Abstract
Chemokines are a superfamily of small structurally related cytokines that have evolved to form a complex network of proteins that typically regulate leucocyte traffic but also carry very diverse sets of immune and non-immune functions. Two general features of cytokines, redundancy and promiscuity, are particularly prominent in chemokines. In part, these properties result from repeated processes of gene duplication and diversification, which has led to the present complex genomic map of chemokines, which contains cases of non-allelic isoforms, copy number polymorphisms and classical allelic variation. This genomic complexity is compounded with pre-translational and post-translational mechanisms resulting in a complex network of proteins whose essential functions are maintained, constituting a remarkable case of robustness reminiscent of crucial metabolic pathways. This reflects the adaptation of a system under strong evolutive pressure, supporting the concept that the chemokine system is essential for the coordination, regulation and fine-tuning of the type of immune response. In this first review, we analyse currently available data on the chemokine superfamily, focusing on its complex genomic organization. Genes encoding essential inflammatory chemokines are grouped into defined chromosomal locations as clusters and miniclusters that, from the genetic point of view, can be considered single entities given their overall functions (many ligands of a cluster bind to a few shared receptors). We will try to interpret this genomic organization of chemokines in relation to the main functions acquired by each individual member or by each cluster. In a second review, we shall focus on the relationship of chemokine variability and disease susceptibility.
Keywords: chemokines, evolution, genome, human, variability
Chemokines: a special case of complexity
Chemokines are a large superfamily of small (approximately 8–15 kDa) structurally related cytokines that regulate cell trafficking of various types of leucocytes to areas of injury and play different roles in both inflammatory and homeostatic processes [1–3]. To date, 42 chemokines and 18 chemokine receptors (CKRs) have been identified in humans, but this number does not take into account several described variants that can increase their effective complexity.
Chemokines share from 20% to 95% of the amino acid sequence identity and have been grouped into four subfamilies, CXC, CC, CX3C and C chemokines, according to their structural cysteine motif found near the NH2 terminus [4]. All of them adopt a similar folding pattern even in cases of low overall sequence identity: the chemokine scaffold consists of a N-terminal region followed and connected via cysteine bonds to its core of three β-sheets and a C-terminal α-helix. Chemokines carry out their biological functions through binding to seven-transmembrane domain G protein-coupled receptors expressed on different leucocytes, endothelial and other cell types [5–7].
Based upon the site and circumstance of their expression, chemokines can generally be classified as either inducible (inflammatory) or constitutive (homeostatic). Inducible chemokines are usually expressed under inflammatory stimuli. In contrast, constitutive chemokines are expressed in the absence of infection or damage, controlling from cell trafficking in the embryo and foetus to homeostatic leucocyte homing for immune surveillance.
In evolution, diversification through the generation of multiple alleles is very common and the immune system contains several groups of genes with prominent allelic variation, the MHC genes being the most extreme example in the human genome. The chemokine system constitutes a less well-studied but very revealing case of how through evolution, a complex network of genes has acquired a very diverse set of related functions. In this review, we analyse the existing correlations between the genomic organization of the different groups of chemokines as clusters and miniclusters and their linked function as we believe that this perspective will help to better understand the sometimes confusing chemokine system.
How the chemokine network conciliates fine-tuning, reliability and robustness with extreme variability and phylogenic plasticity, by using redundancy and promiscuity
To preserve the overall function, the chemokine network is relatively insensitive to alterations of their individual components. In fact, the chemokine superfamily has achieved, in spite of its broad variability, a high degree of robustness, mainly through the redundancy and binding promiscuity of their ligands and receptors [8]. This type of complexity is a general feature of cytokines, but reaches its highest level in the chemokine superfamily. Chemokines are redundant in their effects on the target cells (no chemokine is uniquely active on one leucocyte population and usually a given leucocyte population has receptors for different chemokines), and the interaction with their receptors also has considerable promiscuity (most known receptors interact with multiple ligands and most ligands interact with more than one receptor).
Although redundancy among chemokines is not universal, there is a considerable degree of functional overlapping among some of them (this also applies to CKRs). The chemokine/CKR system robustness makes it possible that when one given chemokine or receptor is defective there is usually an alternative set of chemokines or CKRs that can maintain the main biological functions. This principle has a striking exception: the CXCL12/CXCR4 pair. CXCL12 and CXCR4-knockout mice show embryonic lethality [9,10], providing a clear example of the wide range of biological effects of chemokines, whose functions extend beyond simply attracting leucocytes, taking part in organ development, angiogenesis, angiostasis and immune regulation.
In spite of the fact that one chemokine might act on different cells and, conversely, a single cell responds to many chemokines, the role of chemokines in both inflammatory and homeostatic processes is finely regulated case-by-case by the secretion of specific sets of chemokines and by a spatial and temporal control of chemokine production (as well as regulation of receptor expression on particular leucocyte subtypes). The comparison of the properties of each of the different ligands of a common receptor suggests the existence of additional mechanisms to limit redundancy [11]. Two ligands that share a given receptor may exhibit significant differences in their affinity for the receptor, their ability to bind to glycosaminoglycans (GAGs, see below), their capacity to induce receptor desensitization, their half-life, their susceptibility to protease modifications, or in the territories of distribution (e.g. extracellular matrix, endothelium, etc.). A new level of chemokine regulation has emerged from the discovery that chemokines can also act as receptor antagonists. By this dual-activity, chemokines would attract or activate a population of cells bearing one receptor, while at the same time preventing the recruitment and/or activation of a different cell population via another receptor [12].
Another important mechanism to control the chemokine activity through regulation of chemokine levels in the body is carried out by decoy receptors, defined as cell surface ligand-binding proteins with high affinity and specificity for chemokines, but which are unable to induce downstream signalling [13]. Three of these so-called ‘silent’ receptors have been identified, DARC [14], D6 [15–17] and CCX-CKR [18,19], each of them interacting with a specific set of chemokines. This chemokine scavenging is important for maintaining chemokine homeostasis because efficient recruitment of inflammatory cells requires low chemokine levels in the bloodstream and high chemokine levels in the target tissue.
