Abstract
The carboxy-terminus of the colonic H+,K+-ATPase is required for stable assembly with the β-subunit, translocation to the plasma membrane and efficient function of the transporter. To identify protein-protein interactions involved in the localization and function of HKα2, we selected 84 amino acids in the carboxy-terminus of the α-subunit of mouse colonic H+,K+-ATPase (CT-HKα2) as the bait in a yeast two-hybrid screen of a mouse kidney cDNA library. The longest identified clone was CD63. To characterize interaction of CT-HKα2 with CD63, recombinant CT-HKα2 and CD63 were synthesized in vitro, incubated, and complexes immunoprecipitated. CT-HKα 2 protein (but not CT-HKα1) co-precipitated with CD63, confirming stable assembly of HKα2 with CD63. In HEK-293 transfected with HKα2 plus β1-Na+,K+-ATPase, suppression of CD63 by RNA interference increased cell surface expression of HKα2/NKβ1 and 86Rb+-uptake. These studies demonstrate that CD63 participates in the regulation of the abundance of the HKα2/NKβ1 complex in the cell membrane.
Keywords: CD63, protein assembly, colonic H+, K+-ATPase, cell surface localization,
ABBREVIATIONS: X+, K+-ATPase = family of proteins that includes the gastric H+, K+-ATPase, the colonic X+, K+-ATPase, and the Na+, K+-ATPases, HKα1 = α-subunit of the gastric H+, K+-ATPase, HKα2 = α-subunit of the colonic H+, K+-ATPase, NKα1 = α1-Na+, K+-ATPase, NKα2 = α2-Na+, K+-ATPase, NKα3 = NKα3-Na+, K+-ATPase, NKβ1 = β1-subunit of the Na+, K+-ATPase, NKβ2 = β2-subunit of the Na+, K+-ATPase, NKβ3 = β3-subunit of the Na+, K+-ATPase, βG = β-subunit of the gastric H+, K+-ATPase, PCR = polymerase chain reaction, SDS = sodium dodecyl sulfate, PAGE = polyacrylamide gel electrophoresis, DMSO= dimethyl sulfoxide, PPO = 2, 5-diphenyloxazole, kb = kilo bases, CHAPS = 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate, YPD = yeast peptone dextrose, SD = synthetic dropout
INTRODUCTION
Similarities exist between the Na+,K+-ATPase and the apical colonic H+,K+-ATPase (HKα2). Both pumps are partially sensitive to ouabain and insensitive to Sch-28080 (4, 11, 15), utilize the β1-Na+,K+-ATPase (NKβ1) as the β subunit in complex formation (9, 26), and transport K+ in exchange for Na+ or H+ (12, 14, 19). However, distinct differences in membrane localization are evident; the Na+-pump localizes to the basolateral membrane, whereas the colonic H+,K+-ATPase localizes to the apical membrane (18, 29, 32).
We have previously determined that the 84 amino acid region at the C-terminus of HKα2 is critical for α/β complex translocation to the cell surface. A deletion mutation lacking this region (ΔHKα2) appears poorly protected by NKβ1, and complexes comprised of ΔHKα2 and NKβ1 are retained in the endoplasmic reticulum (ER). Moreover, despite assembly with NKβ1, ΔHKα2 also fails to function, as evidenced by a marked decline in 86Rb+-uptake, Na+-dependent K+-ATPase and cell surface localization (28).
In an attempt to identify additional proteins capable of interacting with and mediating distribution or trafficking of HKα2, we employed the C-terminus of mouse HKα2 as bait using the yeast two-hybrid method to screen a mouse cDNA library. We determined that the tetraspanin protein CD63 associates with the carboxy-terminus of HKα2, but not with the C-terminus of other X+,K+-ATPase α-subunits. The biological relevance of these findings was confirmed by establishing stable assembly of the carboxy-terminus of HKα2 with CD63 in vitro, and by demonstration that selective suppression of CD63 expression by RNA interference increases both cell surface localization of the HKα2/NKβ1 complex and 86Rb+-uptake in HEK-293 cells. Collectively, these findings suggest that CD63 functions as a negative regulator of the colonic H+,K+-ATPase.
MATERIALS AND METHODS
Construct generation
Total RNA was purified from rat kidney, brain, distal colon, or stomach and used to synthesize cDNA using random primers in a reverse transcriptase system (Fisher, Madison, WI). DNA encoding the carboxy-terminus of rat NKα1, NKα2, NKα3, HKα1, HKα2 and mouse HKα2 was subsequently amplified by PCR using primers described in Table 1, and cloned into pGBKT7 containing a cassette for an N-terminal c-myc epitope tag. Similarly, DNA encoding rat NKβ1 from L64 to S304 was amplified by PCR using the sense primer 5′-ACGTCCATGGCACTGAAACCCACGT ACCAGGACCGT-3′ (underlined sequence represents the NcoI site) and antisense primer 5′-ACGTGAATTCTCAGCTCTTAACTT CAATTTTTAC-3′ (underlined sequence represents the EcoRI site), and the resultant product cloned into NcoI/EcoRI-digested pGADT7 plasmid containing an N-terminal HA-epitope cassette. To enable synthesis of m-CD63 in vitro (as described below), DNA encoding m-CD63 in pACT2 (identified in the two-hybrid screen) was excised with SfiI/XhoI and cloned into pGADT7. Generation of siRNA-expressing constructs is described below. Orientation of all cloned inserts was verified by restriction mapping, and sequence verified by sequencing of both strands. All plasmids were amplified using standard protocols (16).
Table 1. Sense and antisense oligonucleotides used to amplify, by PCR, the carboxy-terminus of rat NKα1 (r-NKα1), rat NKα2 (r-NKα2), rat NKα3 (r-NKα3), rat HKα1 (r-HKα1), and rat or mouse HKα2 (r/m-HKα2).
For each sense oligonucleotide, the EcoRI (GAATTC) site used for cloning the insert in pGBKT7 is underlined. For each antisense oligonucleotide, the stop codon is marked with bold characters. The carboxy-terminus of each X+,K+-ATPase family member was defined by amino acid alignment using the PileUp program. The accession number of the DNA sequences used in these experiments are: r-HKα1 = PO9626, r-NKα1 = M14511, r-NKα2 = M14212, r-NKα3 = M14513, r-HKα2 = M90398, m-HKα2 = unpublished observations from our laboratory. The same oligonucleotide pairs were used to amplify the carboxy-terminus of rat and mouse HKα2 (r/m-HKα2.). Abbreviations: R = arginine, N = asparagine.
