Abstract
AD (Alzheimer's disease) is a neurodegenerative disorder characterized by self-assembly and amyloid formation of the 39–43 residue long Aβ (amyloid-β)-peptide. The most abundant species, Aβ(1–40) and Aβ(1–42), are both present within senile plaques, but Aβ(1–42) peptides are considerably more prone to self-aggregation and are also essential for the development of AD. To understand the molecular and pathological mechanisms behind AD, a detailed knowledge of the amyloid structures of Aβ-peptides is vital. In the present study we have used quenched hydrogen/deuterium-exchange NMR experiments to probe the structure of Aβ(1–40) fibrils. The fibrils were prepared and analysed identically as in our previous study on Aβ(1–42) fibrils, allowing a direct comparison of the two fibrillar structures. The solvent protection pattern of Aβ(1–40) fibrils revealed two well-protected regions, consistent with a structural arrangement of two β-strands connected with a bend. This protection pattern partly resembles the pattern found in Aβ(1–42) fibrils, but the Aβ(1–40) fibrils display a significantly increased protection for the N-terminal residues Phe4–His14, suggesting that additional secondary structure is formed in this region. In contrast, the C-terminal residues Gly37–Val40 show a reduced protection that suggests a loss of secondary structure in this region and an altered filament assembly. The differences between the present study and other similar investigations suggest that subtle variations in fibril-preparation conditions may significantly affect the fibrillar architecture.
Keywords: Alzheimer's disease, amyloid-β peptide, atomic force microscopy, hydrogen/deuterium exchange, NMR
Abbreviations: Aβ, amyloid-β; AD, Alzheimer's disease; AFM, atomic force microscopy; H/D, hydrogen/deuterium; HSQC, heteronuclear single-quantum coherence; NOE, nuclear Overhauser effect; NOESY, nuclear Overhauser enhancement spectroscopy
INTRODUCTION
Self-assembly and deposition of proteins into amyloid fibrils and plaques are phenomena that currently have been linked to around 20 different human diseases [1]. The long unbranched fibrils that constitute amyloid typically have a diameter between 50–130 Å (1 Å=0.1 nm) and a characteristic cross-β pattern in which β-strands are arranged perpendicular to the fibrillar axis [2–5]. The best known example of such a disorder is AD (Alzheimer's disease), which is correlated with the aggregation of an endogenic peptide denoted Aβ (amyloid-β)-peptide [6–12]. The Aβ-peptide is a result of proteolytic processing of the membrane-bound amyloid precursor protein. This excision generates an ensemble of peptides with various lengths, where each species exhibits rather distinct biophysical properties. The clinically most relevant fragments include 39–43 residues, of which the Aβ(1–40) and Aβ(1–42) peptides are the most abundant [13]. Although the ratio between Aβ(1–40) and Aβ(1–42) peptides in the human body is about 7:1, the Aβ(1–42) variant is overrepresented in senile plaques, and is also present in the first deposits found during disease development [14,15]. Moreover, overproduction of Aβ(1–42) has been linked to early onset of AD [16,17] and recent experiments in an AD mouse model suggest that selective inhibition of the Aβ(1–42) variant abolishes the disease [18]. Although at present the cytotoxic mechanism in vivo is not completely understood, the correlation with aggregation of the Aβ-peptide is convincing [6–11]. Hence one potential therapeutic approach involves design of inhibitors of the Aβ-assembly. Therefore a thorough knowledge about the molecular architecture of the fibrillar states of Aβ peptides is necessary. In particular, it is of interest to compare the structure of the more aggregation prone Aβ(1–42) variant with its shorter counterparts.
Structural studies of amyloid are hampered by its non-crystalline and solid nature where conventional methods using crystal diffraction and liquid NMR cannot be readily employed. An alternative technique is solid-state NMR, a method that has been used extensively to successfully investigate the structure of fibrils from Aβ(10–35), Aβ(1–40) and Aβ(1–42) peptides [19–24]. More recently, the combined use of quenched H/D (hydrogen/deuterium)-exchange and solution NMR spectroscopy has proven extremely valuable for studies of the structural and dynamic properties of amyloid fibrils [25–31], including fibrils from both the Aβ(1–40) and Aβ(1–42) variants [26,30,31]. With this method, identification of the core region of a fibril is possible since the secondary structure and solvent exclusion in the core protect the labile amide protons from exchanging with the surrounding deuterons. After a designated incubation time in 2H2O the solvent protection is trapped via a rapid conversion of the fibrils into a monomeric and NMR-detectable state during conditions of low back-exchange. By following the post-trap decay of the H/D-exchange the method pinpoints the fibrillar core in a residue-specific and quantitative manner. Applying this method to Aβ(1–42) fibrils we previously identified two solvent-protected core regions, comprising residues Glu11–Gly25 and Lys28–Ala42 [26,30,31]. The residues in between, Ser26 and Asn27, as well as the N-terminal residues, Asp1–Tyr10, were solvent accessible. These findings agree with the most recent fibrillar models derived from solid-state NMR data [23,24], but differ somewhat from similar H/D-exchange NMR studies [26,30,31]. Detailed comparisons of the various studies on Aβ-fibrils are however complex, since recent quenched H/D-exchange NMR and solid-state NMR data indicate that rather subtle changes in fibril-growth conditions significantly affect the fibrillar structure.
In order to identify discriminating features between Aβ(1–40) and Aβ(1–42) fibrils, we have performed quenched H/D-exchange NMR on Aβ(1–40) fibrils prepared under conditions identical to the ones used in our previous investigation on the Aβ(1–42) fibrils [31]. Two highly protected core regions were identified in agreement with our results for the Aβ(1–42) fibrils. However, a significantly higher protection of the N-terminal region, as well as a reduced solvent protection for the C-terminal residues, discriminate the Aβ(1–40) from the Aβ(1–42) fibrillar structure and provide new structural data for current models of Aβ architecture.