In addition to chemokine G protein-coupled receptor interactions, chemokines bind both to soluble glycosaminoglycans (GAGs) and GAGs immobilized on cell surfaces and the extracellular matrix. Chemokine immobilization through the GAG interaction facilitates the formation of haptotactic chemokine gradients and enhances their concentration at the site of production [20]. Interestingly, recent in vivo work exploiting recombinant chemokine mutants suggests that oligomeric chemokine binding to GAGs is crucial for biological responses [21]. Interaction with GAGs may also provide another level of specificity and control to cell migration, beyond that defined by receptor engagement, by selective binding of certain chemokines to different types of GAGs.
In summary, all these mechanisms seem to operate to increase the selectivity of cell recruitment and, in more general terms, to provide mechanisms to exert a fine control of the variability, redundancy and promiscuity of the chemokine network.
Genomic organization of chemokines
Genomic evolution, the first source of variability
Chemokine function probably preceded the origin of the chemokine network as chemokine-like molecules have been detected in sponges [22]. Numerous studies have pointed out that most, if not all, chemokines arose by gene duplication of a single ancestral gene. In fact, chemokine and CKR evolution can be traced through phylogeny from early vertebrates to non-human primates [23] and, overall, chemokines have expanded markedly their role in orchestrating the immune response and in organizing the lymphoid tissue. Co-evolution of pathogens with their hosts has led to adaptive changes where some pathogens, e.g. viruses, encode chemokine homologues as part of their evasion strategy [24,25].
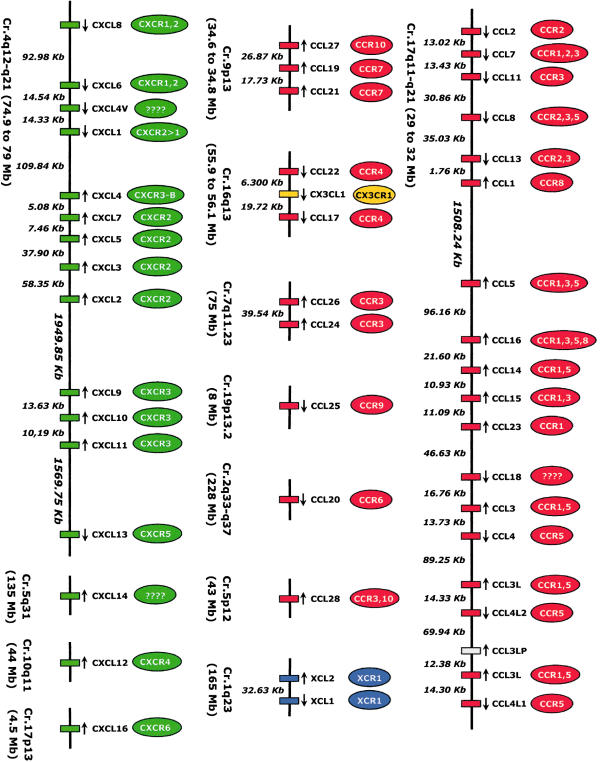
During the evolution, the different options for gene variability (from genomic region duplications to point mutations) have produced the present genomic organization of chemokines in humans [26]. Many chemokines (just as many chemokine receptors) are clustered in defined chromosomal locations. Two main clusters have been recognized, both of them codifying the essential inflammatory chemokines: the CXC cluster, located in chromosome 4q12–21, and the CC cluster, located in chromosome 17q11.2 (Fig. 1). The chemokines that map in the CXC and the CC clusters seem to maintain some specific functions: CXC cluster chemokines recruit mainly neutrophils while CC cluster members typically attract mononuclear cells.
Fig. 1.
Map of genomic organization of human chemokines. CC chemokines in red, CXC chemokines in green, CX3C chemokine in yellow and C chemokines in blue. Distances between genes are expressed in Kb. Gene or cluster chromosomal location is expressed as a distance from the beginning of chromosome (in Mb). Receptors are shown above each ligand. The orientation of each gene is showed by an arrow.
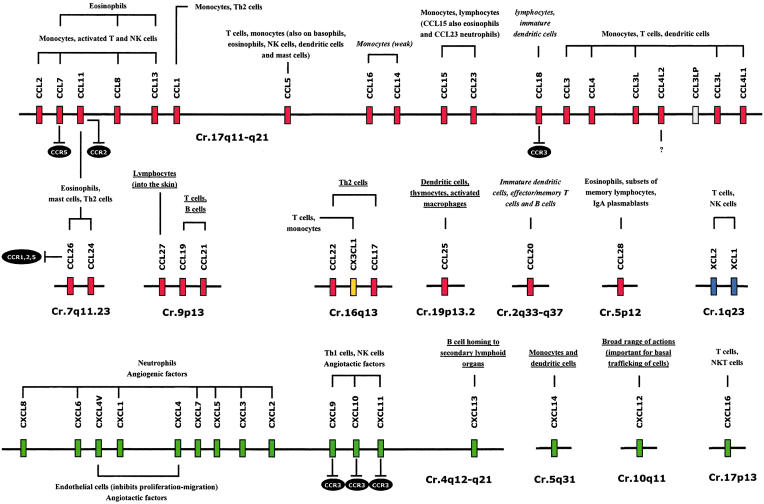
Genes of the more recently identified CC and CXC chemokines tend to be located in different chromosomal locations, far from CC and CXC clusters. These chemokine genes located away from the two major clusters correspond to older genes in evolutionary terms, remaining better conserved among species probably because of their very specific functions (in contrast, the major CXC and CC chemokine clusters were generated more recently). An important characteristic of chemokine genes from the same cluster is that they code for many ligands that interact with a few receptors. Therefore, chemokine clusters are single entities based on their overall function [4] (Fig. 2). For this reason, in this review we have used the genomic organization as the guidance for discussing the chemokine system.
Fig. 2.
