other reagents were of the highest quality available and were used without further purification.
Site-directed mutagenesis and purification of HIV-1 RT
All mutations were introduced into WT HIV-1LAI RT [9] by sitedirected mutagenesis using the QuikChange® mutagenesis kit (Stratagene). Full-length sequencing of mutant RTs was performed to confirm the presence of the desired mutations and to exclude adventitious mutations introduced during mutagenesis. WT and mutant recombinant HIV-1 RTs were overexpressed and purified to homogeneity as described previously [10,11]. For subunit selective mutagenesis, the p66 (66 kDa subunit of RT) and p51 (51 kDa subunit of RT) RT genes were cloned into the pET-DUET vector (Novagen-EMD Biosciences) and enzymes were purified as described previously using a double-tag strategy [12,13]. The RT concentration was determined spectrophotometrically at 280 nm using a molar absorption coefficient (ϵ280) of 260450 M−1 cm−1.
RNA-dependent DNA polymerase assays
Fixed time assays were used for HIV-1 RT-associated RNA-dependent DNA polymerase activity. Briefly, reactions were carried out in 50 mM Tris/HCl (pH 7.8 at 37 °C), 50 mM KCl, 10 mM MgCl2 and 5 μg/ml of poly(rA)-oligo(dT)12–18 containing 20 μM [3H]TTP, and various concentrations of ligand dissolved in DMSO (3% final concentration). Reactions were initiated by the addition of 50 ng of WT RT, incubated for 20 min at 37 °C and then quenched with 250 μl of ice-cold 10% TCA (trichloroacetic acid) containing 20 mM sodium pyrophosphate. Quenched samples were filtered and the extent of radionucleotide incorporation was determined by liquid scintillation spectrometry.
CD spectroscopy
CD spectra were collected at ambient room temperature (24 °C) using an Aviv Instruments Circular Dichroism Spectrometer (Model 202). HIV-1 RT was dialysed into 500 mM sodium phosphate buffer (pH 7.4) just prior to CD analysis.
Yeast two-hybrid assay
Quantification of protein–protein interactions were determined using the β-gal (β-galactosidase) liquid assay performed on permeablized yeast grown from at least three independent transformants, as described previously [14]. Briefly, individual transformants were grown in 1 ml of synthetic complete medium, without histidine and leucine, containing 2% (w/v) glucose (termed SC-His-Leu) at 30 °C with aeration for 16 h before being diluted to an absorbance (at 600 nm) of 0.2 in 2.5 ml of SC-His-Leu. Cells were then allowed to grow with aeration at 30 °C until the absorbance (at 600 nm) reached 0.5–0.8 before they were pelleted and stored at −20 °C. Thereafter they were permeablized in 50 μl of Y-PER™ Yeast Protein Extraction Reagent (Pierce) and assayed for β-gal activity by adding 1 ml of 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 containing 40 mM 2-mercaptoethanol followed by the addition of 200 μl of orthonitrophenyl-β-D-galactopyranoside. Reactions were incubated at 30 °C, quenched by the addition of 0.5 ml of 1 M Na2CO3, and the absorbance was read at 420 nm. β-gal activity (in Miller units) was calculated by using the following equation: Miller units=A420×1000/incubation time (in min).
Protein expression in yeast
To prepare yeast protein extracts for Western blot analysis, yeast transformants were grown and pelleted as described above. To extract protein, the cells were thawed at room temperature in 160 μl of YeastBuster Protein Extraction Reagent (Novagen) containing 1 μg/ml of pepstatin, leupeptin and aprotinin. The cells were vortexed for 10 s and allowed to mix on a rotating wheel for 20 min at room temperature. The supernatant was clarified after centrifugation at 13000 g for 15 min. The total protein concentration for each protein extract was determined using Bradford reagent (Bio-Rad) and 5–15 μg of total protein was loaded per well for Western blot analysis. Fusion protein expression in yeast was determined by Western blot analysis of lysates with Gal4AD polyclonal antibodies (Upstate Biotechnology) and anti-lexA polyclonal antibodies (Invitrogen). Immunodetection was accomplished using ECL®-Plus (Amersham).
