Abstract
β2-ARs (β2-adrenoceptors) become desensitized rapidly upon recruitment of cytosolic β-arrestin. PDE4D5 (family 4 cAMP-specific phosphodiesterase, subfamily D, isoform 5) can be recruited in complex with β-arrestin, whereupon it regulates PKA (cAMP-dependent protein kinase) phosphorylation of the β2-AR. In the present study, we have used novel technology, employing a library of overlapping peptides (25-mers) immobilized on cellulose membranes that scan the entire sequence of β-arrestin 2, to define the interaction sites on β-arrestin 2 for binding of PDE4D5 and the cognate long isoform, PDE4D3. We have identified a binding site in the β-arrestin 2 N-domain for the common PDE4D catalytic unit and two regions in the β-arrestin 2 C-domain that confer specificity for PDE4D5 binding. Alanine-scanning peptide array analysis of the N-domain binding region identified severely reduced interaction with PDE4D5 upon R26A substitution, and reduced interaction upon either K18A or T20A substitution. Similar analysis of the β-arrestin 2 C-domain identified Arg286 and Asp291, together with the Leu215–His220 region, as being important for binding PDE4D5, but not PDE4D3. Transfection with wild-type β-arrestin 2 profoundly decreased isoprenaline-stimulated PKA phosphorylation of the β2-AR in MEFs (mouse embryo fibroblasts) lacking both β-arrestin 1 and β-arrestin 2. This effect was negated using either the R26A or the R286A mutant form of β-arrestin 2 or a mutant with substitution of an alanine cassette for Leu215–His220, which showed little or no PDE4D5 binding, but was still recruited to the β2-AR upon isoprenaline challenge. These data show that the interaction of PDE4D5 with both the N- and C-domains of β-arrestin 2 are essential for β2-AR regulation.
Keywords: β2-adrenoceptor, β-arrestin, cAMP, desensitization, peptide array, phosphodiesterase 4 (PDE4)
Abbreviations: AKAP79, A-kinase-anchoring protein 79; β2-AR, β2-adrenoceptor; ERK, extracellular-signal-regulated kinase; GFP, green fluorescent protein; GPCR, G-protein-coupled receptor; GRK, GPCR kinase; GST, glutathione S-transferase; HEK-293, human embryonic kidney; MEF, mouse embryonic fibroblast; PDE, phosphodiesterase; PKA, cAMP-dependent protein kinase; siRNA, small interfering RNA; VSV, vesicular-stomatitis virus
INTRODUCTION
Plasma-membrane-localized transmembrane GPCRs (G-protein-coupled receptors) provide a pivotal and ubiquitous system for cellular signal transduction as well as targets for therapeutics and understanding the molecular pathology of various diseases (see, e.g., [1–3]). Routinely, the regulation of GPCR functioning is characterized by a rapid and transient desensitization process [4]. This is mediated by their initial phosphorylation through the action of GRKs (GPCR kinases) [5], which allows for the recruitment of cytosolic arrestins that sterically forbids the association of GPCRs with their functional G-protein [4,6,7]. Although multisite phosphorylation of GPCRs by GRK is required for arrestin binding, the prime ‘phosphate sensor’ is a single arginine residue located in a ‘polar core’ located between the arrestin N- and C-domains [6]. Each of these arrestin domains is formed from a β-strand sandwich where two β-strand sheets are annealed through hydrophobic interactions between side chains inside the sandwich [6,8].
Studies of the β2-AR (β2-adrenoceptor) have played a fundamental role in dissecting out the functional significance of arrestin in regulating GPCR signalling [1,4,7]. Indeed, this system provides a critical paradigm given the importance of selective β2-AR agonists in the treatment of inflammatory lung disease, notably asthma [9], and that changes in β2-AR functioning, expression and polymorphisms relate to heart failure [1,2,10,11]. Mechanistically, agonist occupancy of the β2-AR allows it to couple to Gs, which leads to activation of adenylate cyclase and the generation of the key intracellular second messenger, cAMP [1,4]. GRK phosphorylation of the β2-AR initiates recruitment of β-arrestins, which attenuates β2-adrenoceptor–Gs coupling, thereby lowering adenylate cyclase activity [1,4,12].
It is now well appreciated that cAMP signalling in cells is compartmentalized [13–17], with gradients of cAMP shaped through spatially confined degradation achieved by the action of specifically tethered cAMP phosphodiesterases [18,19]. Isoforms encoded by the PDE4 (family 4 cAMP-specific phosphodiesterase) family play a pivotal role in shaping compartmentalized cAMP responses in many cell types, being sequestered to specific signalling complexes and membranes within cells [13,18,20–24]. Thus dominant-negative [25,26], siRNA (small interfering RNA)-mediated knockdown [27] and targeted gene-knockout approaches [28] have demonstrated that PDE4 isoforms and subfamilies can have unique non-redundant functional roles. The key roles that PDE4 isoforms play in regulating key processes in various cell types associated with the immune system and brain, for example, undoubtedly underpins the promising therapeutic use of PDE4-selective inhibitors for treating various inflammatory diseases and depression, and as cognitive enhancers [9,22,29,30].
Four PDE4 genes encode approx. 20 different isoforms [20,22,31,32]. Each isoform is characterized by a unique N-terminal region that has been shown in many instances to be involved in targeting to signalling complexes, membranes or the cytoskeleton [20,23]. It has been shown that PDE4 enzymes can associate with β-arrestin in a variety of cell types [33] and that the β-arrestin–PDE4 complex can be recruited to the β2-AR upon agonist occupancy [34]. The increased level of cAMP, occurring upon agonist occupancy, elicits the PKA (cAMP-dependent protein kinase)-mediated phosphorylation of the β2-AR [35]. Mediating this is a subpopulation of the RII form of PKA that is tethered to the β2-AR itself by sequestration to the AKAP79 (A-kinase-anchoring protein 79) scaffolding protein [27,36]. One functional consequence of PKA phosphorylation of the β2-AR, which is observed in various cell types, is to switch its coupling from the Gs-mediated activation of adenylate cyclase to the Gi-mediated activation of ERK (extracellular-signal-regulated kinase) [35]. β-Arrestin-recruited PDE4 acts to regulate specifically the phosphorylation of the β2-AR by AKAP79-sequestered PKA, presumably by lowering localized cAMP levels below the threshold required for activation by this spatially restricted pool of PKA [13,25,27,33,34]. β-Arrestin-recruited PDE4 also acts to regulate synaptic release probability and presynaptic inhibition by opioids [37] and, although PDE4 inhibitors have potential for treating asthma [22,29,38], β-arrestin 2 has been implicated in the development of allergic asthma [39].
PDE4 isoforms from all four families have the potential to interact with β-arrestin owing to the presence of a common inter-acting site located on the surface of helix-17 of their catalytic unit [33,40]. However, in a variety of cell types that express the PDE4D5 isoform, which include cardiac myocytes, it is this isoform that is found preferentially sequestered to β-arrestin [33]. Underpinning this is the presence of a further binding site for β-arrestin across a broad region of the isoform-unique N-terminal region of PDE4D5 in addition to the common interaction site found in its catalytic unit [33,40]. Thus the presence of these two binding sites presumably allows β-arrestin to straddle PDE4D5 and transport it to the β2-AR upon agonist challenge. Such a delivery allows β-arrestin-recruited PDE4D5 to attenuate PKA phosphorylation of the β2-AR and desensitize Gi signalling to ERK as β-arrestin desensitizes signalling through Gs [13,25].
Previous two-hybrid analyses of β-arrestin 2 truncates, indicated that both the N- and the C-domains of β-arrestin 2 contribute to the binding of PDE4D5 [33]. In the present study, we have explored the interaction of PDE4D5 with β-arrestin and suggest that the common PDE4 catalytic unit interacts with the extreme N-terminal region of β-arrestin, while the N-terminal region of PDE4D5 interacts with the C-domain of β-arrestin.