EXPERIMENTAL
NMR spectroscopy and resonance assignment of Aβ(1–40)
Isotope-enriched chemicals were purchased from Cambridge Isotope Laboratories. Uniformly 15N-labelled Aβ(1–40) was obtained from Alexo-Tech (www.alexo-tech.com). The NMR sample used for resonance assignment contained ∼2 mM recombinant Aβ(1–40) and was prepared in 80% HFIP (1,1,1,3,3,3-hexafluoroisopropanol-D2)/20% H2O and 150 mM NaCl at pH 3.0, as described previously [31]. Homonuclear two-dimensional clean-TOCSY and heteronuclear two-dimensional 15N-HSQC (heteronuclear single-quantum coherence), as well as three-dimensional 15N-DIPSI-HSQC (where DIPSI is decoupling in the presence of scalar interactions) and 15N-NOESY-HSQC (where NOESY is nuclear Overhauser enhancement spectroscopy) experiments were collected at 15 °C on a 600 MHz Bruker AVANCE spectrometer, equipped with a 5 mm triple-resonance, pulsed-field z-gradient cryoprobe. Recorded experiments were processed using NMRPipe [32] and the sequence-specific backbone resonance assignment was determined with Ansig for Windows [33].
H/D-exchange of Aβ(1–40) fibrils
The fibril samples for the H/D-exchange experiments were produced, treated and analysed by NMR in a manner identical to our previous work on Aβ(1–42) fibrils [31]. Briefly, fibrils were grown by incubating a sample of 1 mM 15N-labelled Aβ(1–40) in 5 mM phosphate buffer (pH 7.0) containing 50 mM NaCl, at 37 °C for 5–8 days with agitation at 130 rpm. Immediately after dissolution and prior to fibril formation the peptide displays a CD spectrum characteristic for a primarily random coil conformation (results not shown). The fibril solution was divided into three aliquots and the pellets were recovered by short centrifugations at 13000 g. The H/D-exchange was initiated by diluting the pellets 30 times using a 2H2O-solution and 50 mM NaCl (p2H 6.6). The fibrils were recovered through centrifugation (13000 g for 2 min at 37 °C) and the washing procedure repeated once to remove residual H2O and soluble material. Subsequently, two of the aliquots were incubated in 2H2O for 2 and 24 h respectively, including the period for the buffer-exchange procedure. The third aliquot contained fully protonated fibrils and served as a control, to discriminate between rapid exchange as a result of the experimental procedure and exchange as a result of the preceding incubation in 2H2O. At the end of the incubation period the fibrils in all three fibril samples were rapidly converted into NMR-detectable monomers as described in [31], in 80% HFIP/20% 2H2O and 150 mM NaCl (p2H 2.6), a solution known to induce a significant fraction of α-helicity in Aβ-peptides [34]. Each sample acquired a peptide concentration corresponding to approximately 2 mM monomeric Aβ(1–40). Hydrogen exchange was subsequently monitored by recording a series of heteronuclear two-dimensional 15N-HSQC experiments, typically started 6–8 min after fibril dissolution. The acquisition time for each 15N-HSQC experiment was 10 min using four transients per increment and 128 (t1)×1024 (t2) complex data points. Prior to each 15N-HSQC experiment a one-dimensional proton NMR spectrum was recorded to quantitatively monitor the dissolution of fibrils into monomers.
Data analysis and structural modelling
Processing and analysis of one-dimensional experiments were carried out in TOPSPIN (Bruker Biospin), while processing of the recorded 15N-HSQC-experiment was performed in NMRPipe [32]. Peak volumes in baseline-corrected 15N-HSQC experiments were determined using NMRView software routines [35]. The non-exchangeable methyl region in the recorded series of one-dimensional spectra was integrated and fitted to a single exponential function to determine the relative monomer concentration of the samples and the rates of fibril dissolution. This was taken into account when the intensities of individual amide resonances in the series of 15N-HSQC spectra were fitted to a single exponential decay in the software Grace. By extrapolating the intensities to zero time the signal intensity in the fibrillar state was obtained. Residue-specific protection ratios were determined from the signal intensity ratio of a sample pre-incubated in 2H2O and the fully protonated control. The experimental uncertainty of the protection ratios were determined by propagation of errors using the standard deviations of the fitted exponentials. It is important to stress the significance of analysing the decay of the fully protonated control as this makes it possible to discriminate between exchanging protons in the fibrillar and monomeric state. A detailed description of the analysis procedure is found in our previous study on Aβ(1–42) [31]. Protection ratios were mapped onto a model of the fibrillar structure of Aβ(1–40) using MOLMOL [36]. This model was prepared from the co-ordinates of the recent solid-state NMR model of the Aβ(9–40) fibril, provided by Dr Robert Tycko [19], to which the missing N-terminal residues were added from the co-ordinates of a structure of Aβ(1–16) (PDB code: 1ZE7) [37]. From this new Aβ(1–40) fibrillar model, a model of a Aβ(1–42) fibril was generated by adding the two additional C-terminal residues from our previous Aβ(1–42) model [31] and by placing the two filaments in a recently proposed shifted arrangement [24]. Details about various fibrillar models are described further below in the Discussion section. Modifications and energy minimization of the models were performed in MOLMOL [36] and Swiss-PdbViewer [38].
AFM (atomic force microscopy)
A portion of the Aβ(1–40) fibril solution was diluted in 10 mM phosphate buffer (pH 7.0) containing 50 mM NaCl, to approximately 5 μM peptide solution that was applied onto freshly cleaved ruby red mica (Goodfellow). The solution was allowed to adsorb for 30 s, followed by washing with distilled water three times and air drying. Analysis was performed using a Nanoscope IIIa multimode AFM (Digital Instruments) in tapping Mode™ in air. A silicon probe was oscillated at around 280 kHz, and images were collected at an optimized scan rate corresponding to 1 Hz. The images were flattened and presented in amplitude mode using Nanoscope software (Digital Instruments).