TyHRT reverse transcription assay
TyHRT elements carrying a luciferase reverse transcription reporter gene (TyHRT-lucAI) were used to assay RT activity and inhibitor susceptibility in a yeast strain, GRY1990, where lacZ is constitutively expressed and serves as a normalization control. Assays were performed in a 96 well format by growing cells carrying TyHRT-lucAI elements overnight in 0.5 ml of AA-URA (amino-acid-supplemented synthetic medium lacking uracil)+glucose in 2.2 ml deep-well plates. Then, 50 μl was transferred to 0.5 ml AA-URA+galactose in 2.2 ml deep-well plates and grown for 48 h without shaking at 30 °C. Subsequently, 50 μl was transferred to 200 μl of YPD [1% (w/v) yeast extract/2% (w/v) peptone/2% (w/v) glucose] medium in 96 well round-bottom tissue culture plates and grown for 6 h at 30 °C. Cells were pelleted at 3000 g for 5 min, decanted and placed at −80 °C for more than 30 min to promote cell lysis upon thawing. Cells were resuspended in 160 μl of 1 mM Tris/HCl (pH 7.5) and 0.1 mM EDTA at room temperature. The cell suspension (80 μl) was then transferred to Microlite 1 (Thermo) flat-bottom micotitre plates followed by the addition of 160 μl Enhanced Luciferase Assay reagent (BD Pharmigen). Then 8 μl of cell suspension was transferred to Microlite 1 (Thermo) flat-bottom micotitre plates followed by the addition of 80 μl of Galacto-Star (Applied Biosystem) reagent. Luciferase activity was read at 15 min and β-gal activity was read at 1 h in a 96 well luminometer (Tropix TR717, Applied Biosytems). RT activity was determined by dividing luciferase activity (RT reporter) RLUs (relative light units) by β-gal (cell growth and lysis normalization) RLUs. Drug-resistance assays were carried out in the presence of 0.4% DMSO. Different RT inhibitor concentrations were achieved by 2-fold serial dilutions in AA-URA+galactose medium. In each experiment RT mutations were run in triplicate and the RT assays were repeated at least three times. RT activity assays in the absence of drug were also carried out in a 96 well format with six independent isolates of each variant and 24 WT RT controls per plate.
In some instances, the standard deviations for activities determined in the TyHRT-luciferase assay were high. This can be attributed to the fact that this is an in vivo assay and, accordingly, is subject to multiple variable parameters including the following. (i) Assays are carried in an spt3 deletion strain to prevent expression of endogenous Ty retroelements. Cells lacking the SPT3 protein show pleiotropic phenotypes and the his3Δ200 deletion impacts mitochondrial maintenance, galactose induction and senescence. (ii) TyHRT elements are expressed from a galactose-responsive promoter. Growth and promoter activity in the presence of galactose varies due to genetic (including the spt3 mutation) and environmental factors. (iii) Lysis of cells is achieved via freeze/thaw of cell pellets and the extent of lysis varies within a 96 well plate and from experiment to experiment. (iv) Luciferase is measured in yeast cell extracts in the presence of cellular proteases that degrade luciferase. These parameters combine to contribute to the variability of the TyHRT-luciferase assay.
Antiviral Assays
WT, I132M, and I135M HIV-1LAI virus stocks were made by transfection of HEK-293T (human embryonic kidney 293T) cells with the plasmids using Lipofectamine™ 2000 (Invitrogen). Viruses were titered on GHOST cells expressing CD4 and CXCR4 [15]. TZM indicator cells (formerly called JC53 BL13+ cells) were infected in duplicate with each virus at an equal multiplicity of infection in the presence or absence of 5-fold dilutions of inhibitors, as previously described [16]. Each inhibition assay was performed in two independent experiments.
RESULTS
Residues 132 and 135 form part of the β7–β8 loop (residues 132–140) in HIV-1 RT. In the 66 kDa (p66) subunit of RT, this loop is situated in the fingers domain and resides a substantial distance from both the DNA polymerase active site and NNRTI-BP. In the 51 kDa (p51) subunit of RT, the β7–β8 loop is situated in the dimer interface and contributes to the formation of the base of the NNRTI-BP (Figure 1). However, Ile132 and Ile135 do not directly interact with the bound ligand nor do they form part of the NNRTI-BP (Figure 1). Accordingly, we hypothesized that mutations at residues 132 and 135 in HIV-1 RT might decrease the susceptibility of the enzyme to NNRTI by inducing subtle conformational changes in the β7–β8 loop that influence the ability of other residues to interact optimally with the bound ligand. Therefore in the present study we have characterized mutations at residues 132 (I132A and I132M) and 135 (I135A and I135M) to elucidate their specific roles in NNRTI resistance. We also performed alanine scanning mutagenesis of residues 136–140 to delineate the roles of each of these residues in NNRTI resistance. Furthermore, RT containing E138K or T139V was included in the present study since these mutations have previously been associated with NNRTI resistance [17,18].
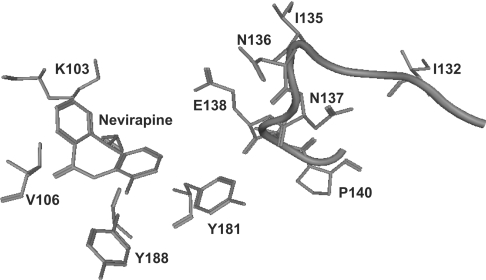
Figure 1. Structural representation of nevirapine bound in the NNRTI-BP of HIV-1 RT.
The NNRTI-BP is formed predominantly from residues in the p66 subunit of HIV-1 RT. These include Lys103, Val106, Tyr181 and Tyr188 (depicted). The β7–β8 loop in the p51 subunit of RT (residues 132–140) is located at the base of the NNRTI-BP. Residues Ile132 and Ile135, however, do not directly interact with the bound ligand. (In this structure, Ile132 and Ile135 are located ∼20 Å and 13 Å from nevirapine.) This Figure was generated using PDB co-ordinates 3HVT and MOE software (Chemical Computing Group).