EXPERIMENTAL
Materials
A rabbit polyclonal antibody against the β2AR and PKA P-β2AR Ser345/346 was from Santa Cruz Biotechnology. Anti-FLAG M2 mouse monoclonal antibody, plus a conjugate of the same antibody to agarose, anti-VSV (vesicular-stomatitis virus) antibody, anti-VSV–agarose antibody and isoprenaline were from Sigma–Aldrich. Protein A beads used in immunoprecipitation reactions were from Invitrogen. Antisera detecting β-arrestin 1/2 and MEFs (mouse embryonic fibroblasts) from animals lacking both β-arrestin 1 and β-arrestin 2 [34,41] were from Dr Robert Lefkowitz (Howard Hughes Medical Center, Duke University, Durham, NC, U.S.A.). HEK-293β2 (human embryonic kidney) cells, which stably express a GFP (green fluorescent protein)- and FLAG-tagged β2-AR, were as described previously [42]. ECL® (enhanced chemiluminescence) kits were from GE Healthcare. cAMP determinations were carried out on cell lysates using the cAMP HTS (high-throughput screening) immunoassay kit from Upstate Biotechnology.
Cell culture
HEK-293 and HEK-293β2 cells were cultured as before [25,27,34,40]. Transfections were performed using Polyfect (Qiagen) according to the manufacturer's instructions. The MEF cell line derived from β-arrestin 1/2 double-knockout animals was transfected using Lipofectamine™ (Gibco-BRL) following the manufacturer's instructions and maintained in DMEM (Dulbecco's modified Eagle's medium) containing 10% (v/v) fetal bovine serum for HEK-293 cells and 10% newborn calf serum for HEK-293β2 cells, both with 1× penicillin/streptomycin. Cells were treated with 10 μM isoprenaline for 5 min, as indicated.
Immunopurification studies
Briefly, detergent-soluble proteins were isolated from cells by disruption in lysis buffer [1% (v/v) Triton X-100, 50 mM Hepes, pH 7.2, 10 mM EDTA and 100 mM NaH2PO4·2H2O] containing Complete™ protease inhibitor cocktail (Roche) to 8% volume. Detergent-insoluble proteins were removed by centrifugation at 10000 g for 10 min, and the soluble fraction was retained. Equal volumes of cell lysate containing 500 μg of protein were cleared by incubation with 30 μl of Protein A slurry. The beads were then removed by centrifugation at 10000 g for 10 min at 4 °C, and cleared lysate was incubated at 4 °C for 2 h with constant agitation with a volume of antiserum determined to immunoprecipitate all β2AR from β-arrestin 1−/−/β-arrestin 2−/− MEFs. Immunoglobulins were then isolated by incubation with Protein A–Sepharose beads for 1 h before retrieval by refrigerated centrifugation at 10000 g for 5 min. A similar protocol was used to isolate FLAG-tagged constructs; however, lysates were pre-cleared with VSV–agarose and immunopurifications were carried out using FLAG-tagged agarose. Immunopurified proteins were run on SDS/PAGE (4–12% NuPage Bis-Tris gradient gels) and immunoblotted as described previously [25,27,34,40].
Site-directed mutagenesis
Site-directed mutagenesis was performed using the circular mutagenesis method. All mutagenesis and deletion constructs were confirmed by DNA sequencing before use.
Mammalian cell expression constructs
Human PDE4D5 cDNA (GenBank® accession number AF012073) with a C-terminal VSV epitope tag, was cloned into pcDNA3 (Invitrogen) as described previously [33,40]. β-Arrestin 2 (GenBank® accession number BC007427 with a C-terminal FLAG epitope tag, was cloned into pcDNA3.
Expression of fusion proteins in Escherichia coli
Full-length PDE4D5 and PDE4D3 were each expressed as N-terminal GST (glutathione S-transferase)-fusion proteins and purified to apparent homogeneity as before [40,43].
SPOT synthesis of peptides and overlay experiments
Peptide libraries were produced by automatic SPOT synthesis [44] and synthesized on continuous cellulose membrane supports on Whatman 50 cellulose membranes using Fmoc (fluoren-9-ylmethoxycarbonyl) chemistry with the AutoSpot-Robot ASS 222 (Intavis Bioanalytical Instruments AG) [44,45]. The interaction of spotted peptides with GST and GST-fusion proteins was determined by overlaying the cellulose membranes with 10 μg/ml recombinant protein. Bound recombinant proteins were detected with specific rabbit antisera and detection was performed with secondary anti-rabbit horseradish-peroxidase-coupled antibody (1:2500 dilution) (Dianova) and visualization by ECL®, as described above.
RESULTS
Probing a β-arrestin 2 peptide array with PDE4D5 and PDE4D3
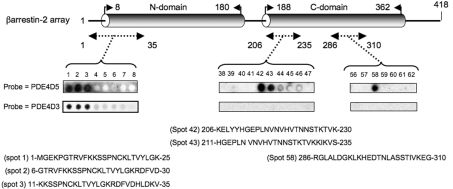
β-Arrestin 2 is a 418-amino-acid protein that consists of two distinct subdomains, called the N-domain and the C-domain, which are linked by a polar core (Figure 1). In co-immunoprecipitation, pull-down and two-hybrid analyses, it has been shown to bind to the PDE4D5 isoform [25,33,40]. In the present study, this interaction was explored using peptide array analysis, which provides a novel and powerful technology for gaining insight into the basis of specific protein–protein interactions [44,45]. In order to do this, a library of overlapping peptides (25-mers), each shifted by five amino acids across the entire sequence of β-arrestin 2, was SPOT-synthesized on cellulose membranes. This immobilized peptide library was probed with purified recombinant GST-fusion proteins of both PDE4D5 and PDE4D3, whose binding was assessed immunologically, with positive interactions identified as dark spots (Figure 1). PDE4D5 and PDE4D3 are long PDE4 isoforms from the same gene family (PDE4D) and differ only in their unrelated isoform-specific N-terminal region, which for PDE4D5 is 88 amino acids long and for PDE4D3 is 15 amino acids long [46]. Probing the β-arrestin 2 peptide library with PDE4D3–GST, positive reactions were only obtained with the first three peptide spots (Figure 1), representing the sequence at the extreme N-terminus of β-arrestin 2 (Met1–Val35). Probing the β-arrestin 2 peptide library with PDE4D5–GST, positive reactions were obtained not only with the first three peptide spots but also with two regions in the C-domain of β-arrestin 2 (Figure 1). One of these additional binding sites was within Lys206–Ser235 and the other within Arg286–Gly310 (Figure 1).
Figure 1. Probing a β-arrestin 2 peptide array with PDE4D5–GST and PDE4D3–GST.
β-Arrestin 2 is shown schematically with its N- and C-domains. Results show immobilized peptide ‘spots’ of overlapping 25-mer peptides each shifted along by five amino acids in the entire β-arrestin 2 sequence probed for interaction with either PDE4D5–GST or PDE4D3–GST and detection by immunoblotting. Positively interacting peptides generate dark spots, while those that do not interact leave white (blank) spots. In all other sections of the array, spots were blank with either probe. Spot numbers relate to peptides in the scanned array and whose sequence is given as indicated. Arrays probed with purified GST did not yield any positively interacting spots. These data are typical of experiments performed three times.
Purified GST did not bind to any spot within the β-arrestin 2 peptide array (results not shown). PDE4D5–GST and PDE4D3–GST failed to bind to any spots other than those shown in Figure 1 within the β-arrestin 2 peptide array (results not shown).