Relationship between genomic organization and function of chemokines. Main cell types (and essential actions) targeted by individual or chemokine cluster are shown. Functions of homeostatic chemokines are underlined and functions of homeostatic/inducible chemokines are in italics. CC chemokines in red, CXC chemokines in green, CX3C chemokine in yellow and C chemokines in blue. Antagonist functions are represented by ⊥.
Human CXC chemokines subfamily
The human CXC chemokines subfamily comprises 15 ligands and six receptors in humans, and their functions extend well beyond the immune system. Most CXC chemokines are evolutionary recent and exclusive of mammals: only CXCL12 (SDF-1) and CXCL14 (BRAK), two homeostatic chemokines, have unambiguous orthologues in fish, suggesting that they are the phylogenetically modern representatives of the ancestral CXC chemokine [27].
Almost all the CXC inflammatory chemokines are located in the chromosome 4 cluster, organized into two subclusters or groups.
The centromeric subcluster contains the genes coding for CXCL1 to CXCL8 (systematic names for GROα, GROβ, GROγ, PF4, ENA-78, GCP-2, NAP-2 and IL-8). All members of this group, with the exception of CXCL4, show the tri-peptide ELR (glutamic-leucine-arginine) motif (next to the first conserved cysteine) implicated in neutrophil chemotaxis through binding to CXCR1 and CXCR2, and also conferring on them pro-angiogenic effects. Despite their similarities, each ELR+ chemokine has distinct effects, due in part to their divergent abilities to bind and activate CXCR1 and CXCR2. The redundancy in ELR+ CXC chemokines and their receptors may provide multiple levels of regulation that allow for specific control of inflammation [28]. Completely different from the other chemokines of this group, CXCL4 is an anti-angiogenic ELR-negative chemokine. CXCL4 (mainly found in platelets) inhibits endothelial cell proliferation/migration and angiogenesis. Growing evidence suggests that CXCL4 biology depends on its unusually high affinity for heparan sulphates, rather than the binding to a well-defined CXCR [29–31]. However, recently, CXCR3-B (an alternatively spliced variant of CXCR3) has been identified as a CXCL4 receptor [32]. Besides, CXCL4 has a non-allelic copy named CXCL4L1 (PF4V1) whose product, although it only differs by three amino acids, has a more potent anti-angiogenic factor [33,34], making it a very interesting molecule for therapy [35].
The telomeric subcluster of inflammatory CXC chemokines in chromosome 4 includes ELR-negative chemokines CXCL9 (Mig), CXCL10 (IP-10) and CXCL11 (I-TAC). These three chemokines bind to CXCR3 (CXCR3-A and CXCR3-B) and attract activated Th1 lymphocytes and NK cells. In contrast to ELR+ chemokines, these three chemokines are potent angiostatic factors. Therefore the CXC chemokines are a unique family of cytokines due to their ability to behave in an opposite manner in the regulation of angiogenesis through the ELR motif and this different activity may have an impact on the pathogenesis of a variety of inflammatory disorders [36]. Additionally, CXCL9, CXCL10 and CXCL11 are natural antagonists for CCR3, suggesting that chemokines that attract Th1 cells via CXCR3 can concomitantly block the migration of Th2 cells in response to CCR3 ligands, thus enhancing the polarization of T cell recruitment [37]. The recently discovered CXCR3 receptor variant CXCR3-B (in addition to the classic form, CXCR3-A) could explain the different functions of CXCL9, CXCL10 and CXCL11 on distinct cell types.
Aside from these two subclusters of CXC inflammatory chemokines but in the same stretch of chromosome 4, we find CXCL13, a homeostatic CXC chemokine constitutively expressed in lymphoid follicles. It is the unique ligand of CXCR5, and essentially contributes to B cell homing and proper positioning of these cells within the microanatomic compartments of secondary lymphoid organs [38] and specifically in defining the B and T cell areas in the lymphoid follicles.
The three remaining CXC chemokines are mapped separately in other chromosomes. The already mentioned CXCL12 (SDF-1, chromosome 10) is a homeostatic CXC chemokine widely expressed and showing a broad range of actions, affecting immune cell chemotaxis to neural development. CXCL12 appears to be particularly important for the regulation of homeostatic traffic and distribution of cells in the different compartments and subcompartments of the immune system, for instance directing naïve T cell traffic through lymph nodes [39,40]. CXCL12 is important for HIV infection because CXCR4, its only known receptor, is also the membrane protein used by HIV T-tropic strains as a co-receptor to entry into target cells [41–43].
CXCL14 (BRAK, chromosome 5) is also a homeostatic CXC chemokine ubiquitously expressed in all normal tissues. The structure of CXCL14 is considerably divergent from all other chemokines, its receptor has not been identified and its activity is not yet well defined. However, it seems that CXCL14 is chemoattractant for monocytes [44] and dendritic cells [45,46] and recent data indicate that CXCL14 expression also inhibits tumour growth [47].
The last CXC chemokine, CXCL16 (chromosome 17) is, in the strictest definition, a member of the CXC family but it is distantly related to all known chemokines and might be phylogenetically closer to CC chemokines. Additionally, its structure is similar to CX3CL1 in having a transmembrane region and a chemokine domain suspended by a mucin-like stalk [48,49]. Thus, CXCL16 not only attracts T cells and NKT cells toward DCs but also supports their firm adhesion to DCs [50]. Interestingly, after proteolytic cleavage by members of the ADAM family of proteases, the chemokine domain of CXCL16 can be released, attracting cells expressing the same receptor [51,52].