RESULTS
Sequence specific backbone assignment of Aβ(1–40) monomers in solution
The recorded spectra were of high quality with good dispersion and few overlapping resonances. The sequence-specific backbone resonance assignment was determined from the NOESY spectrum via a sequential walk between backbone amide resonances and via characteristic α-helical sequential or medium-range NOE (nuclear Overhauser effect) resonances. All of the 39 backbone amide resonances (residues 2–40) could be identified and only two of these, Asn27 and Ile32, showed significant overlap. This ambiguity, however, does not affect the analysis of the H/D-exchange as discussed below. Overall the assignment agreed extremely well with our assignments for Aβ(1–42) [31], and chemical shift differences were mainly detected in the C-terminal region of the peptides.
Fibril formation and AFM analysis of Aβ(1–40)
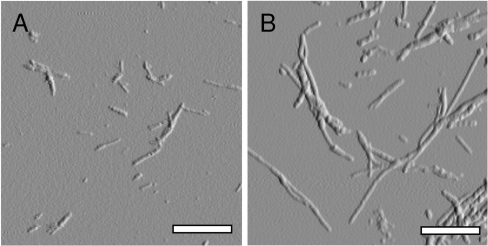
Fibrils were formed by incubating a sample containing 1 mM 15N-labelled Aβ(1–40) peptide in 5 mM phosphate buffer (pH 7.0) containing 50 mM NaCl, for 5–8 days at 37 °C with agitation at 130 rpm. The peptide solution acquired a gel-like appearance and the presence of fibrils was verified by AFM (Figure 1A). The fibrils were of various lengths, 100–500 nm, with a height of 3.5–7 nm and smooth morphology.
Figure 1. Morphologies of Aβ(1–40) and Aβ(1–42) fibrils.
Tapping mode AFM images verifying the presence of fibrillar structures after incubation of recombinant Aβ(1–40) and Aβ(1–42) peptides in 10 mM phosphate buffer (pH 7.0) containing 50 mM NaCl with agitation. Both images were acquired using a 5×5 μm scanning area. The scale bar in each image is 0.5 μm. (A) Aβ(1–40) fibrils with an average height of approximately 3.5–7 nm, a smooth architecture and a length between 100–500 nm. (B) Aβ(1–42) fibrils with a height similar to the fibrils in (A). Aβ(1–42) fibrils often exceeded several μm in length.
Determination of protection ratios of Aβ(1–40) fibrils
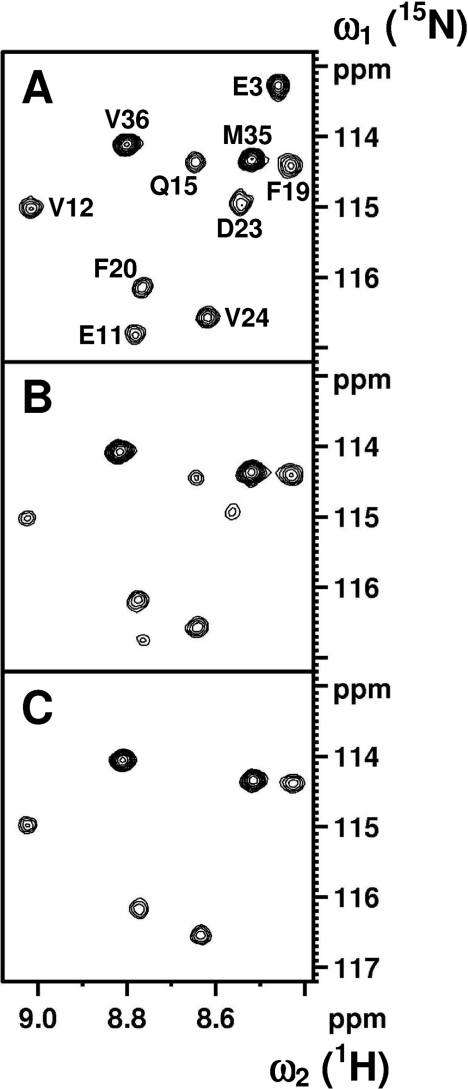
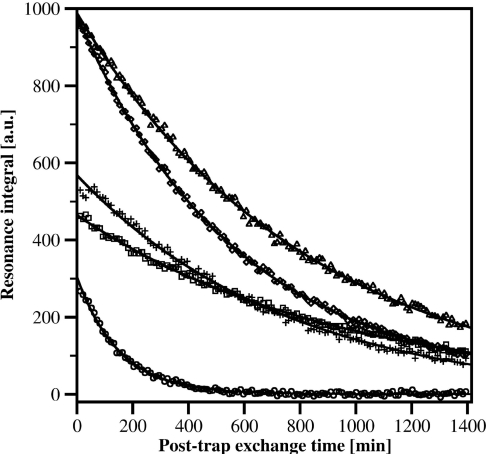
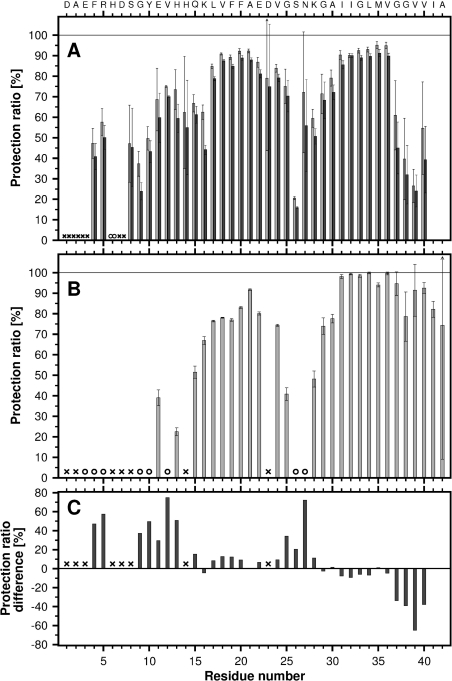
Fibrillar material was collected by centrifugation and H/D-exchange was carried out by resuspension and incubation of the fibrillar pellets in 2H2O. The conversion of fibrils into NMR-detectable monomers followed a single exponential function with an average rate constant of 0.0028 min−1. More than 91% of the total fibril material was dissolved prior to the first 15N-HSQC spectrum. A spectrum of the fully protonated peptide is shown in Figure 2(A) and spectra of 2H2O exchanged fibrils, recorded 11 and 203 min after fibril dissolution, are shown in Figures 2(B) and 2(C) respectively. Analysis of the control sample showed that 36 out of 39 amide resonances could be used as probes for determining the solvent protection of the fibril. Residues Ala2 and Asp7 experience a post-trap exchange rate which is too fast for detection. The minute protection observed for Glu3 is too small and decays too fast to permit a reliable fit. Post-trap decays and curve-fits for five residues, Gln15, Phe20, Val24, Met35 and Val36, are shown in Figure 3. The solvent protection patterns for fibrils that were pre-incubated in 2H2O for 2 or 24 h are shown in Figure 4(A). A total of 35 residues were protected and the protection ratio in general decreased with exchange time, 66% and 60% overall ratio at 2 and 24 h respectively. Two well-protected bell-shaped regions were identified covering residues Ser8–Gly25 and Gly27–Val40, with the strongest protection (close to 90%) for residues Leu17–Gly25 and Ala30–Val36. Notably, Phe4 and Arg5 are partially protected, His6 is unprotected, and Ser26, Gly38 and Val39 are quite poorly protected. Since Asn27 and Ile32 are overlapping, the signal decay rate had to be fitted to a bi-exponential function. Their decay rates were unambiguously assigned through comparisons with the decay rates for these residues in our Aβ(1–42) study [31] and in H/D exchange NMR experiments performed on Aβ(1–40) under slightly different solvent conditions where these resonances were not overlapping (results not shown). The protection ratio determined for Asn27 is less accurate since it has a lower signal intensity and much faster exchange rate than Ile32 (0.0318 compared with 0.0008 min−1). Fast amide proton exchange rates within the monomeric structure are also the origin of experimental uncertainties of the fibrillar protection ratios. Figure 4(B) shows the fibrillar solvent protection pattern for Aβ(1–42), where 35 out of 41 residues were useful as probes. The remaining six residues (Ala2, His6, Asp7, Ser8, His14 and Asp23) experience exchange rates in the monomeric state, which prevent detection [31].