PDE4D5–GST binding to an alanine-scanning substitution array of Gly6–Asp30
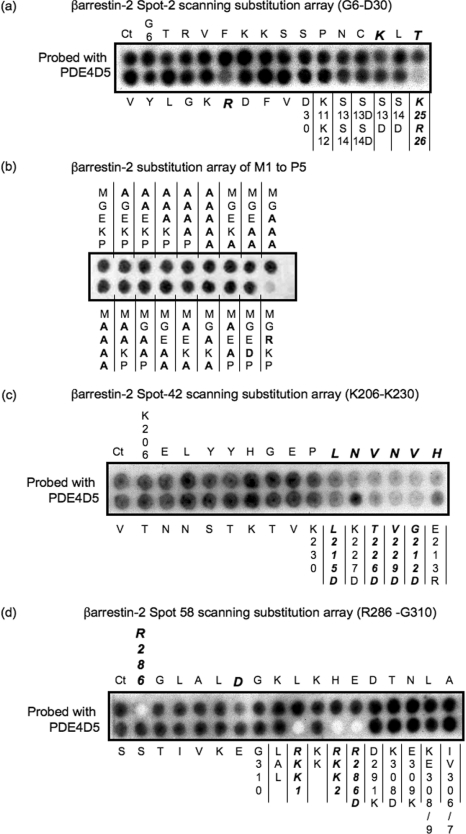
In order to define further the amino acids involved in allowing PDE4D5 to bind to the region of β-arrestin 2 to which PDE4D3 also binds (Figure 1), we screened a family of peptides derived from a 25-mer parent peptide whose sequence reflected Gly6–Asp30 of β-arrestin 2. The 25 progeny of this parent peptide each had a single substitution of alanine for successive amino acids in the sequence to form a scanning peptide array (Figure 2a). A severely reduced interaction was evident for the R26A substitution, with the dual K25A/R26A substitution essentially ablating PDE4D5–GST interaction (Figure 2a). In contrast, paired substitution of alanine for Lys11/Lys12 (K11A/K12A) failed to affect binding of PDE4D5–GST (Figure 2a). We noted reduced interaction of PDE4D5–GST upon substitution of alanine for Lys18 and Thr20 (Figure 2a). No effect on PDE4D5–GST binding was observed upon substitution of either alanine or aspartate for the Ser13/Ser14 pairing, or their individual aspartate substitution (Figure 2a).
Figure 2. Alanine-scanning substitution analysis to probe the binding sites for PDE4D5–GST in the N- and C-domains of β-arrestin 2.
β-Arrestin 2 peptide arrays were probed for PDE4D5–GST binding based upon the indicated 25-mer ‘parent’ β-arrestin 2 peptide, where the indicated amino acids were sequentially and individually replaced by alanine. Ct refers to the native peptide and all other spots reflect peptide ‘progeny’. GST alone did not bind to any peptide spot (results not shown). (a) Alanine substitution array for the Gly6–Asp30 peptide whose sequence is in the N-domain of β-arrestin 2, together with the additional indicated substitutions, which were to either alanine (no label) or aspartate (D). (b) Peptide representing amino acids 1–25 in β-arrestin 2 with changes made solely in the first five amino acids (native sequence MGEKP). The alterations in this portion are indicated using the single-letter amino acid code. These data are typical of experiments performed three times. (c) Alanine substitution array for the Lys206–Lys230 peptide whose sequence is in the C-domain of β-arrestin 2, together with the additional indicated substitutions which were either to aspartate (D) or to arginine (R). (d) Alanine substitution array for the Arg286–Gly310 peptide whose sequence is in the C-domain of β-arrestin 2, together with the additional indicated substitutions which were to either alanine (no label) or aspartate (D) or lysine (K). LAL is L288A/A289D/L290A. RKK1 is R286A/K293A/K295A. RKK2 is R286A/K293A/K295A/K308A. KE is K308A/E309A. IV is I306A/V307A.
Using a 1–25 β-arrestin 2 peptide we also probed the region from Met1 to Pro5 in β-arrestin 2, employing single and multiple replacements with alanine (Figure 2b). However, none of these altered the binding of the PDE4D5 probe. Interestingly, however, charge substitution by Q3R replacement ablated PDE4D5 interaction with the 1–25 β-arrestin 2 peptide, suggesting that, although a negative charge at this position is not required for PDE4D5 binding, the presence of a positive charge is inhibitory. Unfortunately, the crystal structure of β-arrestin 2 has no electron density in this region, which precludes any potential structural insight.
PDE4D5–GST binding to an alanine-scanning substitution array of Lys206–Lys230
To gain insight into the amino acids in the first region of the C-domain of β-arrestin 2 to which PDE4D5 binds specifically, we performed alanine-scanning mutagenesis of the Lys206–Lys230 region (Figure 2c) encompassed by spot 42 (Figure 1). In doing so, we found that substitution of alanine for any single one of a stretch of amino acids from Leu215 to His220 (sequence LNVNVH) severely attenuated/ablated interaction with PDE4D5–GST (Figure 2c). Consistent with this, substitution of aspartate for Leu215 also severely attenuated/ablated interaction with PDE4D5–GST (Figure 2c). However, replacement of the positively charged Lys227 with the negatively charged aspartate, as with alanine substitution, did not affect the interaction with PDE4D5–GST (Figure 2c).
PDE4D5–GST binding to an alanine-scanning substitution array of Arg286–Gly310
In order to gain insight into the amino acids in the second region of the C-domain of β-arrestin 2 to which PDE4D5 binds specifically (Figure 1), we performed alanine-scanning mutagenesis of the Arg286–Gly310 region (Figure 2d) encompassed by spot 58 (Figure 1). In doing so, we found that substitution of alanine for either Arg286 or Asp291 severely attenuated/ablated interaction with PDE4D5–GST (Figure 2d). Dual-alanine mutation of the Lys293/Lys295 pairing, however, had no effect on the PDE4D5–GST interaction (Figure 2d). As with single substitution of alanine for Arg286, peptides with a triple substitution of alanine for either Arg286/Lys293/Lys295 or Arg286/Lys293/Lys308 did not noticeably interact with PDE4D5–GST (Figure 2d). No effect on PDE4D5–GST interaction was observed upon substitution of either aspartate for Lys308 or lysine for Glu309 or upon their dual substitution, K308D/E309K (Figure 2d). Substitution of the Ile306/Val307 pairing with alanine (I306A/V307A) did not prevent interaction with PDE4D5–GST (Figure 2d).
The three regions of β-arrestin 2 identified in peptide array analyses are each involved in conferring PDE4D5 binding
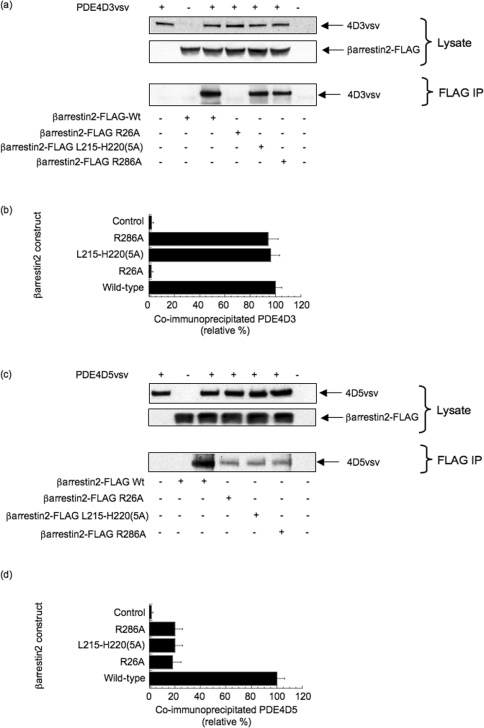
Our peptide array analyses identified three putative sites in β-arrestin 2 involved in conferring specificity for binding to PDE4D5: one in the N-domain deduced as allowing interaction with the PDE4(D) catalytic unit, and two in the C-domain interacting with the unique N-terminal region of PDE4D5. To explore this further, we generated defined mutations in each of these regions where scanning substitution peptide arrays had identified amino acids whose replacement with alanine led to compromised interaction with PDE4D5 (Figure 2). In the first analysis, we co-transfected COS cells with VSV-tagged PDE4D3 and either wild-type or the indicated mutant forms of FLAG-tagged β-arrestin 2. Using an antibody against the FLAG epitope tag, we were able to immunoprecipitate recombinant β-arrestin 2 specifically and determine whether or not recombinant PDE4D3 interacted by immunoblotting for the VSV epitope tag (Figures 3a and 3b). Doing this, we found that VSV–PDE4D3 was co-immunoprecipitated with wild-type β-arrestin 2 and with the R286A mutant and a mutant where the stretch of amino acids from 215 to 220 in β-arrestin 2 was replaced by a five-residue alanine cassette (Figures 3a and 3b). However, it was not co-immunoprecipitated with the R26A mutant form of β-arrestin 2 (Figures 3a and 3b). Repeating this, but co-transfecting with VSV epitope-tagged PDE4D5, we noted that, although VSV–PDE4D5 was co-immunoprecipitated with wild-type β-arrestin 2, its interaction was severely compromised for all of these various mutant forms of β-arrestin 2 (Figures 3c and 3d). These data support the peptide array analyses that indicated that all these three regions were involved in PDE4D5 interaction, whereas only that in the N-domain of β-arrestin 2 was involved in conferring interaction with PDE4D3. This suggests that the isoform-directed specificity of the interaction is conferred or determined by the C-domain of β-arrestin.