Human CC chemokines subfamily (and CX3C)
Within the cluster of pro-inflammatory CC chemokines in chromosome 17, we clearly distinguish two subclusters:
Centromerically, we found CCL2, CCL7, CCL11, CCL8, CCL13 and CCL1 that constitute the ‘MCP’ subcluster. CCL2, CCL8, CCL7 and CCL13 (originally called MCP-1, -2, -3 and -4) are closely related chemoattractants for monocytes, activated T and NK cells; additionally CCL8, CCL7 and CCL13 also attract eosinophils. They bind mainly to CCR2 and CCR3, and may be pathogenic in human diseases characterized by mononuclear and/or eosinophilic infiltration (as atherosclerosis and asthma). Interestingly, CCL7 is a highly potent CCR5 antagonist [53]. CCL11 (eotaxin-1), a potent eosinophil chemoattractant, maps within the MCP cluster, although the two other members of the eotaxin gene family, CCL24 (eotaxin-2) and CCL26 (eotaxin-3), are located together in a minicluster in chromosome 7. These three chemokines recruit and activate CCR3-bearing cells such as eosinophils, mast cells and Th2 lymphocytes, playing a major role in allergic disorders. Interestingly, CCL26 is the first human chemokine that features broad antagonistic activities as a natural antagonist to the ligands of CCR1, CCR2 and CCR5 (CCL11 can also block CCR2 [54]). This suggests that CCL26 may have a modulator rather than an inflammatory function [12,55]. Finally, the last and more telomeric gene codes for CCL1 (I-309). CCL1 has structural features that distinguish it from other chemokines, including an additional pair of cysteines that form a third intramolecular disulphide bond, and a propensity to remain monomeric at concentrations at which many other chemokines dimerize. CCL1 is the only high-affinity ligand for CCR8 and in spite of these peculiarities it has similar functional features to the other members of this subcluster, being also chemotactic for monocytes and Th2 lymphocytes [56].
The second subcluster of pro-inflammatory CC chemokines in chromosome 17 is 1·5 Mb telomeric to the MCP one. To simplify, in this subcluster we find CCR1/CCR5 ligands that we can in turn subdivide into the HCC and MIP groups. (a) The HCC group contains CCL14, CCL15, CCL16 (the systematic names of HCC1, HCC2, HCC4), CCL23 (MPIF) and CCL5 (RANTES). CCL14, CCL15, CCL16 and CCL23 are clustered within a 40-kb long region whereas CCL5 (a very promiscuous chemokine, binding with high affinity to CCR1/CCR3/CCR5) is located approximately 100 Kb away (with several non-related genes within). The HCC genes are relatively large, from 3·1 Kb (CCL14) to 8·8 kb (CCL5), compared with other CC chemokine genes, such as CCL4 and CCL2 (< 2 kb). Moreover, in contrast to most other human CC chemokine genes that comprise three exons, the CCL15 and CCL23 have four exons. In fact, their high nucleotide homology indicates that they have been generated recently by duplication [57]. All these chemokines share the binding to CCR1, even though most of them can bind to other CCRs. This group is functionally more heterogeneous (see Fig. 2) than the MIP group or the MCP subcluster [58–63]. (b) The MIP group is composed of CCL18 (PARC), CCL3 (MIP-1α), CCL3L (LD78β), CCL4 (MIP-1β) and CCL4L. This is a very interesting gene minicluster that could serve as a model of chemokine evolution. CCL3 and CCL4 were formed by duplication of a common ancestral gene [64] and both have a second non-allelic copy, CCL3L and CCL4L, originated by a second duplication of the entire DNA block that contains the two genes. Next to them, can also be found a CCL3 5′-truncated pseudogene, named CCL3LP (LD78γ) [65]. The CCL3L-CCL4L tandem shows also an additional variability: they have a copy number polymorphism (CNP) that means that these genes are present in variable numbers in the genome. Regarding this CNP, there are not only interindividual and interpopulation differences in the copy number of the CCL3L-CCL4L tandem, but, interestingly, there are also haplotypes with different numbers of individual genes [66]. Moreover, a recent study demonstrated that there are two main allelic variants of CCL4L: CCL4L1, the originally described variant, and CCL4L2, in which one base substitution at the acceptor splice site of intron 2 originates a new complex pattern of splicing variants [67]. Functionally, CCL3/CCL3L and CCL4/CCL4L are highly related. They both bind to CCR5, but CCL3 binds also to CCR1 with high affinity [64,68].
The other member of this group and the most centromeric one, CCL18, is larger than average (7·2 kb versus general size of 2–3 kb) as a result of a first intron of 6·0 kb, which contains two pseudo-exons. This feature, together with the presence of two adjacent regions within the CCL18 gene sharing high-sequence similarity with the CCL3 gene, suggests that the CCL18 gene may have been generated by fusion of two CCL3-like genes with deletion and selective use of some exons [69]. CCL18 apparently functions as an inflammatory/inducible or constitutive/homeostatic chemokine depending on the circumstances: it is mainly expressed by a broad range of monocytes/macrophages and DC, and it is constitutively present at high levels in human plasma (probably contributing to the physiological homing of lymphocytes and DC and to the generation of primary immune responses) [70]. Importantly, the CCL18 receptor has not been yet identified. This chemokine exhibits CCR3 antagonistic activity [71].
The rest of the CC chemokines are located in other chromosomes either as a single member or constituting a minicluster. CCL27 (CTACK), CCL19 (ELC) and CCL21 (SLC) are grouped in a minicluster in chromosome 9. CCL19 and CCL21 share only 32% amino acid identity but constitute a genetically and functionally highly related subgroup of homeostatic CC chemokines. They are the only two ligands of CCR7, and they have a potent chemotactic activity for naive T cells and B cells. They clearly play a pivotal role in naive lymphocyte homing and traffic within lymphoid tissues [72,73]. Unrelated to CCL19 and CCL21, CCL27 mediates the migration of lymphocytes into the skin, through binding to the CCR10 [74].
The two closely related chemokines CCL22 (MDC) and CCL17 (TARC) map in chromosome 16 together with CX3CL1 (Fractalkine), the only member of CX3C subfamily. CCL17 and CCL22, the two known ligands of CCR4, are a pair CC chemokines highly expressed in the thymus. It is known that they selectively attract Th2 type memory T cells into the inflammatory sites and regulate Th2-related immune responses [75,76]. Interestingly, the CX3CL1 gene is situated between the CCL17 and CCL22 genes. Although the three-dimensional structure of the chemokine domain of CX3CL1 is different from that of CC chemokines, it was identified thanks to its amino acid homology with members of the CC chemokine family [77]. Unlike other chemokines, CX3CL1 carries the chemokine domain on top of an extended mucin-like stalk and can exist in two forms: as a transmembrane chemokine/mucin hybrid protein and as a soluble chemotactic polypeptide generated by proteolytic cleavage by members of the ADAM family of proteases [78].