Activity of WT and mutant HIV-1 RT
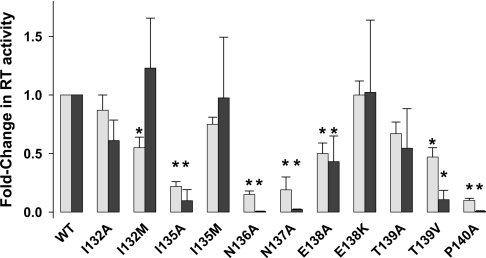
As described above, we generated 11 mutant HIV-1 RT constructs that harboured the following mutations: I132A, I132M, I135A, I135M, N136A, N137A, E138A, E138K, T139A, T139V or P140A. Each of the mutant enzymes was purified to homogeneity and analysed for RNA-dependent DNA polymerase activity. The mutant RT constructs were also introduced into the TyHRT system and their relative reverse transcription activities were assessed using a luciferase-based reverse transcription reporter. In general, the activities of each of the mutant enzymes were similar in both assay systems (Figure 2), although some differences can be noted. These differences may be due to the fact that TyHRT activity is dependent on multiple RT functions whereas only RNA-dependent DNA polymerase activity was assessed for the purified recombinant enzymes. Of note, the substitutions I132A, I132M, I135M and E138K were well tolerated by HIV-1 RT. By contrast, the activities of the I135A, N136A, N137A, E138A, T139A, T139V and P140A enzymes were compromised.
Figure 2. Activity of mutant HIV-1 RT determined using recombinant purified enzymes or the TyHRT assay.
Data are reported as an average±S.D. of at least three separate experiments. The grey and black bars represent the fold-change in RT activity using recombinant purified enzymes (grey bars) or the TyHRT assay (black bars). *P<0.01 compared with WT (Student's t test).
Far-UV CD analyses of WT and mutant HIV-1 RT
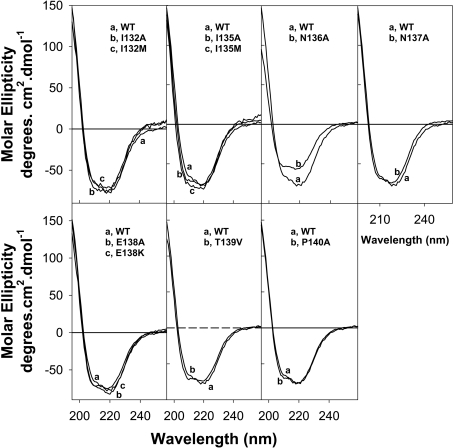
To determine whether the reduced activity of the mutant enzymes was a result of protein misfolding, far-UV CD spectroscopic analyses of WT and mutant HIV-1 RT were performed. The CD spectra for WT and all mutant enzymes, except for the N136A RT, were essentially identical (Figure 3) indicating that these mutations did not impact on the spatial arrangement or secondary structure of the peptide backbone. In regard to N136A HIV-1 RT, a recent study also demonstrated that mutations at this codon perturb the secondary structure of the enzyme [19]. Therefore the loss of reverse transcription activity associated with the N136A mutation may be due to protein-associated conformational changes.
Figure 3. Far-UV CD spectra of WT and mutant HIV-1 RT.
Role of β7–β8 loop in RT heterodimer stability
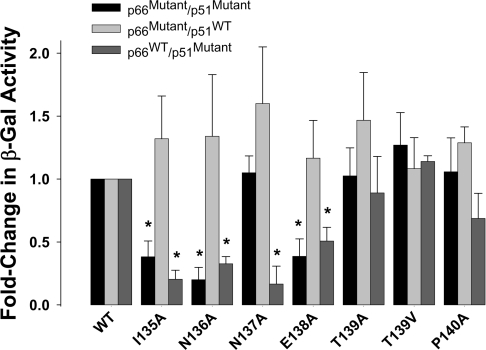
The DNA polymerase activity of HIV-1 RT is tightly coupled to the quaternary structure of the enzyme [20,21]. Because the β7–β8 loop resides in the dimer interface where many of the residues participate in the subunit–subunit interactions of RT [19,22–24], we were interested to determine whether the loss of DNA polymerase activity associated with the I135A, N136A, N137A, E138A, T139A, T139V and P140A mutations was related to defects in subunit dimerization. Accordingly, we analysed the ability of these mutant enzymes to form functional heterodimers using the yeast two-hybrid assay [14] (Figure 4). In this assay system, p66 was fused to the LexA DNA-binding domain ‘the bait’ and p51 was fused to the Gal4 activation domain (GalAD) ‘the prey’. Specific interaction between the two subunits resulted in trans-activation of the Lac Z reporter gene in the yeast strain CTY10–5d, which permits the interaction for the RT subunits to be quantified by assaying for β-gal activity. Initially, β-gal activity in yeast co-expressing mutant p66 bait and p51 prey fusions engineered with the same mutations in each subunit was measured. This analysis revealed that the substitutions I135A, N136A and E138A significantly decreased β-gal activity compared with yeast expressing the WT p66 bait and p51 fusion proteins (Figure 4). To delineate the effects of mutations in each of the subunits on RT dimerization, we performed a subunit-selective analysis. To analyse the effects of mutations in the p66 subunit we co-transformed yeast with mutant p66 bait and WT p51 prey, and to analyse the effects of mutations in the p51 subunit we co-transformed yeast with WT p66 bait and mutant p51 prey. Our data show that the mutations in the p51 subunit were largely responsible for the observed decrease in β-gal activity (Figure 4).