| Forward | Reverse | From-to (amino acid) | Length (amino acids) | |
|---|---|---|---|---|
| r-NKα1 | 5′-ACGTGAATTCGTCTTCCAGCAGGGAATGAAGAAC-3′ | 5′-ACGTCTCGAGCTAGTAGTAGGTTTCCTTCTCCAC-3′ | R941/Y1024 | 84 |
| r-NKα2 | 5′-ACGTGAATTCGTGTTCCAGCAGGGCATGAAGAAC-3′ | 5′-ACGTCTCGAGTCAGTAGTACGTCTCCTTCTCCAC-3′ | N939/Y1020 | 82 |
| r-NKα3 | 5′-ACGTGAATTCGTCTTCCAGCAGGGCATGAAGAAT-3′ | 5′-ACGTCTCGAGTCAATAGTAGGTCTCTTTCTCCAC-3′ | N932/Y1013 | 82 |
| r-HKα1 | 5′-ACGTGAATTCGCTTTCCAGCAGGGATTCTTCAGG-3′ | 5′-ACGTCTCTCAATAGTAGAGTTCCTGGTCCCACCA-3′ | N961/Y1033 | 73 |
| r/m-Kα2 | 5′-ACGAGGAATTCCATCTTTCAGCAGGGGCTCTTCAG-3′ | 5′-ACGTTCTAGATTCAATAATACATGTTCTTATCCC-3′ | N953/Y1036 | 84 |
Transfection of yeast (S. cerevisiae) AH109 with the carboxy-terminus of mouse HKα2
Yeast AH109 was grown in 300 ml of yeast peptone dextrose (YDP) containing 15 μg/ml kanamycin (Clontech, Palo Alto, CA) to an optical density of 0.4–0.5 at 600 nm, then centrifuged at 1,000xg for 5 min at room temperature. The pellet was rinsed with 30 ml sterile water, centrifuged again, and resuspended in 1.5 ml 10 mM TrisHCl, 1 mM EDTA, 100 mM LiAc pH7.5. One hundred microliters of the yeast suspension were added to 10 μg of m-CT-HKα2-pGBKT7 and 100 μg of carrier herring sperm DNA (Clontech, Palo Alto, CA), then to 600 μl 40% PEG-4000 in 10 mM TrisHCl, 1 mM EDTA, and 100 mM LiAc pH7.5. The mixture was incubated for 30 min at 30°C. After addition of 70 μl of dimethylsulfoxide (DMSO), the yeast was incubated at 42°C for 15 min, then chilled in ice, and centrifuged. The resultant pellet was resuspended in 500 μl 10 mM TrisHCl, 1 mM EDTA pH7.5. The transfected yeast was plated on 2% agar in synthetic dropout (SD) (Fisher, Madison, WI) supplemented with all nutrients except tryptophan. The dishes were incubated for 3–4 days at 30°C.
Screening of a mouse kidney cDNA library in pACT2 vector.
A cDNA library made from total RNA purified from mouse kidney was purchased from Clontech (Palo Alto, CA). For screening, one colony expressing m-CT-HKα2 protein fused to GAL4-BD was amplified in 300 ml SD media in the absence of tryptophan until the optical density reached 0.4–0.5. All subsequent steps were as described above, except that competent yeast (1500 μl) was mixed with 200 μg of mouse kidney cDNA library in pACT2 and 1500 μg of herring sperm carrier DNA. The transformed yeast was plated on 20 Petri dishes (15 cm diameter) containing 2% agar in SD supplemented with all amino acids except tryptophan, leucine, histidine, and adenine. The dishes were incubated at 30°C for 3–4 days, and visible colonies were transferred to 5 ml SD lacking the same amino acids and incubated for 2 days at 30°C. Plasmid DNA was purified as described by Hoffman (22). Briefly, the yeast was centrifuged at 1,000xg for 5 min at room temperature, digested with 100 units of lyticase (Sigma, St. Louis, MO.) for 1 h at 37°C in 100 μl 10 mM TrisHCl, 1 mM EDTA pH8.0. Twenty μl 10% SDS was then added, and the samples were frozen at −70°C. One hundred microliters of breaking buffer (4% Triton X-100, 1% SDS, 200 mM NaCl, 10 mM TrisHCl, and 1 mM EDTA pH=8.0) and 300 mg of glass beads (Sigma, St. Louis, MO) were added to thawed samples and vortexed for 2 min. The proteins were extracted with 200 μl H2O-saturated phenol/chloroform/isoamyl alcohol (24:24:1). The aqueous phase was retained, and the RNA digested with 1 mg/ml RNAse A. The samples were diluted to a final volume of 1 ml, and the DNA precipitated with 400 μl 40% PEG-8000/1.5 M NaCl) and resuspended. The isolated DNA was used to transform XL1blue MR E. coli by electroporation, and the transfected E. coli were plated on Petri dishes containing 100 μg/ml ampicillin. Colonies from these dishes were used to purify and sequence the insert (prey) in the mouse kidney library that interacted with m-CT-HKα2.
Assessment of m-CT-HKα2 and m-CD63 interaction in vitro.
m-CT-HKα2 in pGBKT7 and m-CD63 in pGADT7 plasmids were used to synthesize[35S]-methionine-labeled m-CT-HKα2 and m-CD63 proteins using the TnT T7- coupled reticulocyte lysate system (Promega, Madison, WI), as per manufacturer’s instructions. After synthesis, the proteins were mixed and incubated for 1 h at room temperature. The m-CT-HKα2/m-CD63 complex was immunoprecipitated for 1 h at room temperature by adding 10 μl rabbit anti-hemagglutinin polyclonal antibody (Clontech, Palo Alto, CA). The reaction was diluted with 400 μl of buffer (10 mM TrisHCl, pH=8.0, 150 mM NaCl, 1 mM PMSF, 3 mM benzamidine and 1 μg/ml soybean trypsin inhibitor). Five μl of packed agarose-A (Santa Cruz Biotechnology, Santa Cruz) were added and incubated for an additional 1 h at room temperature with continuous shaking. The resin was washed 4 times with 1 ml of 10 mM TrisHCl, 150 mM NaCl, 1 mM PMSF, 3 mM benzamidine, 1 μg/ml soybean trypsin inhibitor and 1% egg albumin, followed by two additional washes with the same buffer without egg albumin. Proteins were extracted with Laemmli sample buffer (27) and resolved on a 10% SDS-PAGE that had been pre-run for 4 hours at 100 volts to increase the resolution of low molecular proteins. The gel was fixed with 50% methanol, washed extensively with DMSO, submerged in a solution of 20% PPO in DMSO for 45 min, dried, and exposed to film for 3 days at −70°C.
Northern analysis
Northern analysis was performed as described previously by our laboratory (16) using a 32P-labelled probe based on the mouse CD63 sequence identified in the yeast two-hybrid screen.
Specific suppression of CD63 protein synthesis in HEK-293 cells by siRNA
A sense oligonucleotide 5′-tcgagGTTCTTGCTCTACGTCCTCCtagtactgaGGAGGACGTAGA GCAAGAACttttt-3′ containing a XhoI site, a 20-nucleotide sequence (GTTCTTGCTCTACGTC CTCC) corresponding to nucleotides 33 to 52 of the open reading frame of the human CD63 (accession number BT008095), a non-relevant sequence (tagtactga), followed by a reverse-complementary sequence of human CD63 (GGAGGACGTAGAGCAAGAAC) and a poly T (ttttt) tail was synthesized. The reverse oligonucleotide (5′-ctagaAAAAGTTCTTGCTCT ACGTCCTCCTCA GTACTAGGAGGACGTAGAGCAAGAACC-3′) containing the 5′ overhanging end of the XbaI site (ctaga, lower-case characters) was also synthesized. The two oligonucleotides were purified then annealed as described previously (1) and cloned into the plasmid pSuppressorNeo (Imgenex, San Diego, CA). The sequence of the insert was verified by double-stranded DNA sequencing. The construct was linearized with BamHI and used (9 μg per 10 cm dishes) to stably transfect HEK-293 cells following the instructions of the manufacturer. Colonies were selected using 250 μg/ml G418 (Invitrogen, Carlsbad, CA), and screened by immunolocalization of CD63 protein in the intracellular compartments with anti-human CD63 monoclonal antibody (Jackson ImmunoResearch, West Grove, PA), as described below.
Intracellular localization of CD63 in HEK-293 cells
HEK-293 cells were washed twice with PBS, fixed with 3.7% formaldehyde in PBS, washed twice with PBS and incubated in 1% BSA plus 0.1% saponin in PBS for 10 min at room temperature. Anti-human CD63 monoclonal antibody (Jackson ImmunoResearch, West Grove, PA), diluted 1:100, was added to the cells for 30 min at room temperature. Cells were washed three times with PBS, after which rhodamine conjugate goat anti-mouse IgG (1:1000) (Jackson ImmunoResearch, West Grove, PA) was added for 30 min at room temperature. Cells were viewed using a Zeiss Axioplan 2 fluorescence microscope equipped with rhodamine filters and recorded using a Zeiss Axiocam CCD camera. Controls experiments were performed by omitting the primary antibody. Colonies growing in presence of G418 that did not express CD63 proteins were designated the experimental group (CD63-knockdown). Colonies growing in the presence of G418 but expressing levels of CD63 protein similar to non-transfected cells were used as controls.