CCL20 (LARC, chromosome 2) is the only chemokine known to interact with CCR6, a property shared with the anti-microbial peptides β-defensins [79]. Like CCL18, CCL20 is considered a constitutive and inducible chemokine. Constitutive expression of CCL20 was originally demonstrated in the liver, mucosa-associated and lymphoid tissues and induction in monocytes as well as many other cells [80].
CCL25 (TECK, chromosome 19) is the only chemokine binding to CCR9. It is constitutively and selectively expressed at high levels in the thymus and small intestine. CCL25 has been reported to chemoattract dendritic cells, thymocytes and activated macrophages and probably plays a significant role in the recruitment of developing thymocytes to discrete compartments within the thymus and, in general, in T lymphocyte development [81,82].
The last CC chemokine, CCL28 (MEC, chromosome 5), is most homologous to CCL27, displaying about 40% identity and sharing binding to CCR10 although CCL28 also binds to CCR3. It is expressed not in the skin as CCL27 but, instead in diverse mucosal tissues, suggesting that CCL28 may play an important role in the physiology and/or recruitment of specialized cells into mucosal tissues [83–85].
Human C chemokines subfamily
The C chemokines subfamily, located in a minicluster in chromosome 1, has only two highly homologous genes encoding XCL1 (lymphotactin-α) and XCL2 (lymphotactin-β). These two chemokines that differ by only two amino acid residues both bind to XCR1, and have some homology with a number of CC chemokines, especially with CCL8 and CCL3. The major structural feature of XCL1/2 is that they lack two of the four-cysteine residues characteristics of the chemokines (and therefore have only one disulphide bond). XCL1/2 are potent chemoattractants of T and NK cells, but not of monocytes or neutrophils, and they are the major products of activated CD8+ T cells [86,87].
Concluding remarks
The human chemokine superfamily forms a complex and robust network of genes and proteins playing very diverse roles but also displaying a lot of functional interactions between them. Current genomic organization of human chemokine genes provides a useful tool to better understand the complexity of this superfamily. Based on an exhaustive genomic map of all human chemokines, we have linked the genetic organization (clusters and miniclusters) with the main functions, focusing on inflammatory chemokines. Future studies on the relationship between genetics and function should focus on other important mechanisms of variability, such as polymorphisms and alternative splicings, as they may lead to disease susceptibility. In this context, it would be important not only to deal with each chemokine separately but also to take into account all genetically and functionally related members (a second review will mainly focus on these aspects).
Acknowledgments
This work was supported by grants from the FIPSE (Fundación para la Investigación y la Prevención del Sida en España) (project 36487/05) and FIS (Fondo de Investigaciones Sanitarias) (project PI 02/0104).
References
- 1.Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76:301–14. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 2.Luster AD. Chemokines – chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338:436–45. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 3.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–8. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 4.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–7. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 5.Murphy PM. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 6.Premack BA, Schall TJ. Chemokine receptors: gateways to inflammation and infection. Nat Med. 1996;2:1174–8. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 7.Zlotnik A, Morales J, Hedrick JA. Recent advances in chemokines and chemokine receptors. Crit Rev Immunol. 1999;19:1–47. [PubMed] [Google Scholar]
- 8.Mantovani A. The chemokine system: redundancy for robust outputs. Immunol Today. 1999;20:254–7. doi: 10.1016/s0167-5699(99)01469-3. [DOI] [PubMed] [Google Scholar]
- 9.Tachibana K, Hirota S, Iizasa H, et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–4. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 10.Ma Q, Jones D, Borghesani PR, et al. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA. 1998;95:9448–53. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devalaraja MN, Richmond A. Multiple chemotactic factors: fine control or redundancy? Trends Pharmacol Sci. 1999;20:151–6. doi: 10.1016/s0165-6147(99)01342-5. [DOI] [PubMed] [Google Scholar]
- 12.Petkovic V, Moghini C, Paoletti S, Uguccioni M, Gerber B. Eotaxin-3/CCL26 is a natural antagonist for CC chemokine receptors 1 and 5. A human chemokine with a regulatory role. J Biol Chem. 2004;279:23357–63. doi: 10.1074/jbc.M309283200. [DOI] [PubMed] [Google Scholar]
- 13.Comerford I, Nibbs RJ. Post-translational control of chemokines: a role for decoy receptors? Immunol Lett. 2005;96:163–74. doi: 10.1016/j.imlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Lee JS, Frevert CW, Wurfel MM, et al. Duffy antigen facilitates movement of chemokine across the endothelium in vitro and promotes neutrophil transmigration in vitro and in vivo. J Immunol. 2003;170:5244–51. doi: 10.4049/jimmunol.170.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fra AM, Locati M, Otero K, et al. Cutting edge: scavenging of inflammatory CC chemokines by the promiscuous putatively silent chemokine receptor D6. J Immunol. 2003;170:2279–82. doi: 10.4049/jimmunol.170.5.2279. [DOI] [PubMed] [Google Scholar]