Figure 4. Effect of mutations on the β-gal readout in the yeast two-hybrid RT dimerization assay.
Yeast reporter strain CTY10-5d was co-transformed with p66 bait and p51 prey constructs containing mutations in β7–β8 and assayed for β-gal activity. β-gal activity (abscissa) represents the average of three transformants from three independent experiments. *P<0.01 compared with WT (Student's t test).
Decreases in β-gal activity, as measured in a yeast two-hybrid assay, may represent a true diminution in RT subunit interaction or may be due to decreased expression levels of either the bait or prey fusions compared with WT protein. To distinguish between these two possibilities we compared the steady-state protein levels in yeast expressing mutant p66 bait and p51 prey fusion proteins with the WT proteins. Yeast protein extracts were prepared from exponentially growing cells and subjected to Western blot analysis. The p66 bait was detected using LexA antibodies and p51 prey detected using Gal4AD antibodies. No differences in the steady-state protein levels of the p66 bait or p51 prey mutants were observed compared with the WT subunits (results not shown). This result indicates that the observed decreases in β-gal activity were entirely due to a diminution in RT subunit interactions.
NNRTI susceptibility of WT and mutant HIV-1 RT
The NNRTI susceptibility of WT and mutant HIV-1 RT to efavirenz, nevirapine and delavirdine was assessed in three complementary assay systems. These included analyses of recombinant purified RT, the TyHRT assay and antiviral assays in TZM indicator cells (Table 1). In each of the assays, the I132M mutation conferred high-level resistance (>than 10-fold) to nevirapine and delavirdine and low-level resistance (∼2-fold) to efavirenz. The I132A mutation conferred low-level resistance to nevirapine and delavirdine in assays that used purified RT. The I135M mutation also conferred low-level resistance (∼2-fold) to all NNRTIs tested in each of the assays, whereas the I135A mutation conferred high-level resistance to nevirapine and delavirdine, but not to efavirenz, only at the enzyme level. By contrast, all other mutations, with the exception of the E138A and E138K mutations, conferred essentially no resistance to each of the NNRTIs tested.
Table 1. NNRTI susceptibility of WT and mutant HIV-1 RT.
The concentrations of NNRTI required to inhibit the activity of purified RT and TyHRT by 50% (IC50) are reported as an average of three independent experiments±S.D. The effective concentrations of NNRTI required to inhibit virus replication by 50% (EC50) are reported as an average of two independent experiments±S.D. Values in parentheses indicate fold-resistance.
| IC50/EC50 | ||||
|---|---|---|---|---|
| RT | Assay | Efavirenz (nM) | Nevirapine (μM) | Delavirdine (μM) |
| WT | Purified RT | 94.16±6.33 (1.0) | 7.22±1.38 (1.0) | 2.18±0.62 (1.0) |
| TyHRT | 1.73±0.64 (1.0) | 1.16±0.55 (1.0) | * | |
| Antiviral | 0.90±0.00 (1.0) | 0.03±0.00 (1.0) | 0.04±0.01 (1.0) | |
| I132A | Purified RT | 90.07±5.11 (1.0) | 18.05±2.35 (2.5) | 8.70±1.02 (4.0) |
| TyHRT | 2.78±0.97 (1.6) | 1.95±0.52 (1.7) | * | |
| I132M | Purified RT | 432.42±11.31 (4.6) | 62.09±3.69 (8.6) | 30.25±3.35 (13.9) |
| TyHRT | 6.63±2.43 (3.7) | 7.10±1.67 (6.1) | * | |
| Antiviral | 2.00±0.00 (2.2) | 1.25±1.10 (42) | 0.80±0.20 (20.0) | |
| I135A | Purified RT | 188.08±9.34 (2.0) | 87.36±4.56 (12.1) | 34.60±5.59 (15.9) |
| TyHRT | 1.62±0.47 (0.9) | 1.63±0.58 (1.4) | * | |
| I135M | Purified RT | 197.09±13.18 (2.1) | 38.99±2.89 (5.4) | 3.92±0.99 (1.8) |
| TyHRT | 2.76±1.15 (1.6) | 2.02±0.50 (1.7) | * | |
| Antiviral | 2.00±0.00 (2.2) | 0.07±0.01 (2.3) | 0.05±0.02 (1.3) | |
| N136A† | ||||
| N137A | Purified RT | 87.35±22.28 (0.9) | 6.50±1.10 (0.9) | 1.52±0.55 (0.7) |
| TyHRT | 1.30±0.14 (0.8) | 1.28±0.37 (1.1) | * | |
| E138A | Purified RT | 188.08±12.11 (2.0) | 13.72±1.57 (1.9) | 7.62±0.91 (3.5) |
| TyHRT | 4.11±2.09 (2.4) | 1.19±1.03 (1.0) | * | |
| E138K | Purified RT | 216.34±12.13 (2.3) | 15.88±2.26 (2.2) | 13.06±1.56 (6.0) |
| TyHRT | 3.52±1.04 (2.0) | 1.51±0.75 (1.3) | * | |
| T139A† | ||||
| T139V | Purified RT | 113.44±9.23 (1.2) | 12.27±1.89 (1.7) | 2.61±0.97 (1.2) |
| TyHRT | 1.74±0.85 (1.0) | 1.85±0.24 (1.6) | * | |
| P140A† | ||||
*Delavirdine is not soluble in yeast media.