Subcellular localization of HKα2/NKβ1 complex in control and CD63-knockdown HEK-293 cells
Whole cell lysate protein was fractionated by discontinuous sucrose gradients as described previously (28) to investigate subcellular localization of HKα2/NKβ1 in transfected control and CD63-knockdown HEK-293 cells. The top fraction (lower sucrose concentration) containing the plasma membrane fraction (28, 34) was diluted 10-fold with 10 mM TrisHCl pH 8.0, 1 mM EDTA, 1 mM PMSF, 3 mM benzamidine and 1 μg/ml soybean trypsin inhibitor and concentrated by centrifugation for 30 min at 4°C at 30,000xg. The pellet was resuspended, protein concentration determined and 50 μg of protein were deglycosylated with glycosidase F for 1 h at 37°C in the presence of 1% CHAPS. Proteins were resolved on a 10% SDS-PAGE and transferred to a nitrocellulose membrane. The upper half of the membrane was probed with the anti-HKα2 polyclonal antibody, and the lower half probed with a polyclonal antibody against rat NKβ1. Equal loading of the SDS-PAGE was monitored by staining the nitrocellulose membranes with Ponceau S.
Miscellaneous methods
HEK-293 cells were grown at 37°C in DMEM containing 10% newborn calf serum (20). 86Rb+-uptake was performed at 37°C for 15 min in presence of KCl (1 mM) and 86Rb+ (3-4x106 cpm) (8). Immunoblot analysis was performed as described previously (10), with intensity of bands quantified using the Image Tools program (10).
RESULTS
m-CD63 interacts with the carboxy-terminus of mouse HKα2 (m-CT-HKα2) in a yeast two-hybrid screen.
Yeast AH109 was transfected with pGBKT7 containing an insert encoding the 84 carboxy-terminal amino acids of HKα2. The yeast was plated on agar dishes depleted of tryptophan. Four days later, one colony was amplified and transfected with a mouse kidney cDNA mouse library in pACT2 and plated on 2% agar in SD supplemented with all the nutrients except methionine, tryptophan, histidine, and adenine. Four days later, 24 individual yeast colonies were isolated and expanded in SD liquid supplemented with all amino acids except tryptophan, leucine, histidine, and adenine. Plasmid DNA was purified and used to electroporate XL1blue MR E. coli. The transformed bacteria were plated on 1% agar dishes containing 100 μg/ml ampicillin and used to isolate the cDNA encoding the prey protein that interacted with m-CT-HKα2. Double-stranded DNA sequencing of the longest insert (colony # 3) demonstrated that the open reading frame of GAL4 AD protein continued into the linker “A ATT CGC GGC CGC GTG GAC”, which was added to the 5′-end of the cDNA when the library was synthesized, and continued into the insert. The insert coded for 238 amino acids, followed by a stop codon, and 3 non-coding sequence ending with a 30 base poly A sequence followed by a XhoI site that was added when the library was synthesized. A BLAST search revealed the sequence to encode all but the first five amino acids of mouse CD63 (accession number S43511). The remaining 23 inserts, all shorter in size, had strong cross-reactivity with colony 3 in southern analysis, and were therefore not characterized.
HKα2 is the only X+,K+-ATPase α-subunit that assembles with CD63.
All experiments described above were performed using m-CT-HKα2 and m-CD63. Results were similar when the rat carboxy-terminus of HKα2 was used as bait (not shown). Additional two-hybrid screens testing whether the carboxy-termini of the different α-subunits from other X+,K+-ATPases (NKα1, NKα2, NKα3 and HKα1) interacted with m-CD63 revealed that the interaction between the carboxy-terminus of HKα2 and CD63 is specific and did not extend to any other members of the X+,K+-ATPase family (data not shown).
Interaction between bait and prey is required for AH109 growth in the absence of tryptophan, leucine, histidine, and adenine.
To further test the specificity of the HKα2/CD63 interaction, yeast was transfected with m-CT-HKα2 in pGBKT7 and plated on SD dishes supplemented with all the nutrients or on SD dishes deficient in tryptophan, or on SD dishes deficient in leucine or on SD dishes deficient in tryptophan, leucine, histidine, and adenine. Results presented in Table 2 demonstrate that yeast grew when all nutrients were present or when tryptophan was omitted (lane 1). Yeast transfected with m-CD63 in pACT2 grew when all the nutrients were added or when leucine was omitted (lane 2). Yeast co-transfected with m-CT- in pGBKT7 and m-CD63 in pACT2 grew under all four conditions (lane 3), as expected when bait and prey interact. Yeast co-transfected with m-M7M8 in pGBKT7 and m-CD63 in pACT2 grew in the presence of all the nutrients or when tryptophan or leucine was omitted. However, they did not grow when all four nutrients were omitted (lane 4), demonstrating that CD63 did not interact with the β-subunit binding sequence of HKα2. As a positive control, yeast co-transfected with m-M7M8 in pGBKT7 and the extracellular domain of rat NKβ1 in pGADT7 grew under all four conditions (lane 5), demonstrating that a region of HKα2 including transmembrane 7 and 8 interacted with the carboxy-terminus of NKβ1, as previously demonstrated by Colonna et al. (13).
Table 2. Summary of protein-protein interactions suggested by yeast two-hybrid analysis.
The experiment demonstrates that co-expression of CD63 and the carboxy-terminus of HKα2 is required to support the growth of yeast in the absence of tryptophan, leucine, histidine, and adenine in the medium. The experiment also demonstrates that the extracellular domain of NKβ1 interacts with the extracellular sequence of HKα2 that extends between transmembrane transmembrane region 7 (M7) and 8 (M8), as predicted by the model of Colonna (13). r-extrNKβ1-pGADT7 represents the extracellular domain of rat NKβ1 that was cloned in the plasmid pGADT7. r-intNKβ1-pGADT7 represents that the intracellular plus transmembrane sequence of rat NKβ1 that was cloned in the plasmid pGADT7.
| Lane | Plasmid | +W,+L,+H,+A | −W,+L,+H,+A | +W, −L, +H, +A | −W, −L, −H,− A |
|---|---|---|---|---|---|
| 1 | m-CT-HKα2-pGBKT7 | + | + | – | – |
| 2 | m-CD63-pACT2 | + | – | + | – |
| 3 | m-CT-HKα2-pGBKT7/m-CD63-pACT2 | + | + | + | + |
| 4 | m-M7M8-pGBKT7/m-CD63-pACT2 | + | + | + | – |
| 5 | m-M7M8-pGBKT7/r-extNKβ1-pGADT7 | + | + | + | + |
| 6 | m-CD63-pACT2/r-extNKβ1-pGADT7 | + | + | + | - |
| 7 | m-CD63-pACT2/r-intNKβ1-pGADT7 | + | + | + | - |
m-CT-HKα2 and m-CD63 form a complex in vitro.
m-CD63 from the plasmid pACT2 was cloned into plasmid pGADT7 as described under Materials and Methods to enable synthesis of both proteins using the rabbit reticulocyte system in the presence of [35S]-methionine. In vitro translated m-CT-HKα2 and m-CD63 proteins were incubated together in vitro and m-CD63 was subsequently immunoprecipitated with anti-hemagglutinin antibody, and immunoprecipiated complexes analyzed by autoradiography. Figure 2 shows the results of a representative experiment. In the absence of m-CD63, immunoprecipitation with anti- anti-hemagglutinin antibody co-precipitated a small amount of m-CT-HKα2, reflecting the non-specific binding of the recombinant m-CT-HKα2 to protein A/G PLUS agarose bits (panel A, lane 1). In the absence of m-CT-HKα2 , immunoprecipitation of m-CD63 was observed (upper panel) while m-CT-HKα2 was not (panel A, lane 2). For incubations including both m-CT-HKα2 and m-CD63 , m-CT-HKα2 co-precipitated with m-CD63 (panel A, lanes 3) such that the intensity of the band representing m-CT-HKα2 was 3-5-fold of that observed in the control (lane 1). This finding demonstrates the capacity of m-CT-HKα2 and m-CD63 to form a complex in vitro.