Figure 2. NMR spectra of Aβ(1–40).
Contour plots from a selected region of a 15N-HSQC spectrum measured on a 2 mM 15N-labelled sample of Aβ(1–40). (A) Fully protonated monomeric Aβ(1–40). (B and C) Spectra of partly deuterated monomeric Aβ(1–40), recorded 11 and 203 min after fibril dissolution in the 2H2O solvent respectively. Prior to dissolution, the fibrils in (B) and (C) were incubated in 2H2O at p2H 6.6 for 2 h in order to exchange solvent-accessible protons in the fibril. Assignments are indicated in (A).
Figure 3. H/D-exchange of Aβ(1–40).
Examples of the measured signal decay for five amide groups within Aβ(1–40) as a result of post-trap exchange with the surrounding 2H2O. ○, Gln15; □, Phe20; +, Val24; ◇, Met35; △, Val36.
Figure 4. Solvent protection for the backbone amide protons of Aβ(1–40) and Aβ(1–42) fibrils.
Protection is defined as the ratio of the observed intensity after a pre-incubation period in 2H2O to the intensity in a completely protonated sample (defined as 100%). (A) Solvent protection in Aβ(1–40) fibrils, where light and dark grey bars indicate the protection after 2 and 24 h of pre-incubation in 2H2O respectively. (B) Solvent protection for Aβ(1–42) fibrils after 2 h of pre-incubation in 2H2O (the data are from [31] and included for comparative reasons). (C) A protection ratio difference plot of Aβ(1–40) and Aβ(1–42) calculated from data shown in (A) and (B). ○ correspond to a protection ratio of 0% and × represent residues which exchange too fast in the monomeric state to enable detection. Error bars show the experimental uncertainty of the measurements, determined by error propagation using S.D.
DISCUSSION
The structural organization of fibrils from either Aβ(1–40) or Aβ(1–42) peptides have been extensively investigated, resulting in several proposed models all with a characteristic cross-β structure (reviewed in [39,40]). Solid-state NMR studies on Aβ fibrils have significantly contributed to the understanding of the fibril architecture, and suggest a fibrillar model in which the Aβ-peptide attains two β-strands that stack perpendicular to the fibrillar axis, forming a filament structure of two separate in-register parallel β-sheets [21,22]. Scanning transmission electron microscopy in combination with solid-state NMR studies furthermore suggests that the smallest fibrillar form under physiological conditions includes two filaments [21] arranged in an anti-parallel fashion [23,24]. Fibril cross-sections describing the suggested molecular structures as well as the filament arrangements for Aβ(1–40) and Aβ(1–42) fibrils respectively, are schematically shown in Figures 5(A) and 5(B). Two alternative models have also been described: the Aβ(1–40) fibril model shown in Figure 5(C) derived through scanning cysteine mutagenesis and threading analysis [41,41a], and the Aβ(1–42) fibril model shown in Figure 5(D) derived from double compensatory mutagenesis in combination with H/D-exchange NMR [30]. Although most of the recently presented structural information on Aβ-fibrils is similar, it is increasingly evident that minor alterations of the solvent conditions and procedures for fibril preparation have a significant impact on the corresponding structures. Interestingly, a recent study clearly establishes a correlation between Aβ-fibrillar structure and neurotoxicity [9]. This observation may in part explain previously diverging results for Aβ toxicity, and it highlights the need for further structural studies. The substantial differences between Aβ(1–40) and Aβ(1–42) with regard to their aggregation propensity and role in AD pathology make it important to identify structural discrepancies in their fibrillar forms. Previous studies are difficult to compare since different fibril growth conditions were used.