†RT activity too low to effectively evaluate NNRTI susceptibility.
To delineate the effects of I132M, I135M and E138K in each of the RT subunits on NNRTI susceptibility, we performed a subunit-selective analysis using purified enzyme that harboured each mutation in either the p66 or p51 subunit. The data (Table 2) clearly demonstrate that the NNRTI resistance is conferred by the mutations present in the p51, and not p66, subunit.
Table 2. NNRTI susceptibility of HIV-1 RT containing subunit specific mutations.
The concentrations of NNRTI required to inhibit the activity of purified RT by 50% (IC50) are reported as an average of three independent experiments±S.D. Values in parentheses indicate fold-resistance.
| IC50 | |||
|---|---|---|---|
| Efavirenz (nM) | Nevirapine (μM) | Delavirdine (μM) | |
| p66WT/p51WT | 15.44±3.16 (1.0) | 5.21±1.08 (1.0) | 1.68±0.50 (1.0) |
| p66I132M/p51I132M | 69.08±9.02 (4.6) | 44.80±4.26 (8.6) | 23.35±2.56 (13.9) |
| p66I132M/p51WT | 15.34±4.46 (1.0) | 5.20±1.05 (1.0) | 1.60±0.85 (1.0) |
| p66WT/p51I132M | 78.19±10.05 (5.2) | 47.93±6.36 (9.2) | 21.33±3.85 (12.7) |
| p66I135M/p51I135M | 32.11±1.09 (2.1) | 28.13±3.36 (5.4) | 3.02±0.45 (1.8) |
| p66I135M/p51WT | 15.37±8.02 (1.0) | 4.91±1.25 (0.9) | 1.70±0.55 (1.0) |
| p66WT/p51I132M | 35.36±1.19 (2.3) | 25.52±2.88 (4.9) | 3.86±0.35 (2.3) |
| p66E138K/p51E138K | 35.17±1.29 (2.3) | 11.46±2.77 (2.2) | 10.08±2.22 (6.0) |
| p66E138K/p51WT | 15.21±7.03 (1.0) | 5.00±1.10 (1.0) | 1.64±0.65 (1.0) |
| p66WT/p51E138K | 36.22±9.02 (2.4) | 12.50±3.20 (2.4) | 10.25±2.70 (6.1) |
NRTI susceptibility of WT and mutant HIV-1 RT
The susceptibility of recombinant purified WT and mutant HIV-1 RT to the NRTI AZT-TP (3′-azido-3′-dideoxythymidine triphosphate), ddTTP (3′-dideoxythymidine triphosphate) and d4T-TP (2′,3′-didehydrothymidine triphosphate) was also assessed. These data (Table 3) demonstrated that none of the mutant enzymes exhibited any changes in susceptibility to each of the nucleoside analogue inhibitors tested.
Table 3. NRTI susceptibility of WT and mutant HIV-1 RT.
The concentrations of NRTI required to inhibit the activity of purified RT by 50% (IC50) are reported as an average of three independent experiments±S.D. Values in parentheses indicate fold-resistance
| IC50 (μM) | |||
|---|---|---|---|
| ddTTP | AZT-TP | d4T-TP | |
| WT | 0.10±0.05 (1.0) | 0.06±0.01 (1.0) | 0.20±0.02 (1.0) |
| I132A | 0.07±0.02 (0.7) | 0.02±0.01 (0.3) | 0.13±0.03 (0.7) |
| I132M | 0.06±0.01 (0.6) | 0.03±0.01 (0.5) | 0.12±0.04 (0.6) |
| I135A | 0.08±0.02 (0.8) | 0.04±0.01 (0.6) | 0.16±0.05 (0.8) |
| I135M | 0.09±0.03 (0.9) | 0.06±0.01 (1.0) | 0.17±0.05 (0.9) |
| N136A | * | ||
| N137A | 0.10±0.04 (1.0) | 0.06±0.02 (1.0) | 0.18±0.04 (0.9) |
| E138A | 0.09±0.02 (0.9) | 0.03±0.01 (0.5) | 0.16±0.06 (0.8) |
| E138K | 0.10±0.03 (1.0) | 0.07±0.02 (1.2) | 0.16±0.06 (0.8) |
| T139A | * | ||
| T139V | 0.10±0.05 (1.0) | 0.06±0.02 (1.0) | 0.14±0.06 (0.7) |
| P140A | * | ||
* RT activity too low to effectively evaluate NRTI susceptibility.