Figure 2. CD63 interacts with the carboxy-terminus of HKα2.
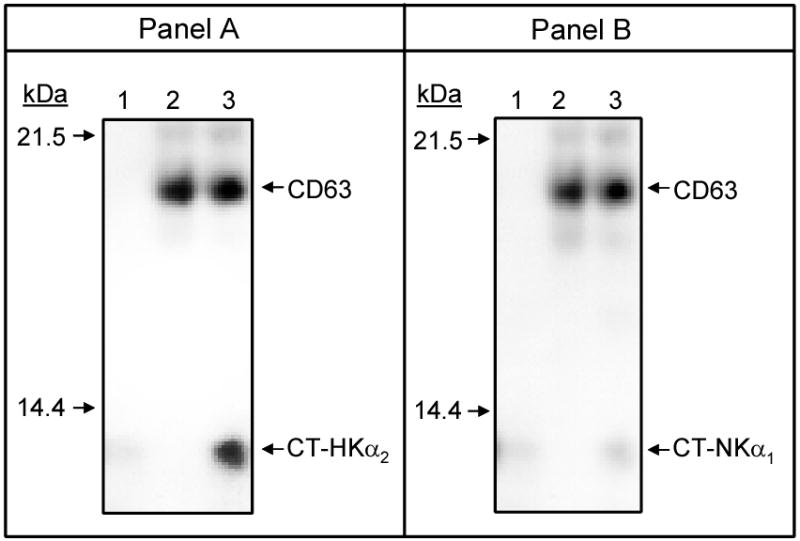
HA-tagged m-CD63, m-CT-HKα2, and r-CT-NKα1 were synthesized and incubated, and immune complexes precipitated using an anti-hemagglutinin polyclonal antibody, as described in Materials and Methods. Panel A: lane 1, m-CT-HKα2 was incubated alone; lane 2, m-CD63 was incubated alone; lane 3, m-CT-HKα2 was incubated with m-CD63. Panel B: lane 1, r-CT-NKα1 was incubated alone; lane 2, m-CD63 was incubated alone; lane 3, r-CT-NKα1 was incubated with m-CD63. Abbreviations used in all figures are the same as in tables 1 and 2 or as indicated in the list of abbreviations. The molecular weights of the different proteins are indicated in kDa. The experiment was repeated three times with similar results.
Additional experiments were performed to assess the capacity of the carboxy-terminus of other α-subunits of the X+,K+-ATPase to associate with m-CD63 in vitro. Results depicted in Panel B of Figure 2 demonstrate that unlike the carboxy-terminus of HKα2, the carboxy-terminus of NKα1 protein fails to complex with m-CD63. Co-precipitation of r-CT-NKα1 with m-CD63 (Panel B, lane 3) was not significantly greater than that observed for immunoprecipates obtained in the absence of m-CD63 (Panel B, lane 1). These findings are consistent with results from our yeast two-hybrid experiments described above and further suggest that CD63 interacts specifically with the carboxy-terminus of HKα2.
CD63 mRNA is expressed in distal colon, renal medulla and HEK-293 cells.
If CD63 functions as a chaperone for HKα2, CD63 should be expressed in mIMCD3 (mouse Inner Medullary Collecting Duct) and mOMCD (mouse Outer Medullary Collecting Duct) cells in culture and in distal colon (31). Total RNA was purified from mIMCD3, mOMCD, distal colon and HEK-293 cells, resolved on agarose gel, transferred to a nylon membrane and probed with the m-CD63 identified using the yeast two-hybrid system. Results displayed in Figure 3 demonstrate that CD63 mRNA is highly expressed in mIMCD, mOMCD and HEK-293 cells in culture and distal colon. CD63 mRNA displays the expected mobility corresponding to 1.2 kb in the four samples tested in our studies. However, one larger band was detected in mRNA isolated from mIMCD-3 or mOMCD. This larger band could represent an alternative splice variant that is expressed at low levels, a CD63 pre-mRNA species, or a cross-reaction with mRNA of other homologous tetraspanin proteins.
Figure 3. CD63 mRNA is expressed in distal colon, renal medulla, and HEK-293 cells.
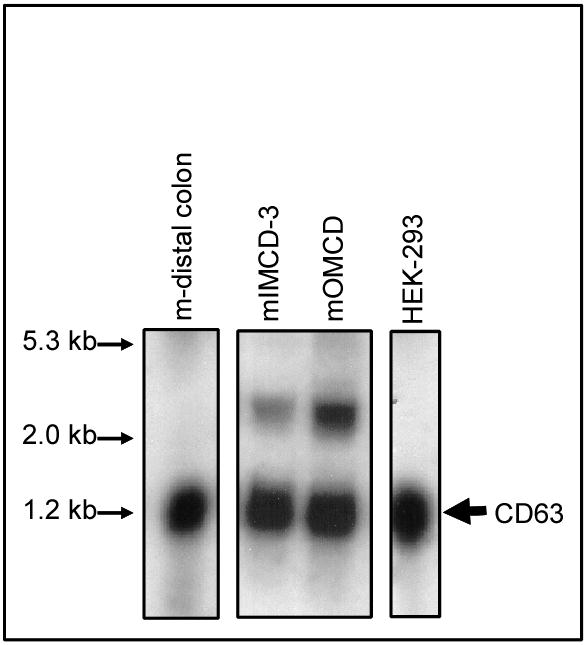
Total RNA (10 μg) from mouse distal colon, mIMCD3, and mOMCD and HEK-293 cells were probed with m-CD63 cDNA. The film was exposed overnight at −70°C. The molecular weight of the mRNA is expressed in kilobases (kb). The experiment was repeated three times with similar results.
Endogenous CD63 protein expressed in HEK-293 cells is abolished by siRNA
Cellular localization of CD63 protein was determined by immunocytochemistry using a monoclonal antibody against human CD63 (Figure 4). CD63 is observed at the cell surface (panel A). However, large quantities of CD63 are also detected in intracellular compartments. Transfection of siRNA targeting CD63 resulted in successful suppression of CD63 expression in numerous cloned lines established by selection with G418 (panel B). The bottom left panel of Figure 4 demonstrates that CD63 was not detected when the primary antibody was omitted in the control cells, and the bottom right panel demonstrates that CD63 was not detected in CD63-knockdown HEK-293.
Figure 4. CD63 protein expression is abolished in HEK-293 cells using siRNA.