- 16.Weber M, Blair E, Simpson CV, et al. The chemokine receptor D6 constitutively traffics to and from the cell surface to internalize and degrade chemokines. Mol Biol Cell. 2004;15:2492–508. doi: 10.1091/mbc.E03-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galliera E, Jala VR, Trent JO, et al. beta-Arrestin-dependent constitutive internalization of the human chemokine decoy receptor D6. J Biol Chem. 2004;279:25590–7. doi: 10.1074/jbc.M400363200. [DOI] [PubMed] [Google Scholar]
- 18.Gosling J, Dairaghi DJ, Wang Y, et al. Cutting edge: identification of a novel chemokine receptor that binds dendritic cell- and T cell-active chemokines including ELC, SLC, and TECK. J Immunol. 2000;164:2851–6. doi: 10.4049/jimmunol.164.6.2851. [DOI] [PubMed] [Google Scholar]
- 19.Townson JR, Nibbs RJ. Characterization of mouse CCX-CKR, a receptor for the lymphocyte-attracting chemokines TECK/mCCL25, SLC/mCCL21 and MIP-3beta/mCCL19: comparison to human CCX-CKR. Eur J Immunol. 2002;32:1230–41. doi: 10.1002/1521-4141(200205)32:5<1230::AID-IMMU1230>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka Y, Adams DH, Hubscher S, Hirano H, Siebenlist U, Shaw S. T-cell adhesion induced by proteoglycan-immobilized cytokine MIP-1 beta. Nature. 1993;361:79–82. doi: 10.1038/361079a0. [DOI] [PubMed] [Google Scholar]
- 21.Proudfoot AE, Handel TM, Johnson Z, et al. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci USA. 2003;100:1885–90. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller WE, Wiens M, Muller IM, Schroder HC. The chemokine networks in sponges: potential roles in morphogenesis, immunity and stem cell formation. Prog Mol Subcell Biol. 2004;34:103–43. doi: 10.1007/978-3-642-18670-7_5. [DOI] [PubMed] [Google Scholar]
- 23.Clay CC, Rodrigues DS, Brignolo LL, et al. Chemokine networks and in vivo T-lymphocyte trafficking in nonhuman primates. J Immunol Meth. 2004;293:23–42. doi: 10.1016/j.jim.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 24.McFadden G, Murphy PM. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr Opin Microbiol. 2000;3:371–8. doi: 10.1016/s1369-5274(00)00107-7. [DOI] [PubMed] [Google Scholar]
- 25.Boomker JM, de Leij LF, The TH, Harmsen MC. Viral chemokine-modulatory proteins: tools and targets. Cytokine Growth Factor Rev. 2005;16:91–103. doi: 10.1016/j.cytogfr.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 26.DeVries ME, Kelvin AA, Xu L, Ran L, Robinson J, Kelvin DJ. Defining the origins and evolution of the chemokine/chemokine receptor system. J Immunol. 2006;176:401–15. doi: 10.4049/jimmunol.176.1.401. [DOI] [PubMed] [Google Scholar]
- 27.Huising MO, Stet RJ, Kruiswijk CP, Savelkoul HF, Lidy Verburg-van Kemenade BM. Molecular evolution of CXC chemokines: extant CXC chemokines originate from the CNS. Trends Immunol. 2003;24:307–13. doi: 10.1016/s1471-4906(03)00120-0. [DOI] [PubMed] [Google Scholar]
- 28.Bizzarri C, Beccari AR, Bertini R, Cavicchia MR, Giorgini S, Allegretti M. ELR+ CXC chemokines and their receptors (CXC chemokine receptor 1 and CXC chemokine receptor 2) as new therapeutic targets. Pharmacol Ther. 2006;112:139–49. doi: 10.1016/j.pharmthera.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Sachais BS, Higazi AA, Cines DB, Poncz M, Kowalska MA. Interactions of platelet factor 4 with the vessel wall. Semin Thromb Hemost. 2004;30:351–8. doi: 10.1055/s-2004-831048. [DOI] [PubMed] [Google Scholar]
- 30.Bikfalvi A. Platelet factor 4: an inhibitor of angiogenesis. Semin Thromb Hemost. 2004;30:379–85. doi: 10.1055/s-2004-831051. [DOI] [PubMed] [Google Scholar]
- 31.Mixon TA, Dehmer GJ. Recombinant platelet factor 4 for heparin neutralization. Semin Thromb Hemost. 2004;30:369–77. doi: 10.1055/s-2004-831050. [DOI] [PubMed] [Google Scholar]
- 32.Lasagni L, Francalanci M, Annunziato F, et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J Exp Med. 2003;197:1537–49. doi: 10.1084/jem.20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green CJ, Charles RS, Edwards BF, Johnson PH. Identification and characterization of PF4varl, a human gene variant of platelet factor 4. Mol Cell Biol. 1989;9:1445–51. doi: 10.1128/mcb.9.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisman R, Surrey S, Ramachandran B, Schwartz E, Poncz M. Structural and functional comparison of the genes for human platelet factor 4 and PF4alt. Blood. 1990;76:336–44. [PubMed] [Google Scholar]
- 35.Struyf S, Burdick MD, Proost P, Van Damme J, Strieter RM. Platelets release CXCL4L1, a nonallelic variant of the chemokine platelet factor-4/CXCL4 and potent inhibitor of angiogenesis. Circ Res. 2004;95:855–7. doi: 10.1161/01.RES.0000146674.38319.07. [DOI] [PubMed] [Google Scholar]
- 36.Belperio JA, Keane MP, Arenberg DA, et al. CXC chemokines in angiogenesis. J Leukoc Biol. 2000;68:1–8. [PubMed] [Google Scholar]
- 37.Loetscher P, Pellegrino A, Gong JH, et al. The ligands of CXC chemokine receptor 3, I–TAC, Mig, and IP10, are natural antagonists for CCR3. J Biol Chem. 2001;276:2986–91. doi: 10.1074/jbc.M005652200. [DOI] [PubMed] [Google Scholar]
- 38.Gunn MD, Ngo VN, Ansel KM, Ekland EH, Cyster JG, Williams LT. A B-cell-homing chemokine made in lymphoid follicles activates Burkitt's lymphoma receptor-1. Nature. 1998;391:799–803. doi: 10.1038/35876. [DOI] [PubMed] [Google Scholar]