DISCUSSION
The detection and characterization of minor drug-resistant variants is important for the clinical management of HIV infection and for studies dissecting the mechanisms of antiretroviral treatment failure. Using the TyHRT phenotypic assay, we detected unusual and rare NNRTI resistance mutations at codons 132 and 135 in RTs (in particular I132M and I135M) from clinical isolates [6–8]. Other studies have also detected NNRTI resistance-associated mutations at codon 135 in HIV-1 RT [18,25]. In the present study we demonstrate that these mutations confer NNRTI resistance both at the virus and enzyme level. In fact, the I132M mutation conferred high-level resistance (>10-fold) to nevirapine and delavirdine in each of the assay systems tested.
HIV-1 RT is a heterodimer composed of a 66 kDa (p66) subunit and a p66-derived 51 kDa (p51) subunit [26]. Our subunit selective mutagenesis studies demonstrated that I132M and I135M confer NNRTI resistance via the p51, and not p66, subunit of HIV-1 RT. In p51, the β7–β8 subunit (residues 132–140) resides in the dimer interface and contributes to the base of the NNRTI-BP (Figure 1). However, Ile132 and Ile135 do not interact with the bound ligand nor do they directly form part of the NNRTI-BP. As described in the Results section, we hypothesized that mutations at Ile132 and Ile135 may decrease NNRTI susceptibility by acting through other residues in the β7–β8 loop. Accordingly, we also carried out alanine-scanning mutagenesis studies of residues 136–140. Interestingly, most of these mutations significantly decreased the activity of RT. CD and yeast two-hybrid analyses suggested that for some enzymes this was due to either a conformational change induced in RT structure (N136A) or a defect in RT heterodimer formation (I135A, N137A and E138A). These results are consistent with previously published studies which have demonstrated a key role for the β7–β8 loop in RT stability [19,22–24]. For the T139A, T139V and P140A mutations, however, the loss of RT activity could not be attributed to either a conformational change in RT structure or heterodimer instability. It should be noted though that previous studies suggested an important role for Thr139 and Pro140 in RT dimerization [23,24]. Despite the discrepancies between the present study and those carried out by Auwerx et al. [23,24], taken together our combined results highlight an important role for these residues in RT structure and function.
Whereas mutations at residues 132 and 135 in RT conferred NNRTI resistance, mutations at residues 137, 139 and 140 had no effect on drug susceptibility. The results are consistent with previous in-depth mutagenesis studies which demonstrated that residues 137, 139 and 140 did not influence NNRTI susceptibility [23,24]. NNRTI resistance mutations, however, have been selected at residue Thr139 but these are mostly associated with resistance to (+)-calanolide A and do not affect the sensitivity of nevirapine, delavirdine and efavirenz [23]. By contrast, mutations at residue 138 can significantly affect NNRTI susceptibility (the present study and [27–29]). Thus it might be possible that mutations at residues 132 and 135 affect NNRTI susceptibility via Glu138. A crystal structure of E138K HIV-1 RT in complex with nevirapine has recently been solved [30]. These data show that Glu138 indirectly interacts with the bound ligand via water molecules, and that the E138K mutation alters these interactions resulting in a small shift in the position of nevirapine, which may influence its ring stacking with Tyr181 [30]. We hypothesize that the I132M and I135M mutations may affect β7–β8 loop positioning and the concomitant interactions of Glu138 with the bound ligand. However, crystal structure analyses of I132M and I135M RTs in complex with NNRTI are required to confirm this hypothesis.