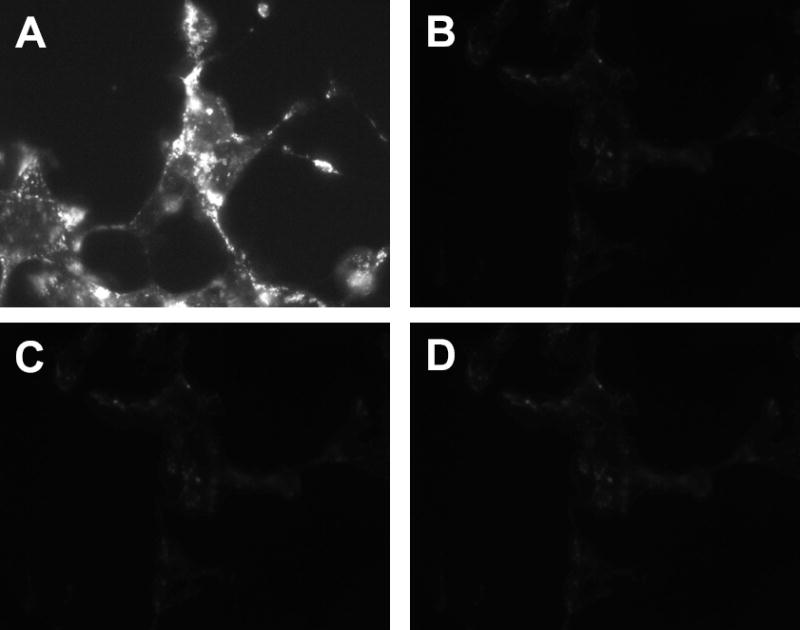
Panel A: Immunocytochemical localization of endogenous CD63 protein in a G418 selected HEK-293 line that expresses CD63. The experiment was performed at room temperature in presence of saponin. Panel B: CD63 expression in a clonal line of HEK-293 transfected with siRNA targeting CD63 then selected with G418, CD63-knockdown). Panel C: The same as in panel A, but with primary antibody omitted. Panel D: The same as in panel B, but with primary antibody omitted. The anti-CD63 monoclonal antibody (primary antibody) was diluted 1:100. The immunocytochemical localization of CD63 shown in panels A to D was repeated 10 times with similar results.
HKα2 protein localization, but not expression, is altered in CD63-specific knockdown HEK-293 cells.
HEK-293 cells (control or CD63-knockdown) were transiently transfected with HKα2 plus NKβ1 in pcDNA3.1(+)-Neo. Forty-eight hours later, the cells were scraped, washed with PBS, and lysed as described in Materials and Methods. One hundred micrograms of protein were deglycosylated with glycosidase F as described previously (3, 9), separated on a 10% SDS-PAGE and transferred to a nitrocellulose membrane. The top half of the membrane was probed with anti-HKα2 antibody (10) and the bottom half probed with anti-NKβ1 antibody. Immunoblot analysis depicted in Figure 5 demonstrates that HKα2, (top panel), as well as of NKβ1 (bottom panel) were expressed to similar levels in control and CD63-knockdown cells.
Figure 5. Expression of HKα2 and NKβ1 protein is not altered by knocking down expression of CD63 with siRNA.
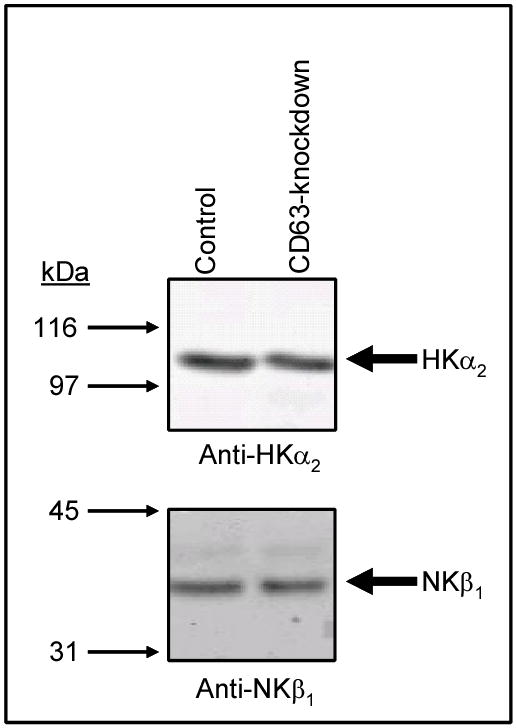
Control HEK-293 cells or CD63-knockdown HEK-293 cells were co-transfected with HKα2 plus NKβ1. Forty-eight hours later, the cells were lysed, and 100 μg of protein deglycosylated with glycosidase F and resolved on a 10% SDS-PAGE, the proteins were transferred to a nitrocellulose membrane and the stained with Ponceau S to verify equal protein loading of the lanes. The immunoblot analyses were performed using an anti-HKα2 (dilution 1:1000) (10) or anti NKβ1 antibody (dilution 1:1000) (29). For these experiments, controls were HEK-293 cells transfected with CD63 siRNA using the plasmid pSuppressorNeo and selected with G418, but expression of CD63 protein was not different than that determined in wild HEK-293 cells. The experiment was repeated 4 times with similar results and the difference between control and CD63-knockdown is not statistically significant.
Suppression of CD63 increases plasma membrane localization of HKα2/NKβ1 but not NKα1/NKβ1
Subcellular localization of HKα2/NKβ1 in controls and CD63-knockdown HEK-293 cells was defined using a discontinuous sucrose gradient as described by Tamarappoo et al. (34) and our laboratory (28). The results of a representative experiment are displayed in Figure 6. Panel A demonstrates that HKα2 migrates to the cell surface (low sucrose concentration, represented by fractions 1–2) or migrates to the bottom of the tube (fraction 6) when control HEK-293 cells were co-transfected with HKα2/NKβ1. Panel B demonstrates that when CD63 expression was suppressed selectively, a large proportion of HKα2 migrates to the plasma membrane, while only a minor proportion of the protein migrates to the high sucrose concentration. Panel C demonstrates a large proportion of NKβ1 accumulates in fraction 3 and some in fraction 2. The proportion of NKβ1 accumulated in fraction 2 increases when the transfection was performed using CD63-knockdown cells (Panel D). Panels D and F demonstrate that the intracellular distribution of NKα1 is independent of the presence or absence of CD63 expression in HEK-293 cells. Immunoblots performed with anti-calnexin demonstrated that the ER migrated to the high sucrose concentration (fraction 6) in all the conditions (results not shown).
Figure 6. Plasma membrane expression of HKα2 increases in CD63-knockdown HEK-293 cells.