Figure 3. Consequences of mutations in the N- and C-domain binding sites in β-arrestin 2, based on peptide array data, for the binding of PDE4D3 and PDE4D5.
(a) HEK-293 cells were co-transfected with VSV-epitope-tagged PDE4D3 (GenBank® accession number L20970) together with the indicated forms of FLAG-tagged β-arrestin 2, namely either wild-type (Wt) or mutations of R26A, R286A and one where the stretch of amino acids from Leu215 to His220 in β-arrestin 2–FLAG was replaced by a five-residue alanine cassette [β-arrestin 2–FLAG L215–H220(5A)]. The lower panel shows the result of immunoprecipitating the indicated β-arrestin 2–FLAG construct with an anti-FLAG antibody and probing for co-immunoprecipitated PDE4D3–VSV by immunoblotting with an anti-VSV antibody. (b) Quantification, by densitometry, of co-immunoprecipitated PDE4D3–VSV from three separate experiments as in (a) with means±S.D. and with the indicated control analysing cells that had not been transfected with β-arrestin 2–FLAG, but which has been transfected with PDE4D3–VSV. (c) As in (a), but co-transfection with VSV-epitope-tagged PDE4D5 (GenBank® accession number AF012073) instead of PDE4D3–VSV. (d) As in (b), but co-transfection with PDE4D5–VSV instead of PDE4D3–VSV.
Probing mutations in the three regions of β-arrestin 2 involved in PDE4D5 interaction for functional consequences
The β2-AR becomes PKA-phosphorylated in MEFs and various other cell types owing to the action of an associated membrane-bound PKA whose activity is attenuated consequently upon the recruitment of PDE4D5, in complex with β-arrestin 2, to the β2-AR [25,27,40]. We exploited this property in order to determine whether the functional impact of PDE4D5 on the β2-AR is attenuated in MEFs transfected with forms of β-arrestin 2 mutated so as to compromise their interaction with PDE4D5.
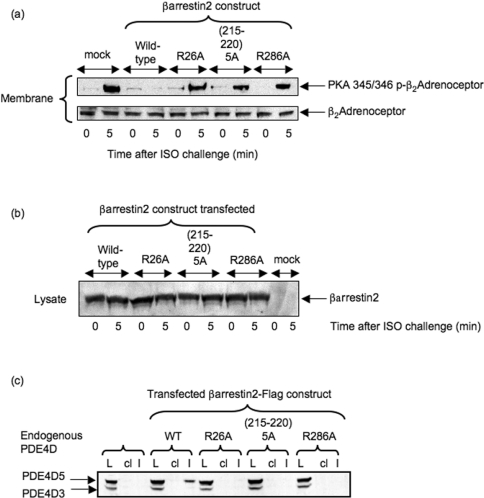
In order to evaluate this, cells were utilized that lack endogenous β-arrestins so as to incorporate mutant recombinant species, and compare their functioning with wild-type recombinant β-arrestin 2 in this system. β-Arrestin 2 is the predominant form of β-arrestin in MEFs [41]. However, MEFs generated from double-knockout mice, by the disruption of genes encoding both β-arrestin 1 and β-arrestin 2 [41], exhibit no immunologically detectable forms of either of these two proteins [34,41]. MEFs from β-arrestin 1−/−/β-arrestin 2−/− double-knockout mice were transfected to allow similar levels of expression with either wild-type β-arrestin 2 or the indicated mutant species (Figure 4b). MEFs express endogenously both PDE4D3 and PDE4D5, as shown from immunoblots of lysates from these cells (Figure 4c). However, as seen with other cells [40], it is PDE4D5 that preferentially interacts with β-arrestin 2, demonstrated here by its selective immunoprecipitation with β-arrestin 2 (Figure 4c). Disruption of any of the three sites on β-arrestin 2, suggested from scanning peptide arrays, by either a R26A mutation or a R286A mutation or replacement of the LNVNVH motif with an alanine cassette, ablated interaction with PDE4D5 as indicated by failure to co-immunoprecipitate (Figure 4c).
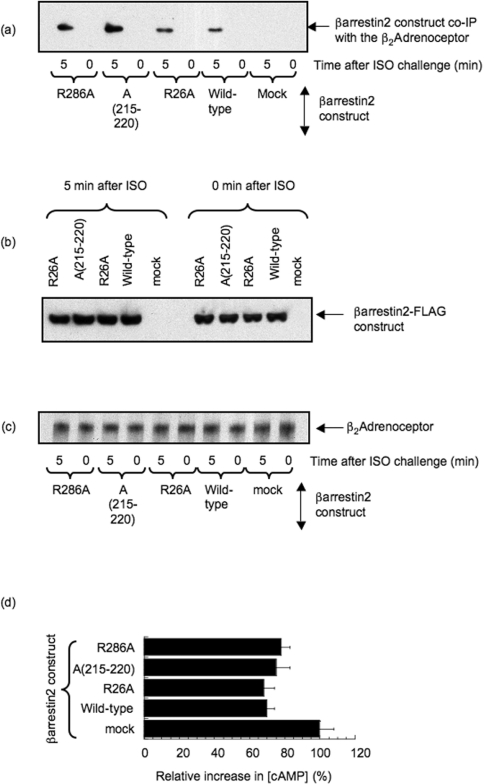
Figure 4. Functional consequences of mutations disrupting N- and C-domain binding sites in β-arrestin 2 for PDE4D5.
(a) Upper panel: PKA-phosphorylation status of the β2-AR, as assessed using a phospho-specific antiserum, in β-arrestin 1−/−/β-arrestin 2−/− double-knockout MEFs treated with 10 μM isoprenaline (ISO) for either 0 or 5 min, as indicated. This was assessed in both mock-transfected cells and also in cells transfected to express either wild-type β-arrestin 2–FLAG or the various β-arrestin 2–FLAG mutants as described in the legend to Figure 3. Lower panel: loading blot for the β2-AR in these cells. (b) Immunoblot of the indicated forms of wild-type and mutant β-arrestin 2 expressed in β-arrestin 1−/−/β-arrestin 2−/− double-knockout MEFs treated with isoprenaline (ISO) as indicated. (c) Immunoblot of endogenous PDE4D species in lysates (L) of MEFs transfected with the indicated wild-type and mutant forms of β-arrestin 2–FLAG. This detects the presence of endogenous PDE4D3 and PDE4D5 in lysates (L) from these cells. Also shown are PDE4D immunoblots of both FLAG immunoprecipitates (I) and bead control immunoprecipitates (cl), where no anti-FLAG antiserum was added. The blots are typical of experiments performed three times.
Challenge of MEFs from β-arrestin 1−/−/β-arrestin 2−/− double-knockout animals with 10 μM isoprenaline for 5 min caused a profound increase in PKA phosphorylation status of the β2-AR as detected using a phospho-antiserum (Figure 4a). However, transfection of these cells with wild-type β-arrestin 2 substantially decreased the PKA phosphorylation status of the β2-AR upon isoprenaline challenge (Figure 4a). In contrast with this, mutations to disrupt the interaction of PDE4D5 with β-arrestin 2, either in its N-domain (R26A) or in the first site in the C-domain (215–220 alanine cassette) or at the second site in the C-domain (R286A), all negated the inhibitory effect seen for wild-type β-arrestin 2 on isoprenaline-stimulated PKA phosphorylation of the β2-AR (Figure 4a).
Thus transfection of wild-type recombinant β-arrestin 2 ablates the isoprenaline-induced PKA-phosphorylation status of the β2-AR by allowing recruitment of PDE4D5. In contrast, such an action is not seen when using forms of β-arrestin 2 engineered so as to compromise their ability to bind PDE4D5 (Figure 4a).