Acknowledgments
This research was supported in part by the NIH (National Institutes of Health) grant R01 GM068406-01 (to N. S.-C.) and with federal funds from the NCI (National Cancer Institute), NIH, under contract N01-CO-12400 (to D. V. N.). G. T. was supported by NHMRC (National Health and Medical Research Council) RD Wright Career Development Award 235102 and K. L. M. by NHMRC Project Grant 381703. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- 1.Tsai C. H., Lee P. Y., Stollar V., Li M. L. Antiviral therapy targeting viral polymerase. Curr. Pharm. Des. 2006;12:1339–1355. doi: 10.2174/138161206776361156. [DOI] [PubMed] [Google Scholar]
- 2.Goody R. S., Muller B., Restle T. Factors contributing to the inhibition of HIV reverse transcriptase by chain-terminating nucleotides in vitro and in vivo. FEBS Lett. 1991;291:1–5. doi: 10.1016/0014-5793(91)81089-q. [DOI] [PubMed] [Google Scholar]
- 3.De Clercq E. The role of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Antiviral Res. 1998;38:153–179. doi: 10.1016/s0166-3542(98)00025-4. [DOI] [PubMed] [Google Scholar]
- 4.Nissley D. V., Garfinkel D. J., Strathern J. N. HIV reverse transcription in yeast. Nature. 1996;380:30. doi: 10.1038/380030a0. [DOI] [PubMed] [Google Scholar]
- 5.Nissley D. V., Boyer P. L., Garfinkel D. J., Hughes S. H., Strathern J. N. Hybrid Ty1/HIV-1 elements used to detect inhibitors and monitor the activity of HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13905–13910. doi: 10.1073/pnas.95.23.13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nissley D. V., Halvas E. K., Hoppman N. L., Garfinkel D. J., Mellors J. W., Strathern J. N. Sensitive phenotypic detection of minor drug-resistant human immunodeficiency virus type 1 reverse transcriptase variants. J. Clin. Microbiol. 2005;43:5696–5704. doi: 10.1128/JCM.43.11.5696-5704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halvas E. K., Aldrovandi G. M., Balfe P., Beck I. A., Boltz V. F., Coffin J. M., Frenkel L. M., Hazelwood J. D., Johnson V. A., Kearney M., et al. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J. Clin. Microbiol. 2006;44:2612–2614. doi: 10.1128/JCM.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flys T., Nissley D. V., Claasen C. W., Jones D., Shi C., Guay L. A., Musoke P., Mmiro F., Strathern J. N., Jackson J. B., et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J. Infect. Dis. 2005;192:24–29. doi: 10.1086/430742. [DOI] [PubMed] [Google Scholar]
- 8a.Nissley D. V., Julias J., Mellors J. W., Hughes S. H., Strathern J. N. Detection and characterization of rare drug resistant HIV-1 RT variants using a sensitive phenotypic assay. Antiviral Ther. 2004;9:S140. [Google Scholar]
- 9.Shi C., Mellors J. W. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 1997;41:2781–2785. doi: 10.1128/aac.41.12.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Grice S. F., Cameron C. E., Benkovic S. J. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 1995;262:130–144. doi: 10.1016/0076-6879(95)62015-x. [DOI] [PubMed] [Google Scholar]
- 11.Le Grice S. F., Gruninger-Leitch F. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem. 1990;187:307–314. doi: 10.1111/j.1432-1033.1990.tb15306.x. [DOI] [PubMed] [Google Scholar]
- 12.Maier G., Dietrich U., Panhans B., Schroder B., Rubsamen-Waigmann H., Cellai L., Hermann T., Heumann H. Mixed reconstitution of mutated subunits of HIV-1 reverse transcriptase coexpressed in Escherichia coli-two tags tie it up. Eur. J. Biochem. 1999;261:10–18. doi: 10.1046/j.1432-1327.1999.00304.x. [DOI] [PubMed] [Google Scholar]
- 13.Miranda L. R., Gotte M., Liang F., Kuritzkes D. R. The L74V mutation in human immunodeficiency virus type 1 reverse transcriptase counteracts enhanced excision of zidovudine monophosphate associated with thymidine analog resistance mutations. Antimicrob. Agents Chemother. 2005;49:2648–2656. doi: 10.1128/AAC.49.7.2648-2656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tachedjian G., Aronson H. E., Goff S. P. Analysis of mutations and suppressors affecting interactions between the subunits of the HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6334–6339. doi: 10.1073/pnas.97.12.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y. J., Dragic T., Cao Y., Kostrikis L., Kwon D. S., Littman D. R., KewalRamani V. N., Moore J. P. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J. Virol. 1998;72:9337–9344. doi: 10.1128/jvi.72.11.9337-9344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambrose Z., Boltz V., Palmer S., Coffin J. M., Hughes S. H., Kewalramani V. N. In vitro characterization of a simian immunodeficiency-human immunodeficiency virus (HIV) chimera expressing HIV Type 1 reverse transcriptase to study antiviral resistance in pigtail macaques. J. Virol. 