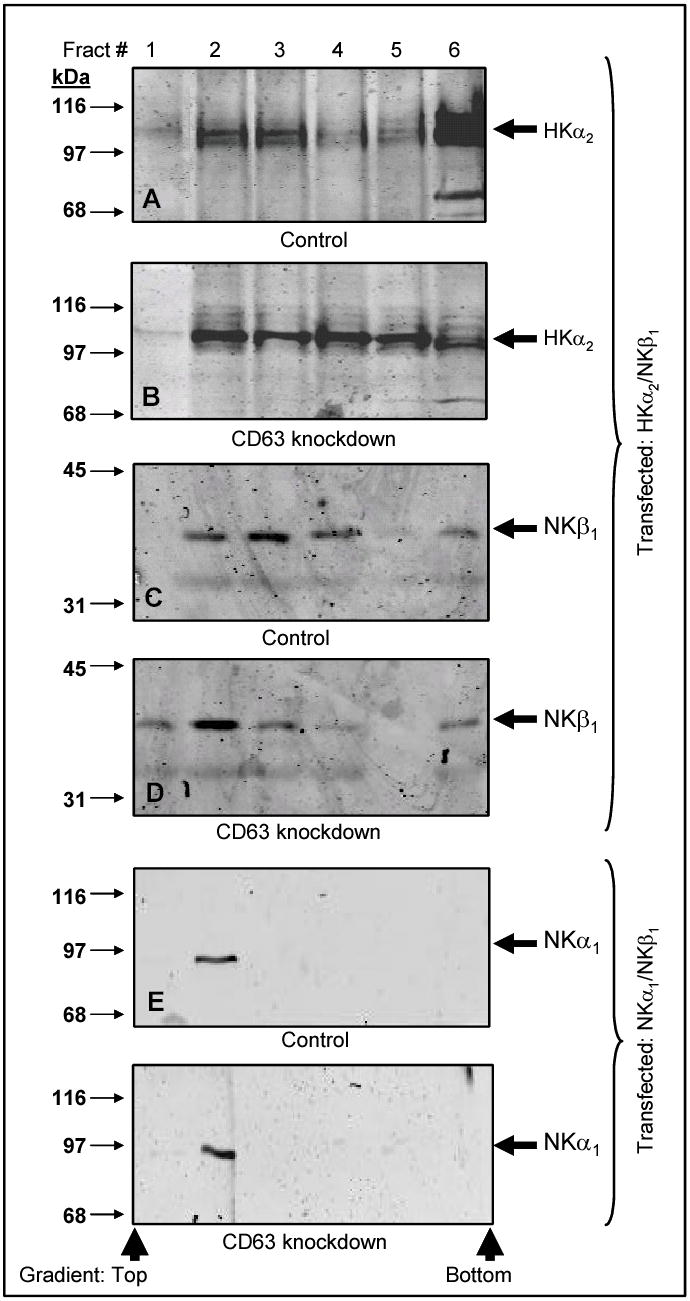
Control HEK-293 cells were co-transfected with HKα2 plus NKβ1. Forty eight hours later, the cells were lysed as described in Materials and Methods. The membranes were fractionated using a discontinuous sucrose gradient (28, 34). The proteins of the different fractions were processed as described in Materials and Methods and probed with anti-HKα2 antibody (panel A) or anti-NKα1 antibody (panel C). The results demonstrated that HKα2 and NKβ1 migrate to the plasma membrane (fractions 1 and 2) or remain in the heavy sucrose fraction (fraction 6). Panels B and D: The same as in panels A and C but the experiment was performed in CD63-knockdown HEK-293 cells. Panels E and F demonstrate that the presence or absence of CD63 does not alter the pattern of migration of NKα1 in the sucrose gradient when the cells were co-transfected with NKα1/NKβ1. Immunoblot analysis performed with anti-calnexin (antibody commercially available from Santa Cruz Biotechnology, Santa Cruz, CA) demonstrated that the ER components accumulated in fraction 6. The experiment was performed three times with similar results. The quantity of HKα2 and NKβ1 that accumulates in the top of the gradient is statistically greater in CD63-knockdown HEK-293 cells vs. the controls (p<0.05).
Suppression of CD63 increases 86Rb+-uptake by HKα2/NKβ1 but not by NKα1/NKβ1
To determine whether the effect of reduced CD63 protein expression on HKα2/NKβ1 plasma membrane localization was associated with alterations in complex function, control and CD63-knockdown HEK-293 cells were co-transfected with HKα2/NKβ1, NKα1/NKβ1 or pcDNA/NKβ1 and assays of 86Rb+-uptake were performed. Results displayed in Figure 7 demonstrate that 86Rb+-uptake mediated by co-expressed HKα2 and NKβ1 was significantly increased by suppression of CD63. Conversely, 86Rb+-uptake, sensitive to 2 mM ouabain, in cells co-expressing NKα1 and NKβ1 was not significantly different between control and CD63-knockdown lines. In addition, suppression of CD63 protein expression did not alter 86Rb+-uptake sensitive to low concentrations (10 μM) of ouabain, supporting our view that CD63 does not interact with NKα1. Collectively, these data suggest that the increase in HKα2/NKβ1 expression at the plasma membrane conferred by a reduction in CD63 expression translates into increased transporter activity.
Figure 7. 86Rb+-uptake by HKα2/NKβ1 complex but not by NKα1/NKβ1 complex is increased in CD63-knockdown cells.
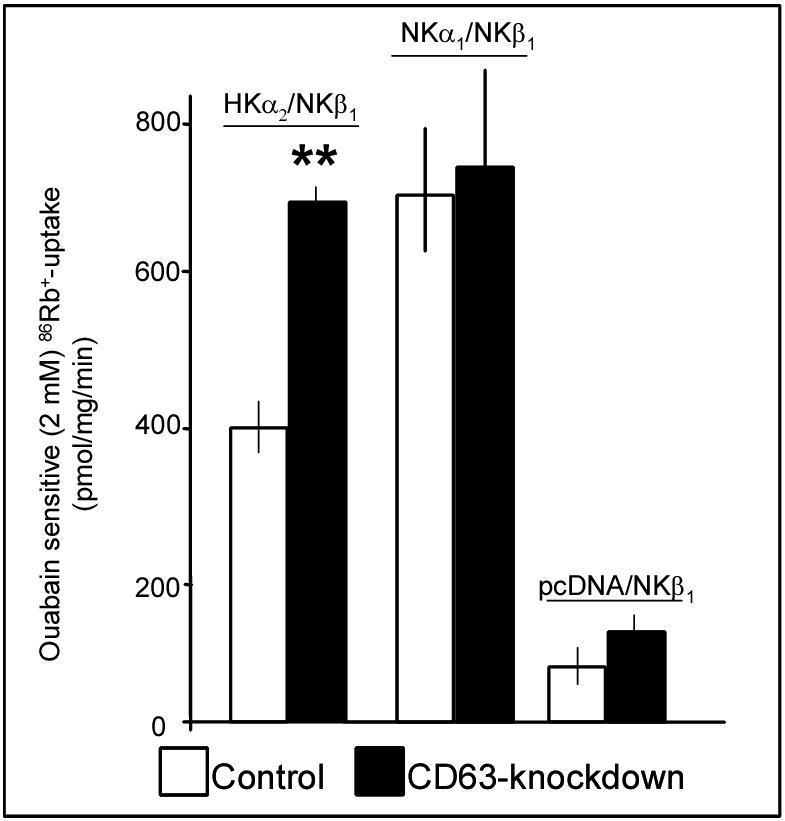
Control HEK-293 cells (open bars) or CD63-knockdown HEK-293 cells (solid bars) were co-transfected with HKα2 plus NKβ1, NKα1 plus NKβ1, or pcDNA vector plus NKβ1. Ouabain (10 μ M) was used to block endogenous Na+,K+-ATPase of HEK-293 cells and 2 mM ouabain was used to block the transport activity of the transfected rat HKα2 or rat NKα1. Results (mean ± S.E.M.) represent the difference in 86Rb+-uptake when the experiments were performed with 10 μM ouabain vs. 2 mM ouabain. ** = 86Rb+-uptake in control HEK-293 cell is statistically different (P<0.01) from that in the CD63-knockdown cells for HKα2/NKβ1 -transfected cells. No statistical difference was observed between control and CD63-knockdown cells when the cells were co-transfected with NKα1/NKβ1 or pcDNA plus NKβ1. The experiment was repeated three times with similar results.
DISCUSSION
Our results demonstrate that CD63 interacts in a specific manner with the carboxy-terminus of HKα2. Association between CD63 and HKα2 was indicated by four independent lines of investigation: 1) the yeast two-hybrid system (Table 2); 2) co-immunoprecipitation of proteins synthesized in vitro using rabbit reticulocyte lysate (figure 2); 3) immunoblot analysis in membrane fractions to localize the HKα2/NKβ1 complex in HEK 293 cells, when expression of CD63 was suppressed selectively by RNA interference (Figure 6); and 4) 86Rb+(K+)-uptake in control and CD63-knockdown HEK-293 transfectants (Figure 7).