- 39.Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184:1101–9. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aiuti A, Webb IJ, Bleul C, Springer T, Gutierrez-Ramos JC. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J Exp Med. 1997;185:111–20. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bleul CC, Farzan M, Choe H, et al. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–33. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 42.Oberlin E, Amara A, Bachelerie F, et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–5. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 43.Moore JP, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–62. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 44.Kurth I, Willimann K, Schaerli P, Hunziker T, Clark-Lewis I, Moser B. Monocyte selectivity and tissue localization suggests a role for breast and kidney-expressed chemokine (BRAK) in macrophage development. J Exp Med. 2001;194:855–61. doi: 10.1084/jem.194.6.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shellenberger TD, Wang M, Gujrati M, et al. BRAK/CXCL14 is a potent inhibitor of angiogenesis and a chemotactic factor for immature dendritic cells. Cancer Res. 2004;64:8262–70. doi: 10.1158/0008-5472.CAN-04-2056. [DOI] [PubMed] [Google Scholar]
- 46.Shurin GV, Ferris R, Tourkova IL, et al. Loss of new chemokine CXCL14 in tumor tissue is associated with low infiltration by dendritic cells (DC), while restoration of human CXCL14 expression in tumor cells causes attraction of DC both in vitro and in vivo. J Immunol. 2005;174:5490–8. doi: 10.4049/jimmunol.174.9.5490. [DOI] [PubMed] [Google Scholar]
- 47.Schwarze SR, Luo J, Isaacs WB, Jarrard DF. Modulation of CXCL14 (BRAK) expression in prostate cancer. Prostate. 2005;64:67–74. doi: 10.1002/pros.20215. [DOI] [PubMed] [Google Scholar]
- 48.Matloubian M, David A, Engel S, Ryan JE, Cyster JG. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol. 2000;1:298–304. doi: 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 49.Wilbanks A, Zondlo SC, Murphy K, et al. Expression cloning of the STRL33/BONZO/TYMSTR ligand reveals elements of CC, CXC, and CX3C chemokines. J Immunol. 2001;166:5145–54. doi: 10.4049/jimmunol.166.8.5145. [DOI] [PubMed] [Google Scholar]
- 50.Shimaoka T, Nakayama T, Fukumoto N, et al. Cell surface-anchored SR-PSOX/CXC chemokine ligand 16 mediates firm adhesion of CXC chemokine receptor 6-expressing cells. J Leukoc Biol. 2004;75:267–74. doi: 10.1189/jlb.1003465. [DOI] [PubMed] [Google Scholar]
- 51.Gough PJ, Garton KJ, Wille PT, Rychlewski M, Dempsey PJ, Raines EW. A disintegrin and metalloproteinase 10-mediated cleavage and shedding regulates the cell surface expression of CXC chemokine ligand 16. J Immunol. 2004;172:3678–85. doi: 10.4049/jimmunol.172.6.3678. [DOI] [PubMed] [Google Scholar]
- 52.Abel S, Hundhausen C, Mentlein R, et al. The transmembrane CXC-chemokine ligand 16 is induced by IFN-gamma and TNF-alpha and shed by the activity of the disintegrin-like metalloproteinase ADAM10. J Immunol. 2004;172:6362–72. doi: 10.4049/jimmunol.172.10.6362. [DOI] [PubMed] [Google Scholar]
- 53.Blanpain C, Migeotte I, Lee B, et al. CCR5 binds multiple CC-chemokines: MCP-3 acts as a natural antagonist. Blood. 1999;94:1899–905. [PubMed] [Google Scholar]
- 54.Ogilvie P, Bardi G, Clark-Lewis I, Baggiolini M, Uguccioni M. Eotaxin is a natural antagonist for CCR2 and an agonist for CCR5. Blood. 2001;97:1920–4. doi: 10.1182/blood.v97.7.1920. [DOI] [PubMed] [Google Scholar]
- 55.Ogilvie P, Paoletti S, Clark-Lewis I, Uguccioni M. Eotaxin-3 is a natural antagonist for CCR2 and exerts a repulsive effect on human monocytes. Blood. 2003;102:789–94. doi: 10.1182/blood-2002-09-2773. [DOI] [PubMed] [Google Scholar]
- 56.Zingoni A, Soto H, Hedrick JA, et al. The chemokine receptor CCR8 is preferentially expressed in Th2 but not Th1 cells. J Immunol. 1998;161:547–51. [PubMed] [Google Scholar]
- 57.Nomiyama H, Fukuda S, Iio M, Tanase S, Miura R, Yoshie O. Organization of the chemokine gene cluster on human chromosome 17q11.2 containing the genes for CC chemokine MPIF-1, HCC-2, HCC-1, LEC, and RANTES. J Interferon Cytokine Res. 1999;19:227–34. doi: 10.1089/107999099314153. [DOI] [PubMed] [Google Scholar]
- 58.Schulz-Knappe P, Magert HJ, Dewald B, et al. HCC-1, a novel chemokine from human plasma. J Exp Med. 1996;183:295–9. doi: 10.1084/jem.183.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howard OM, Dong HF, Shirakawa AK, Oppenheim JJ. LEC induces chemotaxis and adhesion by interacting with CCR1 and CCR8. Blood. 2000;96:840–5. [PubMed] [Google Scholar]
- 60.Nomiyama H, Hieshima K, Nakayama T, et al. Human CC chemokine liver-expressed chemokine/CCL16 is a functional ligand for CCR1, CCR2 and CCR5, and constitutively expressed by hepatocytes. Int Immunol. 2001;13:1021–9. doi: 10.1093/intimm/13.8.1021. [DOI] [PubMed] [Google Scholar]
- 61.Pardigol A, Forssmann U, Zucht HD, et al. HCC-2, a human chemokine: gene structure, expression pattern, and biological activity. Proc Natl Acad Sci USA. 1998;95:6308–13. doi: 10.1073/pnas.95.11.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Youn BS, Zhang SM, Broxmeyer HE, et al. Characterization of CKbeta8 and CKbeta8–1: two alternatively spliced forms of human beta-chemokine, chemoattractants for neutrophils, monocytes, and lymphocytes, and potent agonists at CC chemokine receptor 1. Blood. 1998;91:3118–26. [PubMed] [Google Scholar]