It is possible that the differences between wild-type β-arrestin 2 and the various engineered species could also have arisen if such mutations had ablated their isoprenaline-dependent recruitment to the β2-AR. Addressing this, we have shown that wild-type recombinant FLAG-tagged β-arrestin 2 is recruited to the β2-AR upon challenge of MEFs with isoprenaline and that similar, or even greater, levels of recruitment are evident with the various mutants examined here (Figure 5a). Such analyses were carried out with similar levels of expression of the various recombinant β-arrestin 2 forms (Figure 5b) and analysis of immunoprecipitates containing equal amounts of β2-AR (Figure 5c). A key function of β-arrestin 2 is to cause desensitization of β2-AR-stimulated adenylate cyclase, thereby attenuating cAMP generation [4,7,47]. We can see that transfection of MEFs with wild-type recombinant β-arrestin 2 does indeed attenuate cAMP generation after a 5 min challenge with isoprenaline and that the various mutant forms of β-arrestin 2 act similarly (Figure 5d). Thus our various β-arrestin 2 mutants, although unable to bind PDE4D5, appear to both be recruited similarly to the β2-AR and similarly desensitize isoprenaline-stimulated adenylate cyclase activity.
Figure 5. Consequences of mutations in the N- and C-domain binding sites in β-arrestin 2, based on peptide array data, for the binding of PDE4D3 and PDE4D5.
(a) β-Arrestin 1−/−/β-arrestin 2−/− double-knockout MEFs were transfected to express either wild-type β-arrestin 2–FLAG, the various β-arrestin 2–FLAG mutants as described in the legend to Figure 3 or transfected with vector alone (mock). Cells were then challenged with 10 μM isoprenaline (ISO) for the indicated time before disruption for immunoprecipitation of the β2-AR. Immunoprecipitates from equal quantities of lysate protein were immunoblotted with an anti-FLAG antibody to detect associated β-arrestin 2–FLAG. (b) FLAG immunoblots of equal quantities of lysate protein from the transfection studies described in (a) using the indicated β-arrestin 2–FLAG constructs. (c) As in (b), but immunoblotting for the β2-AR. (d) Increase in intracellular cAMP level in β-arrestin 1−/−/β-arrestin 2−/− double-knockout MEFs transfected to express either wild-type β-arrestin 2–FLAG, the various β-arrestin 2–FLAG mutants as described in the legend to Figure 3 or transfected with vector alone (mock), after a 5 min challenge with isoprenaline. For comparison, the increase in intracellular cAMP is shown relative to that seen in mock-transfected cells, which lack both β-arrestin 1 and β-arrestin 2. Results are typical of three experiments given, in (d), as means±S.D. and where mock-transfected cells had 0.96±0.05 pmol of cAMP/μg of cell lysate protein, rising to 4.28±0.19 pmol of cAMP/μg of cell lysate protein after a 5 min challenge with isoprenaline.
DISCUSSION
β-Arrestins, as well as causing desensitization by binding to GRK-phosphorylated GPCRs, can bind various other proteins involved in mediating cell signalling actions [4,6,7,12], indicating that β-arrestins act as signal scaffolding proteins. Indeed, it now appears that far more proteins have the potential to interact with β-arrestin than could simply be accommodated upon any one molecule, suggesting competition for binding. This warrants determination of the ‘footprint’ that individual partners place upon the surface of β-arrestins in order to gain insight into which combinations of proteins may be allowed to scaffold with β-arrestins in any one complex.
Members of the 20+-strong family of PDE4s play a key role in underpinning compartmentalized cAMP signalling [13,18,24,27,48]. The first demonstration that a particular PDE isoform could exert a specific functional role in cells came from the use of dominant-negative and siRNA-knockdown strategies targeted at PDE4D5 [25–27,48]. These uncovered the ability of β-arrestin-complexed PDE4D5 to control the phosphorylation of β2-AR via a spatially restricted PKA population. One functional consequence of this is to allow switching of β2-AR signalling to ERK activation [35]. This process, which we have characterized in both HEK-293 cells and primary cardiac myocytes [25], depends upon the recruitment of a cytosolic β-arrestin–PDE4D5 complex to the plasma membrane-located β2-AR. There the local delivery of cAMP hydrolysing PDE regulates a PKA subpopulation, spatially constrained through tethering to the β2-AR by the scaffold protein AKAP79 [27].
PDE4D5 selectively interacts with β-arrestins in cells where it is expressed [33]. Such selectivity occurs because PDE4D5 not only has a β-arrestin-binding site on its conserved catalytic unit, through which all PDE4 isoforms can potentially interact with β-arrestin, but also has an additional β-arrestin-binding site over an extended surface of its isoform-specific N-terminal region [40]. We used a novel peptide array technology to identify these binding sites, which were confirmed by biochemical analysis [40]. We used such an approach to gain insight into how PDE4D5 interacts with β-arrestin 2. In order to do this, we probed a library of overlapping 25-mer peptides that scanned the entire sequence of β-arrestin 2 with not only PDE4D5, but also the cognate PDE4D3 long isoform. Through this we anticipated being able to identify regions on β-arrestin 2 that are involved in anchoring the ‘common’ PDE4(D) catalytic site, as well as those involved in anchoring the isoform-specific N-terminal region of PDE4D5.
Both PDE4D3 and PDE4D5 probes recognized spots 1–3, encompassing the first 35 amino acids of β-arrestin 2 (Figure 1). This sequence contributes β-strands 1 and 2 in a surface-exposed β-sheet motif within the arrestin N-domain [49], and its recognition by both PDE4D3 and PDE4D5 indicates that it is the conserved catalytic unit of PDE4 that interacts with this sequence. Focusing on peptide-2 (Gly6–Asp30), we generated a family of peptides in which successive amino acids in the sequence were singly substituted by alanine so as to form a scanning peptide array (Figure 2a). Analysis of this array identified reduced interaction of the PDE4D5 probe upon substitution of alanine for three amino acids, namely Lys18, Thr20 and Arg26, suggesting them to be of potential importance in conferring interaction with the PDE4 catalytic unit (Figure 2a). Alanine substitution of the positively charged Lys25, adjacent to Arg26, by itself had no effect on PDE4D binding (Figure 2a). The X-ray crystal structure of bovine β-arrestin 2, which reflects its basal conformation, being associated neither with PDE4 nor GPCR [49], and shows that Lys18 and Thr20 are located on β-strand 2 proximal to the β-1/2 hairpin and that both residues are prominently surface-exposed (Figure 6). Lys25 and Arg26, which are also surface-exposed, lie on a loop C-terminal to β-strand 2 and, in the crystal structure, the side chains of Lys25 and Arg26 are presented in diametrically opposed directions that, if mimicked in the peptide, may contribute to the selectivity of PDE4D interaction with these adjacent positively charged species. However, we note that the dual K25A/R26A substitution completely ablated interaction with PDE4D (Figure 2a), which suggests that it is likely that conformational flexibility at the C-terminal end of peptide-2 may allow Lys25 to facilitate, to some small extent, the interaction with PDE4D that Arg26 provides. We substantiated the role of Arg26 in full-length β-arrestin 2, where mutation to alanine ablated interaction with PDE4D5, as evinced by the failure of the two proteins to co-immunoprecipitate (Figures 3c and 3d).
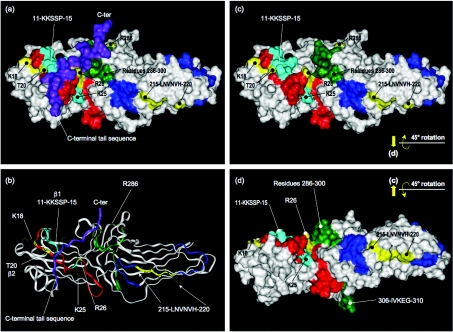
Figure 6. Structure of bovine β-arrestin 2 and location of residues implicated in PDE4D binding.
(a) Residues implicated in PDE4D binding mapped on to the solvent-accessible surface of bovine β-arrestin 2 in its basal conformation (PDB 1JSY); numbering used is taken from human β-arrestin 2 sequence. N-terminal residues 1–6 and residues 356–382 (preceding the C-terminal sequence) are disordered. Red (residues 7–35) corresponds to array peptides-1 to -3; dark blue (residues 206–235) corresponds to array peptides-42 and -43; green (residues 286–310) corresponds to array peptide-58; the arrestin C-terminal sequence is shown purple. Residues whose individual mutation to alanine significantly compromises PDE4D binding are highlighted in yellow. Lys25 and the K11KSSP15 sequence within peptide-2 are highlighted in light blue. (b) Ribbon representation of structure viewed as in (a). (c) As (a), but with the C-terminal sequence displaced. (d) As (c) but rotated 45° about the horizontal axis; phospho-GPCR docks from the rear of the structure as viewed.