2004;78:13553–13561. doi: 10.1128/JVI.78.24.13553-13561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balzarini J., Karlsson A., Sardana V. V., Emini E. A., Camarasa M. J., De Clercq E. Human immunodeficiency virus 1 (HIV-1)-specific reverse transcriptase (RT) inhibitors may suppress the replication of specific drug-resistant (E138K)RT HIV-1 mutants or select for highly resistant (Y181C→C181I)RT HIV-1 mutants. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6599–6603. doi: 10.1073/pnas.91.14.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y., Paxinos E., Galovich J., Troyer R., Baird H., Abreha M., Kityo C., Mugyenyi P., Petropoulos C., Arts E. J. Characterization of a subtype D human immunodeficiency virus type 1 isolate that was obtained from an untreated individual and that is highly resistant to nonnucleoside reverse transcriptase inhibitors. J. Virol. 2004;78:5390–5401. doi: 10.1128/JVI.78.10.5390-5401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balzarini J., Auwerx J., Rodriguez-Barrios F., Chedad A., Farkas V., Ceccherini-Silberstein F., Garcia-Aparicio C., Velazquez S., De Clercq E., Perno C. F., et al. The amino acid Asn136 in HIV-1 reverse transcriptase (RT) maintains efficient association of both RT subunits and enables the rational design of novel RT inhibitors. Mol. Pharmacol. 2005;68:49–60. doi: 10.1124/mol.105.012435. [DOI] [PubMed] [Google Scholar]
- 20.Restle T., Muller B., Goody R. S. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. A target for chemotherapeutic intervention. J. Biol. Chem. 1990;265:8986–8988. [PubMed] [Google Scholar]
- 21.Tachedjian G., Radzio J., Sluis-Cremer N. Relationship between enzyme activity and dimeric structure of recombinant HIV-1 reverse transcriptase. Proteins. 2005;60:5–13. doi: 10.1002/prot.20480. [DOI] [PubMed] [Google Scholar]
- 22.Pandey P. K., Kaushik N., Talele T. T., Yadav P. N., Pandey V. N. The β7-β8 loop of the p51 subunit in the heterodimeric (p66/p51) human immunodeficiency virus type 1 reverse transcriptase is essential for the catalytic function of the p66 subunit. Biochemistry. 2001;40:9505–9512. doi: 10.1021/bi002872j. [DOI] [PubMed] [Google Scholar]
- 23.Auwerx J., Rodriguez-Barrios F., Ceccherini-Silberstein F., San-Felix A., Velazquez S., De Clercq E., Camarasa M. J., Perno C. F., Gago F., Balzarini J. The role of Thr139 in the human immunodeficiency virus type 1 reverse transcriptase sensitivity to (+)-calanolide A. Mol. Pharmacol. 2005;68:652–659. doi: 10.1124/mol.105.012351. [DOI] [PubMed] [Google Scholar]
- 24.Auwerx J., Van Nieuwenhove J., Rodriguez-Barrios F., de Castro S., Velazquez S., Ceccherini-Silberstein F., De Clercq E., Camarasa M. J., Perno C. F., Gago F., Balzarini J. The N137 and P140 amino acids in the p51 and the P95 amino acid in the p66 subunit of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase are instrumental to maintain catalytic activity and to design new classes of anti-HIV-1 drugs. FEBS Lett. 2005;579:2294–2300. doi: 10.1016/j.febslet.2005.02.077. [DOI] [PubMed] [Google Scholar]
- 25.Leigh-Brown A. J., Precious H. M., Whitcomb J. M., Wong J. K., Quigg M., Huang W., Daar E. S., D'Aquila R. T., Keiser P. H., Connick E., et al. Reduced susceptibility of human immunodeficiency virus type 1 (HIV-1) from patients with primary HIV infection to nonnucleoside reverse transcriptase inhibitors is associated with variation at novel amino acid sites. J. Virol. 2000;74:10269–10273. doi: 10.1128/jvi.74.22.10269-10273.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 27.Sato A., Hammond J., Alexander T. N., Graham J. P., Binford S., Sugita K., Sugimoto H., Fujiwara T., Patick A. K. In vitro selection of mutations in human immunodeficiency virus type 1 reverse transcriptase that confer resistance to capravirine, a novel nonnucleoside reverse transcriptase inhibitor. Antiviral Res. 2006;70:66–74. doi: 10.1016/j.antiviral.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Hazen R. J., Harvey R. J., St Clair M. H., Ferris R. G., Freeman G. A., Tidwell J. H., Schaller L. T., Cowan J. R., Short S. A., Romines K. R., et al. Anti-human immunodeficiency virus type 1 activity of the nonnucleoside reverse transcriptase inhibitor GW678248 in combination with other antiretrovirals against clinical isolate viruses and in vitro selection for resistance. Antimicrob. Agents Chemother. 2005;49:4465–4473. doi: 10.1128/AAC.49.11.4465-4473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferris R. G., Hazen R. J., Roberts G. B., St Clair M. H., Chan J. H., Romines K. R., Freeman G. A., Tidwell J. H., Schaller L. T., Cowan J. R., et al. Antiviral activity of GW678248, a novel benzophenone nonnucleoside reverse transcriptase inhibitor. Antimicrob. Agents Chemother. 2005;49:4046–4051. doi: 10.1128/AAC.49.10.4046-4051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren J., Nichols C. E., Stamp A., Chamberlain P. P., Ferris R., Weaver K. L., Short S. A., Stammers D. K. Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138. FEBS J. 2006;273:3850–3860. doi: 10.1111/j.1742-4658.2006.05392.x. [DOI] [PubMed] [Google Scholar]