- 63.Appay V, Rowland-Jones SL. RANTES: a versatile and controversial chemokine. Trends Immunol. 2001;22:83–7. doi: 10.1016/s1471-4906(00)01812-3. [DOI] [PubMed] [Google Scholar]
- 64.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–81. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 65.Hirashima M, Ono T, Nakao M, et al. Nucleotide sequence of the third cytokine LD78 gene and mapping of all three LD78 gene loci to human chromosome 17. DNA Seq. 1992;3:203–12. doi: 10.3109/10425179209034019. [DOI] [PubMed] [Google Scholar]
- 66.Townson JR, Barcellos LF, Nibbs RJ. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur J Immunol. 2002;32:3016–26. doi: 10.1002/1521-4141(2002010)32:10<3016::AID-IMMU3016>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 67.Colobran R, Adreani P, Ashhab Y, et al. Multiple products derived from two CCL4 loci: high incidence of a new polymorphism in HIV+ patients. J Immunol. 2005;174:5655–64. doi: 10.4049/jimmunol.174.9.5655. [DOI] [PubMed] [Google Scholar]
- 68.Maurer M, von Stebut E. Macrophage inflammatory protein-1. Int J Biochem Cell Biol. 2004;36:1882–6. doi: 10.1016/j.biocel.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 69.Tasaki Y, Fukuda S, Iio M, et al. Chemokine PARC gene (SCYA18) generated by fusion of two MIP-1alpha/LD78alpha-like genes. Genomics. 1999;55:353–7. doi: 10.1006/geno.1998.5670. [DOI] [PubMed] [Google Scholar]
- 70.Schutyser E, Richmond A, Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J Leukoc Biol. 2005;78:14–26. doi: 10.1189/jlb.1204712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nibbs RJ, Salcedo TW, Campbell JD, et al. C-C chemokine receptor 3 antagonism by the beta-chemokine macrophage inflammatory protein 4, a property strongly enhanced by an amino-terminal alanine-methionine swap. J Immunol. 2000;164:1488–97. doi: 10.4049/jimmunol.164.3.1488. [DOI] [PubMed] [Google Scholar]
- 72.Gunn MD, Tangemann K, Tam C, Cyster JG, Rosen SD, Williams LT. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc Natl Acad Sci USA. 1998;95:258–63. doi: 10.1073/pnas.95.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim CH, Pelus LM, White JR, Applebaum E, Johanson K, Broxmeyer HE. CK beta-11/macrophage inflammatory protein-3 beta/EBI1-ligand chemokine is an efficacious chemoattractant for T and B cells. J Immunol. 1998;160:2418–24. [PubMed] [Google Scholar]
- 74.Homey B, Alenius H, Muller A, et al. CCL27–CCR10 interactions regulate T cell-mediated skin inflammation. Nat Med. 2002;8:157–65. doi: 10.1038/nm0202-157. [DOI] [PubMed] [Google Scholar]
- 75.Yamashita U, Kuroda E. Regulation of macrophage-derived chemokine (MDC, CCL22) production. Crit Rev Immunol. 2002;22:105–14. [PubMed] [Google Scholar]
- 76.Imai T, Baba M, Nishimura M, Kakizaki M, Takagi S, Yoshie O. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J Biol Chem. 1997;272:15036–42. doi: 10.1074/jbc.272.23.15036. [DOI] [PubMed] [Google Scholar]
- 77.Mizoue LS, Bazan JF, Johnson EC, Handel TM. Solution structure and dynamics of the CX3C chemokine domain of fractalkine and its interaction with an N-terminal fragment of CX3CR1. Biochemistry. 1999;38:1402–14. doi: 10.1021/bi9820614. [DOI] [PubMed] [Google Scholar]
- 78.Bazan JF, Bacon KB, Hardiman G, et al. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–4. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 79.Yang D, Chertov O, Bykovskaia SN, et al. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–8. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 80.Schutyser E, Struyf S, Van Damme J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 2003;14:409–26. doi: 10.1016/s1359-6101(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 81.Kunkel EJ, Campbell JJ, Haraldsen G, et al. Lymphocyte CC chemokine receptor 9 and epithelial thymus-expressed chemokine (TECK) expression distinguish the small intestinal immune compartment: epithelial expression of tissue-specific chemokines as an organizing principle in regional immunity. J Exp Med. 2000;192:761–8. doi: 10.1084/jem.192.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vicari AP, Figueroa DJ, Hedrick JA, et al. TECK: a novel CC chemokine specifically expressed by thymic dendritic cells and potentially involved in T cell development. Immunity. 1997;7:291–301. doi: 10.1016/s1074-7613(00)80531-2. [DOI] [PubMed] [Google Scholar]
- 83.Lazarus NH, Kunkel EJ, Johnston B, Wilson E, Youngman KR, Butcher EC. A common mucosal chemokine (mucosae-associated epithelial chemokine/CCL28) selectively attracts IgA plasmablasts. J Immunol. 2003;170:3799–805. doi: 10.4049/jimmunol.170.7.3799. [DOI] [PubMed] [Google Scholar]
- 84.Pan J, Kunkel EJ, Gosslar U, et al. A novel chemokine ligand for CCR10 and CCR3 expressed by epithelial cells in mucosal tissues. J Immunol. 2000;165:2943–9. doi: 10.4049/jimmunol.165.6.2943. [DOI] [PubMed] [Google Scholar]
- 85.Wang W, Soto H, Oldham ER, et al. Identification of a novel chemokine (CCL28), which binds CCR10 (GPR2) J Biol Chem. 2000;275:22313–23. doi: 10.1074/jbc.M001461200. [DOI] [PubMed] [Google Scholar]
- 86.Yoshida T, Ishikawa I, Ono Y, Imai T, Suzuki R, Yoshie O. An activation-responsive element in single C motif-1/lymphotactin promoter is a site of constitutive and inducible DNA–protein interactions involving nuclear factor of activated T cell. J Immunol. 1999;163:3295–303. [PubMed] [Google Scholar]
- 87.Kelner GS, Kennedy J, Bacon KB, et al. Lymphotactin: a cytokine that represents a new class of chemokine. Science. 1994;266:1395–9. doi: 10.1126/science.7973732. [DOI] [PubMed] [Google Scholar]