In the β-arrestin 2 basal conformation (Figures 6a and 6b), Arg26 is partially occluded by folding of the arrestin C-terminal tail across the surface of β-strands 1 and 2. Indeed, the guanidinium functionality of Arg26 interacts with three residues in this tail, forming a salt bridge to Glu389 and hydrogen bonds to the main chain carbonyl groups of Phe391 and Ala392. Significantly, the recruitment of β-arrestin to phospho-GPCR is associated with a hinged re-orientation of the β-arrestin N- and C-domains in a conformational change that requires displacement of the C-terminal sequence from being folded over β-strands 1 and 2 in the basal state [6,12,47]. We suggest that the β-arrestin C-terminal sequence may be similarly displaced upon β-arrestin–PDE4D5 interaction and, in this way, the side chain of Arg26 might be more accessible for interaction with PDE4 (Figure 6c).
The difference between peptide-3, to which β-arrestin 2 binds, and peptide-4, to which it does not, is the sequence KKSSP (Figure 2a). This sequence is surface-exposed in the β-arrestin 2 crystal structure. However, mutation of any of these amino acids individually to alanine did not affect β-arrestin 2 binding (Figure 2a). Additionally, the dual K11A/K12A substitution had no effect, eliminating these positively charged residues from a potential interaction with the PDE4D catalytic unit. Indeed, Lys11 and Lys12 are thought to ‘guide’ the GRK-phosphorylated receptor to the key phosphate sensor provided by Arg170 located in a buried polar core within β-arrestin 2 [6,12,47]. Similarly, substitution of Ser13 and Ser14, either singly or together, with either alanine or aspartate, had little evident effect (Figure 2a) on the interaction of peptide-3 with PDE4D5. The importance of the KKSSP sequence in peptide-3 therefore probably arises from its impact on the peptide's conformation, as these residues contribute to β-strand 1 and pair in an antiparallel fashion with the sequence containing Lys18/Thr20 in β-strand 2. Thus, although individual residues within the KKSSP sequence may not interact directly with the PDE4D catalytic unit, the removal of this sequence in peptide-4 may destroy conformational control over the presentation of the Lys18/Thr20 side chains and/or the organization of these two residues relative to Arg26.
We propose that the PDE4D catalytic unit forms a contact surface spanning Lys18–Arg26 and running approximately along the axis of β-strand 2 of β-arrestin 2. It is conceivable that other residues on β-strands 1 and 2 may contribute direct interactions with the enzyme's surface, but that the extent of the interaction is insufficient to compromise binding of PDE4D5 to peptide-2 when residues are replaced individually by alanine. Given the potential importance of Lys18 and Arg26 for conferring binding to the PDE4 catalytic unit, we note from analysis of the PDE4D catalytic unit the presence of a negatively charged glutamate in the motif FXFELXL that is required for β-arrestin 2 binding [40]. It is tempting to suggest that this glutamate residue in the PDE4D catalytic unit may form an ion pair with residues in the β-arrestin 2 N-domain when the proteins interact. However, we also note that a cluster of negatively charged amino acids that abuts this motif in the PDE4 catalytic unit, as in FQFELTLEED (negatively charged residues are underlined), and these may provide the locus of a second negative charge that is able to accommodate the positively charged pairing of Lys18 and Arg26 in the N-domain of β-arrestin 2.
Analysis of the β-arrestin 2 peptide arrays identified two sets of PDE4D5-specific interacting peptides in the protein's C-domain (Figure 1). One was spot 58 (Arg286–Gly310) and the other was encompassed by spots 42 and 43 (Lys206–Ser235). The selectivity for PDE4D5 suggests that it is the unique N-terminal sequence of PDE4D5 that binds to the β-arrestin 2 C-domain. Scanning alanine substitution with peptide-42 showed that the motif L215NVNVH220 may be of importance for conferring β-arrestin 2 binding because single substitution of any of these residues reduced interaction with PDE4D5 (Figure 2c). The LNVNVH sequence is located on the convex surface of the C-domain β-sandwich (Figure 6). Significantly, this is the opposite face to that implicated in the recruitment of β-arrestin to GRK-phosphorylated receptors [12], consistent with the ability of β-arrestin to associate simultaneously with both the phospho-receptor and PDE4D5. Of the LNVNVH sequence, the side chains of Asn216, Asn218 and His220 are surface-exposed and are thus most likely to interact with PDE4D5. The side chains of Leu215, Val217 and Val219 are oriented into the core structure and form a hydrophobic interface with the inner side of the β-sheet that forms the concave surface of the protein's C-domain. Direct interaction of these side chains with the PDE4D5 N-terminus is difficult to envisage without invoking major structural reorganization of β-arrestin.
Scanning analysis of peptide-58 identified Arg286, whose mutation to either alanine or aspartate ablated interaction with PDE4D5 (Figure 2d). This contrasted with substitutions of alanine for the other positively charged residues in this peptide (Lys293, Lys295 and Lys308), which had no effect on PDE4D5 interaction (Figure 2d). The sequence corresponding to peptide-58 threads through the core structure with residues 286–300 surface-exposed on one side (proximal to Arg26) and residues 306–310 surface-exposed on the other (Figure 6d). The Arg286 side chain is prominently exposed, does not engage other residues in β-arrestin 2 and probably presents an excellent target for interaction with PDE4D5.
Interestingly, peptide-57 (spanning residues 281–305) failed to produce a positive spot with the PDE4D5 probe. This peptide encompasses the region containing Arg286 and the surface-exposed residue group from peptide-58 proximal to Arg26. However, peptide-57 lacks residues I306VKEG310 that form the second exposed surface in the sequence corresponding to peptide-58. This suggested that residues within this locus may also be involved in binding the PDE4D5 N-terminal sequence. However, the double substitution, I306A/V307A, and charge reversal substitutions (K308D and E309K) within this region of peptide-58 failed to affect its interaction with the PDE4D5 probe.
Scanning substitution of peptide-58 also identified Asp291, whose mutation to alanine attenuated interaction with the PDE4D5 probe. This residue is not solvent-exposed in the β-arrestin basal conformation, but is considered to ion pair with the protein's key phosphate sensor, Arg170, in its polar core [6,12,47]. Disruption of this salt bridge by pairing Arg170 with receptor phosphates acts as a switch that allows the conformational transition of arrestin into its high-affinity receptor-binding state. Arg170 must become accessible in order to interact with receptor phosphates. Thus, conceivably, Asp291 may become exposed upon activation of this switch, facilitating PDE4D5 binding to receptor-sequestered β-arrestin 2.
Residues within two distinct regions of the β-arrestin C-domain, Leu215–His220 and Arg286/Asp291, are implicated in the interaction with the PDE4D5 N-terminal sequence. Given the possibility that conformational changes may occur in β-arrestin upon association with PDE4, the relative disposition of these regions, which are well spaced when mapped on to the β-arrestin basal conformation (Figure 6), may well alter. However, it is interesting that scanning peptide array analysis with the unique 88-amino-acid N-terminal region of PDE4D5 suggests that an extensive surface from this sequence is involved in binding to β-arrestin 2 [40]. This would be consistent with the present study, which indicates that two, spatially discrete, surfaces on the β-arrestin 2 C-domain are required for the specific binding of PDE4D5 (Figure 1). Significantly, the location of the β-arrestin C-domain sites for interaction with the PDE4D5 N-terminus, as well as the protein's N-domain surface for interaction with the conserved PDE4 catalytic unit, require that PDE4D5 docks to a distinct face of β-arrestin from that involved in interaction with phospho-GPCR. It is intriguing that, in binding to the protein's N-domain, the PDE4 catalytic unit interacts with a region of β-arrestin 2 immediately adjacent to residues (Lys11/Lys12) believed to be involved in guiding the GRK-phosphorylated receptor to a phosphate sensor at Arg170 in β-arrestin 2 [6,12,47]. The involvement of Asp291 from the phosphate switch, and the spatially proximal Arg286 in binding the N-terminal region of PDE4D5 may be significant. Given that β-arrestins can tether a variety of proteins with distinct functional roles [4,7,47], it is tempting to speculate that the binding of PDE4D5 to β-arrestin 2 may influence the fidelity of interaction of β-arrestin 2 with other partner proteins.