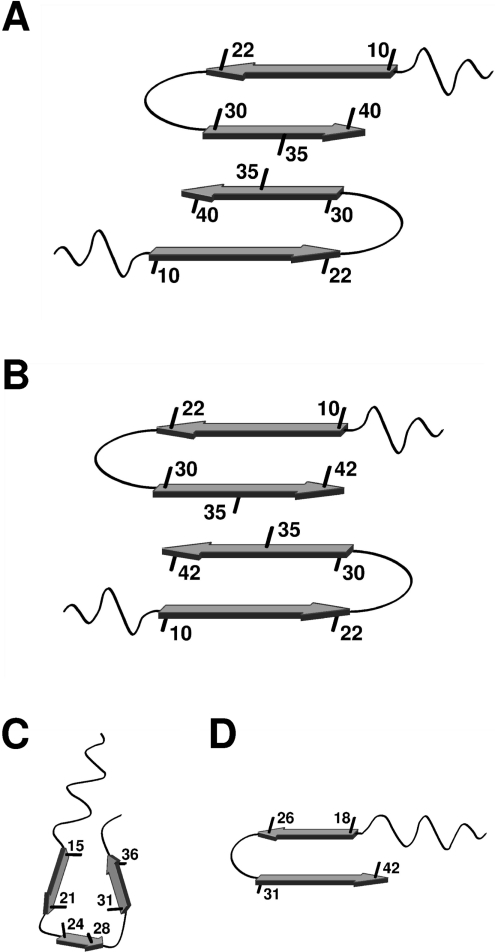
Figure 5. Schematic models of Aβ(1–40) and Aβ(1–42) fibrils.
Four schematic models of an Aβ-fibril, showing the starting point of β-strands and orientations of selected side-chains. (A) Model of the Aβ(1–40) fibril derived by Petkova et al. [23]. (B) Model of the Aβ(1–42) fibril built from the model in (A) together with recent solid-state NMR data on Aβ(1–42) fibrils by Sato et al. [24] and our H/D-exchange NMR data [31]. (C) Model of the Aβ(1–40) fibril proposed by Guo et al. [41a]. (D) Model of the Aβ(1–42) fibril derived by Lührs et al. [30].
In the present study we have determined the solvent protection pattern of fibrils from Aβ(1–40), see Figure 4(A). These fibrils display two well-protected bell-shaped regions, Ser8–Gly25 and Gly27–Val40, and a poorly protected residue, Ser26, consistent with a structural arrangement of two β-strands connected by a turn, in agreement with a current solid-state NMR model (Figure 5A) [42]. Furthermore, the partially protected N-terminal residues, in particular Phe4 and Arg5, indicate the presence of additional secondary structure in this region. This observation is consistent with results from a limited proteolysis study, where approximately 20% of the total sample was resistant to proteolytic digest in the N-terminal region [43]. The bell-shaped protection pattern for Ser8–Gly25 and partial protection for residues Phe4 and Arg5 suggest a possible extension of the first β-strand (comprising residues 10–22 in the model, Figure 5A) towards the N-terminus. However, the unprotected His6 residue indicates an interruption of the secondary structure. The data therefore imply that the two residues, Phe4 and Arg5, are involved in a new structural element which forms additional intra- or inter-molecular hydrogen bonds. Residual structures in the N-terminal region of monomeric Aβ(1–16) and Aβ(1–40) have previously been identified in aqueous solution from NOE data and secondary chemical shifts [37,44]. According to our results these structures are stabilized within the ordered environment of a fibril. The N-terminal region is known to bind divalent metal ions, such as copper and zinc, and has a propensity to form a secondary structure in which metals are co-ordinated by the side chains of His6, His13, His14 and possibly Tyr10 or Glu11 [37,45]. To verify that our results were not influenced by trace amounts of divalent metals, peptide purification and H/D-exchange NMR analysis were repeated in the presence of 2 mM EDTA. The results showed virtually identical protection patterns (results not shown). The partial protection observed for the C-terminal residues of the Aβ(1–40) peptide, Gly37–Val40, is indicative of a less structured C-terminus. A fairly exposed C-terminus in Aβ(1–40) fibrils is supported by several investigations where quenched H/D-exchange [26], proteolytic digests in combination with MS [43], proline and cysteine scanning mutagenesis [41,46], as well as solid-state NMR study [9] were used. Overall, the protection ratios across the peptide sequence show very little additional decay during a 24 h incubation time compared with 2 h. This is particularly true for the most protected residues in the β-sheet region, suggesting that they constitute a stable core of the fibril. Since the exchange rates of the amide protons may contain additional information about the intrinsic quaternary structure of the fibril, we are currently performing a detailed residue-specific analysis of the H/D-exchange kinetics.
Similar to our findings, the quenched H/D-exchange NMR study on Aβ(1–40) fibrils by Whittemore et al. [26], also identified Gln15–Asp23 as highly, Gly37–Val40 as partially and Ser26 as poorly protected residues. However, the present study identifies most N-terminal residues prior to position 15 as partially protected, while the Whittemore study only detected two partially protected residues, Glu11 and Val12 within this region. Discrepancies are also found in the C-terminal region, comprising residues Asn27–Val36, which is well-protected in the present study but displays an alternating pattern with protected and exposed residues in the Wittemore study. Since the fibrils used in these studies were prepared using different solvents and agitation, the most likely cause for the discrepancies is the different preparation methods.