CD63, a member of the tetraspanin superfamily of proteins, contains four hydrophobic transmembrane sequences, a large extracellular region of 95 amino acids between transmembrane segments 3 and 4, and is heavily glycosylated. This protein is expressed in numerous tissues and cells in culture (24, 30). In the present study, we show that CD63 is expressed in established cell lines of renal medullary origin, known to express HKα2 (31). In endothelial cells, CD63 has been shown to traffic from the plasma membrane to late endosomes, then to Weibel-Palade bodies to recycle to the plasma membrane (25, 35). CD63 has also been proposed as a molecular modulator of transporter function (5, 6, 36). Therefore, one implication of the CD63/HKα2 interaction revealed by our study is that CD63 may participate importantly in the internalization of HKα2 from the apical membrane. Nevertheless, a complete understanding of the participation of CD63 in the intracellular trafficking of HKα2 will require future studies.
Several recent studies have suggested that interacting proteins may play a key role in the migration of ion transporters to or from the plasma membrane. Staub et al. (33) demonstrated that Nedd4 is required for internalization and proteasomal degradation of the amiloride-sensitive epithelial Na+-channel (ENaC). Hebert (21) demonstrated that Barttin was required for migration of ClC-Ka and ClC-Kb channels to the plasma membrane in the thick ascending limb and in marginal cells of the inner ear. NHE-3 requires glycophorin A to migrate successfully to the apical membrane of the β-intercalated cell in the renal medulla (2, 23). AE1 interacts with kanadaptin (7). Finally, mutations in these interacting proteins can manifest abnormalities in transport function. Therefore, the growing appreciation of such protein-protein interactions suggests a widespread phenomenon in transport physiology.
Our studies are consistent with the possibility that CD63, by interacting with the carboxy-terminus of HKα2, facilitates internalization of the HKα2/NKβ1 complex from the cell surface. Specifically, when CD63 expression is inhibited by siRNA in transfected cells, the accumulation of HKα2/NKβ1 protein at the cell surface and 86Rb+-uptake are both increased. A somewhat analogous mechanism of interaction has been advanced by the studies of Duffield et al. (17). These investigators demonstrated that CD63 association with the β-subunit of the gastric H+,K+-ATPase (HKβG) facilitated internalization of the gastric H+,K+-ATPase heterodimer. In the present study, however, we demonstrate that CD63 does not interact with the carboxy terminus of HKα1. The biological implications of CD63 interacting with NKβG or HKα2 is not yet fully appreciated.
Our studies demonstrate that the interaction of CD63 with the carboxy-terminus of HKα2 regulates membrane expression of CD63 and transport function as monitored by 86Rb+-uptake. Because all the members of the tetraspanin protein share a large number of common properties (30), it is logical to speculate that other tetraspanin proteins may interact with HKα2 or other α-subunits of the X+,K+-ATPase family. It is also possible that HKα2 affects the function of CD63. If CD63 is regulated by HKα2, such regulation may occur in the distal colon where both HKα2 (10) and CD63 are expressed abundantly (see Figure 3). In the renal medulla, HKα2 is expressed at low levels in animals with a normal plasma potassium concentration. In contrast, HKα2 is upregulated robustly by chronic potassium depletion (10). Therefore, if HKα2 regulates the activity of CD63 in the renal medulla, it seems likely that this phenomenon would be observed during chronic hypokalemia.
In conclusion, our results are in agreement and extend recent findings indicating that CD63 participates in the internalization of certain members of the H+,K+-ATPase family. We have found that CD63 specifically interacts with the C-terminus of HKα2 , but not with other members of the X+,K+-ATPase family of membrane transport proteins. This interaction serves to reduce plasma membrane expression of the HKα2/NKβ1 complex, suggesting a mechanism by which CD63 functions as a negative modulator of H+,K+-ATPase.
Figure 1. Predicted topology of HKα2.
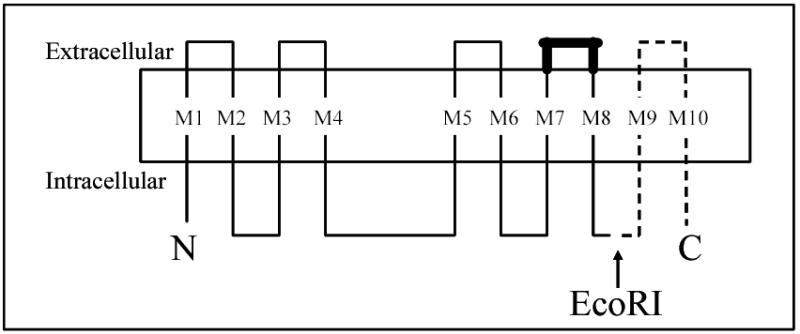
The carboxy-terminus of HKα2 used to screen the mouse kidney cDNA library is represented by a dashed line and extends from the EcoRI site of HKα2 to the stop codon (84 carboxy-terminus amino acids). The bold line between transmembrane M7 and M8 of HKα2 represents the binding site to the β-subunit. N = amino-terminus, C = carboxy-terminus, M1-M10 = predicted transmembrane regions.
Acknowledgments
This work was supported, in part, by a National Institutes of Health (National Institute of Diabetes, Digestive, and Kidney Diseases) Grant, R01 DK-30603 (T.D.B.). The authors thank Dr. Mark C. Willingham (Wake Forest University School of Medicine) for valued suggestions on the microscopy and immunolocalization experiments using anti-CD63 monoclonal antibody and Dr. Raymond Penn (Wake Forest University School of Medicine) for advice during the preparation of this manuscript.
References
- 1.Abramowitz J, Mattera R, Liao CF, Olate J, Perez-Ripoll E, Birnbaumer L, Codina J. Screening of cDNA libraries with oligonucleotides as applied to signal transducing G proteins, receptors and effectors. J Recept Res. 1988;8:561–588. doi: 10.3109/10799898809049012. [DOI] [PubMed] [Google Scholar]
- 2.Alper SL. Genetic diseases of acid-base transporters. Annu Rev Physiol. 2002;64:899–923. doi: 10.1146/annurev.physiol.64.092801.141759. [DOI] [PubMed] [Google Scholar]
- 3.Arystarkhova E, Sweadner KJ. Tissue-specific expression of the Na+,K+-ATPase β3-subunit. The presence of β3 in lung and liver addresses the problem of the missing subunit. J Biol Chem. 1997;272:22405–22408. doi: 10.1074/jbc.272.36.22405. [DOI] [PubMed] [Google Scholar]
- 4.Asano S, Hoshina S, Nakaie Y, Watanabe T, Sato M, Suzuki Y, Takeguchi N. Functional expression of putative H+,K+-ATPase from guinea pig distal colon. Am J Physiol. 1998;275:C669–674. doi: 10.1152/ajpcell.1998.275.3.C669. [DOI] [PubMed] [Google Scholar]
- 5.Berditchevski F, Gilbert E, Griffiths MR, Fitter S, Ashman L, Jenner SJ. Analysis of the CD151-α3β1 integrin and CD151-tetraspanin interactions by mutagenesis. J Biol Chem. 2001;276:41165–41174. doi: 10.1074/jbc.M104041200. [DOI] [PubMed] [Google Scholar]
- 6.Berditchevski F, Tolias KF, Wong K, Carpenter CL, Hemler ME. A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J Biol Chem. 1997;272:2595–2598. doi: 10.1074/jbc.272.5.2595. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Vijayakumar S, Li X, Al-Awqati Q. Kanadaptin is a protein that interacts with the kidney but not the erythroid form of band 3. J Biol Chem. 1998;273:1038–1043. doi: 10.1074/jbc.273.2.1038. [DOI] [PubMed] [Google Scholar]