In order to explore the possible functional significance of the three regions identified here to the interaction between PDE4D5 and β-arrestin 2, we generated a representative mutation from each region in β-arrestin 2 and analysed these for their ability to interact with PDE4D5 (Figures 3c and 3d). The importance of all three regions in the binding of β-arrestin 2 to PDE4D5 in cells is shown from co-immunoprecipitation studies where the interaction was dramatically attenuated/ablated by disruption of either the N-domain site with R26A, or the two C-domain sites with either the R286A mutation for one site or the replacement of LNVNVH with an alanine cassette at the other (Figures 3c and 3d). In marked contrast, interaction of PDE4D3 with β-arrestin 2 was only ablated upon disruption of the N-domain site using the R26A mutation (Figures 3a and 3b). These data are consistent with the peptide arrays studies, indicating that the extreme N-terminus of β-arrestin 2 contains a site that allows binding of the common PDE4 catalytic unit, whereas the C-domain contains two sites that are involved in conferring specific interaction with the unique N-terminal region of PDE4D5.
We wished to determine whether forms of β-arrestin 2 mutated to compromise their interaction with PDE4D5 would have functional consequences on compartmentalized cAMP signalling, specifically on the PKA subpopulation associated with the β2-AR whose spatially localized action can be assessed by monitoring the PKA-phosphorylation status of the β2-AR subsequent to agonist challenge with isoprenaline [27]. To evaluate this we required cells that lack endogenous β-arrestins so as to be able to incorporate mutant species. Thus MEFs from β-arrestin 1−/−/β-arrestin 2−/− double-knockout mice were transfected to express similar levels of either wild-type β-arrestin 2 or mutant species (Figure 5b). In these double-knockout cells, isoprenaline caused a profound PKA phosphorylation of the β2-AR (Figure 4a). However, while expression of wild-type β-arrestin 2 profoundly attenuated this action, this was not evident in β-arrestin 2 mutants unable to bind PDE4D5 (Figure 4a), despite such mutants being recruited to the β2-AR subsequent to isoprenaline challenge (Figure 4a). Compromising PDE4D5 binding to β-arrestin 2 thus has profound functional significance regarding the phosphorylation of the β2-AR by the PKA subpopulation tethered to it [27,36].
In the present study, we have used novel scanning peptide array technology to show that the extreme N-domain of β-arrestin contains a binding site for the conserved PDE4 catalytic unit, whereas it is the C-domain of β-arrestin that contains two surfaces conferring selectivity for PDE4D5. Thus the combined N- and C-domains of β-arrestin 2 act to straddle PDE4D5 in order to form a functional complex that can be recruited to the agonist-occupied β2-AR. The activity of such spatially localized PDE4D5 influences local cAMP levels so as to regulate the phosphorylation of the β2-AR by AKAP79-tethered PKA. Together, these proteins form a spatially constrained module that reflects a defined facet of compartmentalized cAMP signalling in cells. Future structural and other studies of PDE4–β-arrestin 2 complexes are required to provide further insight into this system and to determine whether, as suggested here, the binding of PDE4D5 engenders a conformational change in β-arrestin 2 and affects its ability to bind other partner proteins.
Acknowledgments
This work was supported by Medical Research Council (U.K.) grants G8604010 (to M. D. H.) and G0400053 (to M. D. H. and G. M.), by the Leducq Foundation (to M. D. H.), by National Institutes of Health Grant R01-GM58553 (to G. B. B.), by the Deutsche Forschungsgemeinschaft grant Kl1415/2 (to E. K.) and by European Union Grant 037189 (to E. K. and M. D. H.).
References
- 1.Hill S. J. G-protein-coupled receptors: past, present and future. Br. J. Pharmacol. 2006;147(Suppl. 1):S27–S37. doi: 10.1038/sj.bjp.0706455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang C. M., Insel P. A. GPCR expression in the heart: “new” receptors in myocytes and fibroblasts. Trends Cardiovasc. Med. 2004;14:94–99. doi: 10.1016/j.tcm.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Torrecilla I., Tobin A. B. Co-ordinated covalent modification of G-protein coupled receptors. Curr. Pharm. Des. 2006;12:1797–1808. doi: 10.2174/138161206776873716. [DOI] [PubMed] [Google Scholar]
- 4.Lefkowitz R. J., Shenoy S. K. Transduction of receptor signals by β-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 5.Penela P., Ribas C., Mayor F., Jr Mechanisms of regulation of the expression and function of G protein-coupled receptor kinases. Cell. Signalling. 2003;15:973–981. doi: 10.1016/s0898-6568(03)00099-8. [DOI] [PubMed] [Google Scholar]
- 6.Gurevich V. V., Gurevich E. V. The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol. Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald P. H., Lefkowitz R. J. β-Arrestins: new roles in regulating heptahelical receptors' functions. Cell. Signalling. 2001;13:683–689. doi: 10.1016/s0898-6568(01)00203-0. [DOI] [PubMed] [Google Scholar]
- 8.Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C. Crystal structure of β-arrestin at 1.9 Å: possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 9.Barnes P. J. New drugs for asthma. Nat. Rev. Drug Discov. 2004;3:831–844. doi: 10.1038/nrd1524. [DOI] [PubMed] [Google Scholar]
- 10.Baker J. G., Hall I. P., Hill S. J. Agonist and inverse agonist actions of β-blockers at the human β2-adrenoceptor provide evidence for agonist-directed signaling. Mol. Pharmacol. 2003;64:1357–1369. doi: 10.1124/mol.64.6.1357. [DOI] [PubMed] [Google Scholar]
- 11.Brodde O. E., Leineweber K. β2-Adrenoceptor gene polymorphisms. Pharmacogenet. Genomics. 2005;15:267–275. doi: 10.1097/01213011-200505000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Gurevich V. V., Gurevich E. V. The molecular acrobatics of arrestin activation. Trends Pharmacol. Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Baillie G. S., Houslay M. D. Arrestin times for compartmentalised cAMP signalling and phosphodiesterase-4 enzymes. Curr. Opin. Cell. Biol. 2005;17:129–134. doi: 10.1016/j.ceb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Fischmeister R. Is cAMP good or bad?. Depends on where it's made. Circ. Res. 2006;98:582–584. doi: 10.1161/01.RES.0000215564.22445.7e. [DOI] [PubMed] [Google Scholar]
- 15.Tasken K., Aandahl E. M. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol. Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 16.Wong W., Scott J. D. AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 17.Zaccolo M., Pozzan T. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 2002;295:1711–1715. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- 18.Mongillo M., McSorley T., Evellin S., Sood A., Lissandron V., Terrin A., Huston E., Hannawacker A., Lohse M. J., Pozzan T., et al. Fluorescence resonance energy transfer-based analysis of cAMP dynamics in live neonatal rat cardiac myocytes reveals distinct functions of compartmentalized phosphodiesterases. Circ. Res. 2004;95:67–75. doi: 10.1161/01.RES.0000134629.84732.11. [DOI] [PubMed] [Google Scholar]
- 19.Mongillo M., Tocchetti C. G., Terrin A., Lissandron V., Cheung Y. F., Dostmann W. R., Pozzan T., Kass D. A., Paolocci N., Houslay M. D., Zaccolo M. Compartmentalized phosphodiesterase-2 activity blunts β-adrenergic cardiac inotropy via an NO/cGMP-dependent pathway. Circ. Res. 2006;98:226–234. doi: 10.1161/01.RES.0000200178.34179.93. [DOI] [PubMed] [Google Scholar]
- 20.Houslay M. D., Adams D. R. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houslay M. D., Milligan G. Tailoring cAMP-signalling responses through isoform multiplicity. Trends Biochem. Sci. 1997;22:217–224. doi: 10.1016/s0968-0004(97)01050-5. [DOI] [PubMed] [Google Scholar]