A direct comparison of our data on Aβ(1–40) and Aβ(1–42) fibrils is now possible since both studies were carried out using identical methods and fibril-forming conditions. AFM analysis displayed an overall similar ultrastructural morphology, where the filament height varied between 3.5 and 7 nm due to the occurrence of laterally assembled filaments. However, while the Aβ(1–40) fibrils were between 100–500 nm in length the fibrils of the Aβ(1–42) variant often exceeded several μm in length (Figures 1A and 1B). The solvent protection patterns of the two peptides showed clear discrepancies in both their N- and C-terminal regions (see Figures 4A–4C). In comparison with Aβ(1–42), the N-terminal residues of Aβ(1–40), in particular Phe4 and Arg5, are significantly more protected, showing that the additional C-terminal residues, Ile41 and Val42 in Aβ(1–42) fibrils, affect the formation of secondary structure and possibly metal binding in the N-terminal region. A speculative explanation for the lack of protection in the N-terminal part of Aβ(1–42) is that the two additional residues Ile41 and Val42 interact with the N-terminal part of the first β-strand and lock it in a position so it cannot participate in the formation of additional secondary structures in the N-terminal. The C-terminal residues Gly37–Val40 of Aβ(1–40) show a clear reduction of solvent protection, indicative of a more buried C-terminus in the Aβ(1–42) fibrils. These results are in line with previous studies where Aβ(1–42) fibrils, in comparison with Aβ(1–40) fibrils, show a higher sensitivity to proline substitutions in the C-terminal region [47]. A recent mutational analysis of the Aβ-sequence suggests that the main determinant for the aggregation propensity of Aβ(1–42) lies in the hydrophobicity of residues Ile41 and Ala42 [48]. These results support our previous suggestion that addition of the two most C-terminal residues of Aβ(1–42) may act as a molecular zipper between the cross-β units along the fibril axis, by adding additional hydrogen bonds to the Gly37–Ala42 structural region [31].
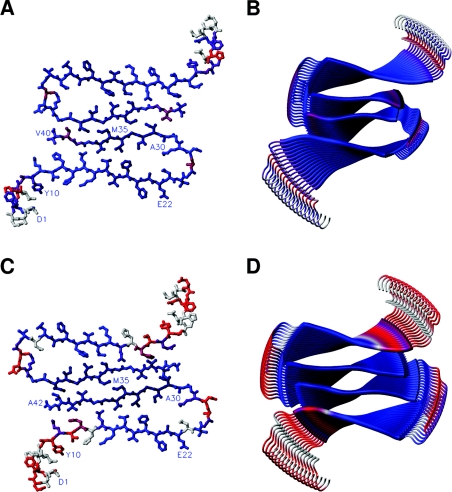
We mapped the solvent protection ratios in Aβ(1–40) fibrils onto the model in Figure 5(A), the model that best agrees with our data, see Figure 6(A) and 6(B). There is also reasonable agreement between the protection ratios for Aβ(1–42) fibrils [31] and the Aβ(1–40) model, except for the C-terminal residues. To fully explain this protection pattern, it seems that the subunits within the Aβ(1–42) fibrils must shift with respect to each other as suggested in a recent solid-state NMR study (Figure 5B) [23,24]. The protection ratios for Aβ(1–42) [31] mapped onto the model in Figure 5(B) are shown in Figures 6(C) and 6(D). A shifted assembly of the filaments of Aβ(1–42) fibrils positions the C-terminal region of Aβ(1–42) in a significantly more solvent-protected environment than that of the C-terminal of Aβ(1–40) (compare Figures 4A and 4B), creating a tightly packed hydrophobic core (Figures 6C and 6D). This model fully accounts for our H/D-exchange NMR data on Aβ(1–42) fibrils [31].
Figure 6. Mapping of the observed protection ratios onto a fibril model.
The solvent protection ratios determined for residues within Aβ(1–40) and Aβ(1–42) fibrils are mapped onto corresponding models of the fibrils. The colour code is varied between the following extremes: navy blue for complete and red for no solvent protection. Residues with no protection ratios available are depicted in grey. Main-chain hydrogen bonds are directed along the fibril axis, perpendicular to the plane of the paper. (A and C) Ball-and-stick models showing a dimer of two cross-β units taken from a cross-section of the Aβ(1–40) and Aβ(1–42) fibril models respectively. Assignments are indicated in some positions with their one-letter amino acid codes. (B and D) Models of the fibrillar assembly for Aβ(1–40) and Aβ(1–42) respectively. The model is based on the structural model by Tycko and co-workers [23], the solution structure of Aβ(1–16) [37], our previous study [31], and the recently proposed filament packing arrangement [24]. The image was prepared in MOLMOL [36].
In conclusion, the present study presents the solvent protection pattern of Aβ(1–40) fibrils at a residue-specific level, relates the results to current models of Aβ-amyloid, and compares the data with similar NMR studies, in particular our previous work on the more aggregation prone Aβ(1–42) variant. Most notably, the results show that the N-terminal region of Aβ(1–40) comprising residues Phe4–His14 is far better protected than in Aβ(1–42) fibrils, indicating formation of additional secondary structure in this part of the peptide. In contrast, the reduced protection of the C-terminal residues Gly37–Val40 indicates a loss of secondary structure and suggests a shift in the filament assembly.
Acknowledgments
We thank Dr Robert Tycko (Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, U.S.A.) for providing us with the co-ordinates of the Aβ(9–40) amyloid model. This work was supported by Hjärnfonden, Magn. Bergvalls Foundation, Carl Trygger Foundation, Insamlingsstiftelsen Umeå Universitet, Åke Wibergs Foundation, Socialstyrelsen, Svenska Lundbecksstiftelsen, Swedish Research Science Council, patients' association FAMY-Norrbotten, AMYL-foundation and Gustafsson Foundation.