- 8.Codina J, Cardwell J, Gitomer JJ, Cui Y, Kone BC, Dubose TD., Jr Sch-28080 depletes intracellular ATP selectively in mIMCD-3 cells. Am J Physiol Cell Physiol. 2000;279:C1319–1326. doi: 10.1152/ajpcell.2000.279.5.C1319. [DOI] [PubMed] [Google Scholar]
- 9.Codina J, Delmas Mata JT, DuBose TD., Jr The α-subunit of the colonic H+,K+-ATPase assembles with β1-Na+,K+-ATPase in kidney and distal colon. J Biol Chem. 1998;273:7894–7899. doi: 10.1074/jbc.273.14.7894. [DOI] [PubMed] [Google Scholar]
- 10.Codina J, Delmas-Mata JT, DuBose TD., Jr Expression of HKα2 protein is increased selectively in renal medulla by chronic hypokalemia. Am J Physiol. 1998;275:F433–440. doi: 10.1152/ajprenal.1998.275.3.F433. [DOI] [PubMed] [Google Scholar]
- 11.Codina J, Kone BC, Delmas Mata JT, DuBose TD., Jr Functional expression of the colonic H+,K+-ATPase α-subunit. Pharmacologic properties and assembly with X+, K+-ATPase β-subunits. J Biol Chem. 1996;271:29759–29763. doi: 10.1074/jbc.271.47.29759. [DOI] [PubMed] [Google Scholar]
- 12.Codina J, Pressley TA, DuBose TD., Jr The Colonic H+,K+-ATPase functions as a Na+-dependent K+(NH4+)-ATPase in apical membranes from rat distal colon. J Biol Chem. 1999;274:19693–19698. doi: 10.1074/jbc.274.28.19693. [DOI] [PubMed] [Google Scholar]
- 13.Colonna TE, Huynh L, Fambrough DM. Subunit interactions in the Na+,K+-ATPase explored with the yeast two-hybrid system. J Biol Chem. 1997;272:12366–12372. doi: 10.1074/jbc.272.19.12366. [DOI] [PubMed] [Google Scholar]
- 14.Cougnon M, Bouyer P, Planelles G, Jaisser F. Does the colonic H+,K+-ATPase also act as an Na+,K+-ATPase? Proc Natl Acad Sci U S A. 1998;95:6516–6520. doi: 10.1073/pnas.95.11.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cougnon M, Planelles G, Crowson MS, Shull GE, Rossier BC, Jaisser F. The rat distal colon P-ATPase α-subunit encodes a ouabain-sensitive H+,K+-ATPase. J Biol Chem. 1996;271:7277–7280. doi: 10.1074/jbc.271.13.7277. [DOI] [PubMed] [Google Scholar]
- 16.DuBose TD, Jr, Codina J, Burges A, Pressley TA. Regulation of H+,K+-ATPase expression in kidney. Am J Physiol. 1995;269:F500–507. doi: 10.1152/ajprenal.1995.269.4.F500. [DOI] [PubMed] [Google Scholar]
- 17.Duffield A, Kamsteeg EJ, Brown AN, Pagel P, and Caplan MJ. The tetraspanin CD63 enhances the internalization of the H+,K+-ATPase β-subunit. Proc Natl Acad Sci U S A, 2003. [DOI] [PMC free article] [PubMed]
- 18.Gallardo P, Cid LP, Vio CP, Sepulveda FV. Aquaporin-2, a regulated water channel, is expressed in apical membranes of rat distal colon epithelium. Am J Physiol Gastrointest Liver Physiol. 2001;281:G856–863. doi: 10.1152/ajpgi.2001.281.3.G856. [DOI] [PubMed] [Google Scholar]
- 19.Grishin AV, Caplan MJ. ATP1AL1, a member of the non-gastric H+,K+-ATPase family, functions as a sodium pump. J Biol Chem. 1998;273:27772–27778. doi: 10.1074/jbc.273.43.27772. [DOI] [PubMed] [Google Scholar]
- 20.Guntupalli J, Onuigbo M, Wall S, Alpern RJ, DuBose TD., Jr Adaptation to low-K+ media increases H+,K+-ATPase but not H+- ATPase-mediated pHi recovery in OMCD1 cells. Am J Physiol. 1997;273:C558–571. doi: 10.1152/ajpcell.1997.273.2.C558. [DOI] [PubMed] [Google Scholar]
- 21.Hebert SC. Bartter syndrome. Curr Opin Nephrol Hypertens. 2003;12:527–532. doi: 10.1097/00041552-200309000-00008. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman CS. Current Protocols in Molecular Biology. John Wiley and Sons, Inc: 13.11.11-13-11.14, 1997.
- 23.Karet FE, Gainza FJ, Gyory AZ, Unwin RJ, Wrong O, Tanner MJ, Nayir A, Alpay H, Santos F, Hulton SA, Bakkaloglu A, Ozen S, Cunningham MJ, di Pietro A, Walker WG, Lifton RP. Mutations in the chloride-bicarbonate exchanger gene AE1 cause autosomal dominant but not autosomal recessive distal renal tubular acidosis. Proc Natl Acad Sci U S A. 1998;95:6337–6342. doi: 10.1073/pnas.95.11.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennel SJ, Lankford PK, Foote LJ, Davis IA. Monoclonal antibody to rat CD63 detects different molecular forms in rat tissue. Hybridoma. 1998;17:509–515. doi: 10.1089/hyb.1998.17.509. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi T, Vischer UM, Rosnoblet C, Lebrand C, Lindsay M, Parton RG, Kruithof EK, Gruenberg J. The tetraspanin CD63/lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol Biol Cell. 2000;11:1829–1843. doi: 10.1091/mbc.11.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraut JA, Hiura J, Shin JM, Smolka A, Sachs G, Scott D. The Na+,K+-ATPase β1-subunit is associated with the HKα2 protein in the rat kidney. Kidney Int. 1998;53:958–962. doi: 10.1111/j.1523-1755.1998.00841.x. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Codina J, Petroske E, Werle MJ, DuBose TD, Jr . The carboxy-terminus of the colonic H+,K+-ATPase α-subunit is required for stable β-subunit assembly and function. Kidney Int. 2004;65:1301–1310. doi: 10.1111/j.1523-1755.2004.00507.x. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Codina J, Petroske E, Werle MJ, Willingham MC, DuBose TD., Jr The effect of β-subunit assembly on function and localization of the colonic H+,K+-ATPase α-subunit. Kidney Int. 2004;66:1068–1075. doi: 10.1111/j.1523-1755.2004.00856.x. [DOI] [PubMed] [Google Scholar]
- 30.Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: molecular facilitators. Faseb J. 1997;11:428–442. [PubMed] [Google Scholar]
- 31.Ono S, Guntupalli J, DuBose TD., Jr Role of H+,K+-ATPase in pHi regulation in inner medullary collecting duct cells in culture. Am J Physiol. 1996;270:F852–861. doi: 10.1152/ajprenal.1996.270.5.F852. [DOI] [PubMed] [Google Scholar]
- 32.Sangan P, Rajendran VM, Mann AS, Kashgarian M, Binder HJ. Regulation of colonic H+,K+-ATPase in large intestine and kidney by dietary Na+-depletion and dietary K+-depletion. Am J Physiol. 1997;272:C685–696. doi: 10.1152/ajpcell.1997.272.2.C685. [DOI] [PubMed] [Google Scholar]
- 33.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. Embo J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 34.Tamarappoo BK, Verkman AS. Defective aquaporin-2 trafficking in nephrogenic diabetes insipidus and correction by chemical chaperones. J Clin Invest. 1998;101:2257–2267. doi: 10.1172/JCI2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vischer UM, Wagner DD. CD63 is a component of Weibel-Palade bodies of human endothelial cells. Blood. 1993;82:1184–1191. [PubMed] [Google Scholar]
- 36.Yauch RL, Hemler ME. Specific interactions among transmembrane 4 superfamil (TM4SF) proteins and phosphoinositide 4-kinase. Biochem J 351 Pt. 2000;3:629–637. [PMC free article] [PubMed] [Google Scholar]