- 22.Houslay M. D., Schafer P., Zhang K. Y. Phosphodiesterase-4 as a therapeutic target. Drug Discov. Today. 2005;10:1503–1519. doi: 10.1016/S1359-6446(05)03622-6. [DOI] [PubMed] [Google Scholar]
- 23.Huston E., Houslay T. M., Baillie G. S., Houslay M. D. cAMP phosphodiesterase-4A1 (PDE4A1) has provided the paradigm for the intracellular targeting of phosphodiesterases, a process that underpins compartmentalized cAMP signalling. Biochem. Soc. Trans. 2006;34:504–509. doi: 10.1042/BST0340504. [DOI] [PubMed] [Google Scholar]
- 24.Baillie G. S., Scott J. D., Houslay M. D. Compartmentalisation of phosphodiesterases and protein kinase A: opposites attract. FEBS Lett. 2005;579:3264–3270. doi: 10.1016/j.febslet.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 25.Baillie G. S., Sood A., McPhee I., Gall I., Perry S. J., Lefkowitz R. J., Houslay M. D. β-Arrestin-mediated PDE4 cAMP phosphodiesterase recruitment regulates β-adrenoceptor switching from Gs to Gi. Proc. Natl. Acad. Sci. U.S.A. 2003;100:940–945. doi: 10.1073/pnas.262787199. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.McCahill A., McSorley T., Huston E., Hill E. V., Lynch M. J., Gall I., Keryer G., Lygren B., Tasken K., van Heeke G., Houslay M. D. In resting COS1 cells a dominant negative approach shows that specific, anchored PDE4 cAMP phosphodiesterase isoforms gate the activation, by basal cyclic AMP production, of AKAP-tethered protein kinase A type II located in the centrosomal region. Cell. Signalling. 2005;17:1158–1173. doi: 10.1016/j.cellsig.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Lynch M. J., Baillie G. S., Mohamed A., Li X., Maisonneuve C., Klussmann E., van Heeke G., Houslay M. D. RNA silencing identifies PDE4D5 as the functionally relevant cAMP phosphodiesterase interacting with β-arrestin to control the protein kinase A/AKAP79-mediated switching of the β2-adrenergic receptor to activation of ERK in HEK293β2 cells. J. Biol. Chem. 2005;280:33178–33189. doi: 10.1074/jbc.M414316200. [DOI] [PubMed] [Google Scholar]
- 28.Ariga M., Neitzert B., Nakae S., Mottin G., Bertrand C., Pruniaux M. P., Jin S. L., Conti M. Nonredundant function of phosphodiesterases 4D and 4B in neutrophil recruitment to the site of inflammation. J. Immunol. 2004;173:7531–7538. doi: 10.4049/jimmunol.173.12.7531. [DOI] [PubMed] [Google Scholar]
- 29.Huang Z., Ducharme Y., Macdonald D., Robichaud A. The next generation of PDE4 inhibitors. Curr. Opin. Chem. Biol. 2001;5:432–438. doi: 10.1016/s1367-5931(00)00224-6. [DOI] [PubMed] [Google Scholar]
- 30.O'Donnell J. M., Zhang H. T. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4) Trends Pharmacol. Sci. 2004;25:158–163. doi: 10.1016/j.tips.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Bolger G. B. Molecular biology of the cyclic AMP-specific cyclic nucleotide phosphodiesterases: a diverse family of regulatory enzymes. Cell. Signalling. 1994;6:851–859. doi: 10.1016/0898-6568(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 32.Conti M., Richter W., Mehats C., Livera G., Park J. Y., Jin C. Cyclic AMP-specific PDE4 Phosphodiesterases as critical components of cyclic AMP signaling. J. Biol. Chem. 2003;278:5493–5496. doi: 10.1074/jbc.R200029200. [DOI] [PubMed] [Google Scholar]
- 33.Bolger G. B., McCahill A., Huston E., Cheung Y. F., McSorley T., Baillie G. S., Houslay M. D. The unique amino-terminal region of the PDE4D5 cAMP phosphodiesterase isoform confers preferential interaction with β-arrestins. J. Biol. Chem. 2003;278:49230–49238. doi: 10.1074/jbc.M303772200. [DOI] [PubMed] [Google Scholar]
- 34.Perry S. J., Baillie G. S., Kohout T. A., McPhee I., Magiera M. M., Ang K. L., Miller W. E., McLean A. J., Conti M., Houslay M. D., Lefkowitz R. J. Targeting of cyclic AMP degradation to β2-adrenergic receptors by β-arrestins. Science. 2002;298:834–836. doi: 10.1126/science.1074683. [DOI] [PubMed] [Google Scholar]
- 35.Lefkowitz R. J., Pierce K. L., Luttrell L. M. Dancing with different partners: protein kinase a phosphorylation of seven membrane-spanning receptors regulates their G protein-coupling specificity. Mol. Pharmacol. 2002;62:971–974. doi: 10.1124/mol.62.5.971. [DOI] [PubMed] [Google Scholar]
- 36.Fraser I. D., Cong M., Kim J., Rollins E. N., Daaka Y., Lefkowitz R. J., Scott J. D. Assembly of an A kinase-anchoring protein–β2-adrenergic receptor complex facilitates receptor phosphorylation and signaling. Curr. Biol. 2000;10:409–412. doi: 10.1016/s0960-9822(00)00419-x. [DOI] [PubMed] [Google Scholar]
- 37.Bradaia A., Berton F., Ferrari S., Luscher C. β-Arrestin2, interacting with phosphodiesterase 4, regulates synaptic release probability and presynaptic inhibition by opioids. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3034–3039. doi: 10.1073/pnas.0406632102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipworth B. J. Phosphodiesterase-4 inhibitors for asthma and chronic obstructive pulmonary disease. Lancet. 2005;365:167–175. doi: 10.1016/S0140-6736(05)17708-3. [DOI] [PubMed] [Google Scholar]
- 39.Walker J. K., Fong A. M., Lawson B. L., Savov J. D., Patel D. D., Schwartz D. A., Lefkowitz R. J. β-Arrestin-2 regulates the development of allergic asthma. J. Clin. Invest. 2003;112:566–574. doi: 10.1172/JCI17265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolger G. B., Baillie G. S., Li X., Lynch M. J., Herzyk P., Mohamed A., Mitchell L. H., McCahill A., Hundsrucker C., Klussmann E., et al. Scanning peptide array analyses identify overlapping binding sites for the signalling scaffold proteins, β-arrestin and RACK1, in cAMP-specific phosphodiesterase PDE4D5. Biochem. J. 2006;398:23–36. doi: 10.1042/BJ20060423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohout T. A., Lin F. S., Perry S. J., Conner D. A., Lefkowitz R. J. β-Arrestin 1 and 2 differentially regulate heptahelical receptor signaling and trafficking. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1601–1606. doi: 10.1073/pnas.041608198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLean A. J., Milligan G. Ligand regulation of green fluorescent protein-tagged forms of the human β1- and β2-adrenoceptors: comparisons with the unmodified receptors. Br. J. Pharmacol. 2000;130:1825–1832. doi: 10.1038/sj.bjp.0703506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yarwood S. J., Steele M. R., Scotland G., Houslay M. D., Bolger G. B. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J. Biol. Chem. 1999;274:14909–14917. doi: 10.1074/jbc.274.21.14909. [DOI] [PubMed] [Google Scholar]
- 44.Kramer A., Schneider-Mergener J. Synthesis and screening of peptide libraries on continuous cellulose membrane supports. Methods Mol. Biol. 1998;87:25–39. doi: 10.1385/0-89603-392-9:25. [DOI] [PubMed] [Google Scholar]
- 45.Frank R. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports: principles and applications. J. Immunol. Methods. 2002;267:13–26. doi: 10.1016/s0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 46.Bolger G. B., Erdogan S., Jones R. E., Loughney K., Scotland G., Hoffmann R., Wilkinson I., Farrell C., Houslay M. D. Characterization of five different proteins produced by alternatively spliced mRNAs from the human cAMP-specific phosphodiesterase PDE4D gene. Biochem. J. 1997;328:539–548. doi: 10.1042/bj3280539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gurevich E. V., Gurevich V. V. Arrestins: ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terrin A., Di Benedetto G., Pertegato V., Cheung Y. F., Baillie G., Lynch M. J., Elvassore N., Prinz A., Herberg F. W., Houslay M. D., Zaccolo M. PGE1 stimulation of HEK293 cells generates multiple contiguous domains with different [cAMP]: role of compartmentalized phosphodiesterases. J. Cell. Biol. 2006;175:441–451. doi: 10.1083/jcb.200605050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milano S. K., Pace H. C., Kim Y. M., Brenner C., Benovic J. L. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–3328. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]