References
- 1.Pepys M. B. Amyloidosis. Annu. Rev. Med. 2006;57:223–241. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- 2.Sunde M., Serpell L. C., Bartlam M., Fraser P. E., Pepys M. B., Blake C. C. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 3.Kirschner D. A., Abraham C., Selkoe D. J. X-ray diffraction from intraneuronal paired helical filaments and extraneuronal amyloid fibers in Alzheimer disease indicates cross-beta conformation. Proc. Natl. Acad. Sci. U.S.A. 1986;83:503–507. doi: 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Erratum. Proc. Natl. Acad. Sci. U.S.A. 1986;8:2776. [Google Scholar]
- 4.Blake C., Serpell L. Synchrotron X-ray studies suggest that the core of the transthyretin amyloid fibril is a continuous β-sheet helix. Structure. 1996;4:989–998. doi: 10.1016/s0969-2126(96)00104-9. [DOI] [PubMed] [Google Scholar]
- 5.Malinchik S. B., Inouye H., Szumowski K. E., Kirschner D. A. Structural analysis of Alzheimer's β(1–40) amyloid: protofilament assembly of tubular fibrils. Biophys. J. 1998;74:537–545. doi: 10.1016/S0006-3495(98)77812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pike C. J., Burdick D., Walencewicz A. J., Glabe C. G., Cotman C. W. Neurodegeneration induced by β-amyloid peptides in vitro: the role of peptide assembly state. J. Neurosci. 1993;13:1676–1687. doi: 10.1523/JNEUROSCI.13-04-01676.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busciglio J., Lorenzo A., Yankner B. A. Methodological variables in the assessment of β amyloid neurotoxicity. Neurobiol. Aging. 1992;13:609–612. doi: 10.1016/0197-4580(92)90065-6. [DOI] [PubMed] [Google Scholar]
- 8.Puzzo D., Arancio O. Fibrillar β-amyloid impairs the late phase of long term potentiation. Curr. Alzheimers Res. 2006;3:179–183. doi: 10.2174/156720506777632871. [DOI] [PubMed] [Google Scholar]
- 9.Petkova A. T., Leapman R. D., Guo Z., Yau W. M., Mattson M. P., Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 10.Klein W. L. Aβ toxicity in Alzheimer's disease: globular oligomers (ADDLs) as new vaccine and drug targets. Neurochem. Int. 2002;41:345–352. doi: 10.1016/s0197-0186(02)00050-5. [DOI] [PubMed] [Google Scholar]
- 11.Wogulis M., Wright S., Cunningham D., Chilcote T., Powell K., Rydel R. E. Nucleation-dependent polymerization is an essential component of amyloid-mediated neuronal cell death. J. Neurosci. 2005;25:1071–1080. doi: 10.1523/JNEUROSCI.2381-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masters C. L., Simms G., Weinman N. A., Multhaup G., McDonald B. L., Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. U.S.A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinkai Y., Yoshimura M., Ito Y., Odaka A., Suzuki N., Yanagisawa K., Ihara Y. Amyloid β-proteins 1–40 and 1–42(43) in the soluble fraction of extra- and intracranial blood vessels. Ann. Neurol. 1995;38:421–428. doi: 10.1002/ana.410380312. [DOI] [PubMed] [Google Scholar]
- 14.Iwatsubo T., Odaka A., Suzuki N., Mizusawa H., Nukina N., Ihara Y. Visualization of Aβ42(43) and Aβ40 in senile plaques with end-specific Aβ monoclonals: evidence that an initially deposited species is Aβ42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 15.Lemere C. A., Blusztajn J. K., Yamaguchi H., Wisniewski T., Saido T. C., Selkoe D. J. Sequence of deposition of heterogeneous amyloid β-peptides and APO E in Down syndrome: implications for initial events in amyloid plaque formation. Neurobiol. Dis. 1996;3:16–32. doi: 10.1006/nbdi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki N., Cheung T. T., Cai X. D., Odaka A., Otvos L., Jr, Eckman C., Golde T. E., Younkin S. G. An increased percentage of long amyloid β protein secreted by familial amyloid βprotein precursor (β APP717) mutants. Science. 1994;264:1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- 17.Eckman C. B., Mehta N. D., Crook R., Perez-tur J., Prihar G., Pfeiffer E., Graff-Radford N., Hinder P., Yager D., Zenk B., et al. A new pathogenic mutation in the APP gene (I716V) increases the relative proportion of Aβ42(43) Hum. Mol. Genet. 1997;6:2087–2089. doi: 10.1093/hmg/6.12.2087. [DOI] [PubMed] [Google Scholar]
- 18.McGowan E., Pickford F., Kim J., Onstead L., Eriksen J., Yu C., Skipper L., Murphy M. P., Beard J., Das P., et al. Aβ42 is essential for parenchymal and vascular amyloid deposition in mice. Neuron. 2005;47:191–199. doi: 10.1016/j.neuron.2005.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petkova A. T., Ishii Y., Balbach J. J., Antzutkin O. N., Leapman R. D., Delaglio F., Tycko R. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benzinger T. L., Gregory D. M., Burkoth T. S., Miller-Auer H., Lynn D. G., Botto R. E., Meredith S. C. Two-dimensional structure of β-amyloid(10–35) fibrils. Biochemistry. 2000;39:3491–3499. doi: 10.1021/bi991527v. [DOI] [PubMed] [Google Scholar]
- 21.Antzutkin O. N., Leapman R. D., Balbach J. J., Tycko R. Supramolecular structural constraints on Alzheimer's β-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry. 2002;41:15436–15450. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- 22.Balbach J. J., Petkova A. T., Oyler N. A., Antzutkin O. N., Gordon D. J., Meredith S. C., Tycko R. Supramolecular structure in full-length Alzheimer's β-amyloid fibrils: evidence for a parallel β-sheet organization from solid-state nuclear magnetic resonance. Biophys. J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petkova A. T., Yau W. M., Tycko R. Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato T., Kienlen-Campard P., Ahmed M., Liu W., Li H., Elliott J. I., Aimoto S., Constantinescu S. N., Octave J. N., Smith S. O. Inhibitors of amyloid toxicity based on β-sheet packing of Aβ40 and Aβ42. Biochemistry. 2006;45:5503–5516. doi: 10.1021/bi052485f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrescu A. T. An NMR-based quenched hydrogen exchange investigation of model amyloid fibrils formed by cold shock protein. A. Pac. Symp. Biocomput. 2001;6:67–78. doi: 10.1142/9789814447362_0008. [DOI] [PubMed] [Google Scholar]
- 26.Whittemore N. A., Mishra R., Kheterpal I., Williams A. D., Wetzel R., Serpersu E. H. Hydrogen-deuterium (H/D) exchange mapping of Aβ 1–40 amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44:4434–4441. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- 27.Olofsson A., Ippel J. H., Wijmenga S. S., Lundgren E., Öhman A. Probing solvent accessibility of transthyretin amyloid by solution NMR spectroscopy. J. Biol. Chem. 2004;279:5699–5707. doi: 10.1074/jbc.M310605200. [DOI] [PubMed] [Google Scholar]
- 28.Ippel J. H., Olofsson A., Schleucher J., Lundgren E., Wijmenga S. S. Probing solvent accessibility of amyloid fibrils by solution NMR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8648–8653. doi: 10.1073/pnas.132098999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshino M., Katou H., Hagihara Y., Hasegawa K., Naiki H., Goto Y. Mapping the core of the β(2)-microglobulin amyloid fibril by H/D exchange. Nat. Struct. Biol. 2002;9:332–336. doi: 10.1038/nsb792. [DOI] [PubMed] [Google Scholar]
- 30.Lührs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Döbeli H., Schubert D., Riek R. 3D structure of Alzheimer's amyloid-β(1–42) fibrils. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olofsson A., Sauer-Eriksson A. E., Öhman A. The solvent protection of alzheimer amyloid-β-(1–42) fibrils as determined by solution NMR spectroscopy. J. Biol. Chem. 2006;281:477–483. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- 32.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Helgstrand M., Kraulis P., Allard P., Härd T. Ansig for Windows: an interactive computer program for semiautomatic assignment of protein NMR spectra. J. Biomol. NMR. 2000;18:329–336. doi: 10.1023/a:1026729404698. [DOI] [PubMed] [Google Scholar]
- 34.Crescenzi O., Tomaselli S., Guerrini R., Salvadori S., D'Ursi A. M., Temussi P. A., Picone D. Solution structure of the Alzheimer amyloid β-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur. J. Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 35.Johnson B. A., Blevins R. A. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 36.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 37.Zirah S., Kozin S. A., Mazur A. K., Blond A., Cheminant M., Segalas-Milazzo I., Debey P., Rebuffat S. Structural changes of region 1–16 of the Alzheimer disease amyloid β-peptide upon zinc binding and in vitro aging. J. Biol. Chem. 2006;281:2151–2161. doi: 10.1074/jbc.M504454200. [DOI] [PubMed] [Google Scholar]
- 38.Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 39.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q. Rev. Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 40.Morgan C., Colombres M., Nunez M. T., Inestrosa N. C. Structure and function of amyloid in Alzheimer's disease. Prog. Neurobiol. 2004;74:323–349. doi: 10.1016/j.pneurobio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 41.Shivaprasad S., Wetzel R. Scanning cysteine mutagenesis analysis of Aβ-(1–40) amyloid fibrils. J. Biol. Chem. 2006;281:993–1000. doi: 10.1074/jbc.M505091200. [DOI] [PubMed] [Google Scholar]
- 41a.Guo J.-T., Wetzel R., Xu Y. Molecular modeling of the core of Aβ amyloid fibrils. Proteins. 2004;57:357–364. doi: 10.1002/prot.20222. [DOI] [PubMed] [Google Scholar]
- 42.Paravastu A. K., Petkova A. T., Tycko R. Polymorphic fibril formation by residues 10–40 of the Alzheimer's β-amyloid peptide. Biophys. J. 2006;90:4618–4629. doi: 10.1529/biophysj.105.076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kheterpal I., Williams A., Murphy C., Bledsoe B., Wetzel R. Structural features of the Aβ amyloid fibril elucidated by limited proteolysis. Biochemistry. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 44.Riek R., Güntert P., Döbeli H., Wipf B., Wüthrich K. NMR studies in aqueous solution fail to identify significant conformational differences between the monomeric forms of two Alzheimer peptides with widely different plaque-competence, Aβ(1–40)(ox) and Aβ(1–42)(ox) Eur. J. Biochem. 2001;268:5930–5936. doi: 10.1046/j.0014-2956.2001.02537.x. [DOI] [PubMed] [Google Scholar]
- 45.Smith D. P., Smith D. G., Curtain C. C., Boas J. F., Pilbrow J. R., Ciccotosto G. D., Lau T. L., Tew D. J., Perez K., Wade J. D., et al. Copper-mediated amyloid-β toxicity is associated with an intermolecular histidine bridge. J. Biol. Chem. 2006;281:15145–15154. doi: 10.1074/jbc.M600417200. [DOI] [PubMed] [Google Scholar]
- 46.Williams A. D., Portelius E., Kheterpal I., Guo J. T., Cook K. D., Xu Y., Wetzel R. Mapping Aβ amyloid fibril secondary structure using scanning proline mutagenesis. J. Mol. Biol. 2004;335:833–842. doi: 10.1016/j.jmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 47.Morimoto A., Irie K., Murakami K., Masuda Y., Ohigashi H., Nagao M., Fukuda H., Shimizu T., Shirasawa T. Analysis of the secondary structure of β-amyloid (Aβ42) fibrils by systematic proline replacement. J. Biol. Chem. 2004;279:52781–52788. doi: 10.1074/jbc.M406262200. [DOI] [PubMed] [Google Scholar]
- 48.Kim W., Hecht M. H. Sequence determinants of enhanced amyloidogenicity of Alzheimer Aβ42 peptide relative to Aβ40. J. Biol. Chem. 2005;280:35069–35076. doi: 10.1074/jbc.M505763200. [DOI] [PubMed] [Google Scholar]