Abstract
The bovine form of the autosomal recessive neurodegenerative disease spinal muscular atrophy (SMA) shows striking similarity to the human form of the disease. It has, however, been mapped to a genomic region not harboring the bovine orthologue of the SMN gene, mutation of which causes human SMA. After refinement of the mapping results we analyzed positional and functional candidate genes. One of three candidate genes, FVT1, encoding 3-ketodihydrosphingosine reductase, which catalyzes a crucial step in the glycosphingolipid metabolism, showed a G-to-A missense mutation that changes Ala-175 to Thr. The identified mutation is limited to SMA-affected animals and carriers and always appears in context of the founder haplotype. The Ala variant found in healthy animals showed the expected 3-ketodihydrosphingosine reductase activity in an in vitro enzyme assay. Importantly, the Thr variant found in SMA animals showed no detectable activity. Surprisingly, in an in vivo assay the mutated gene complements the growth defect of a homologous yeast knockout strain as well as the healthy variant. This finding explains the viability of affected newborn calves and the later neuron-specific onset of the disease, which might be due to the high sensitivity of these neurons to changes in housekeeping functions. Taken together, the described mutation in FVT1 is a strong candidate for causality of SMA in cattle. This result provides an animal model for understanding the underlying mechanisms of the development of SMA and will allow efficient selection against the disease in cattle.
Keywords: cattle, fine-mapping, genetics, motor neuron, neurodegenerative disease
Spinal muscular atrophy (SMA) is a recessive neurodegenerative disease that is characterized by severe loss of motor neurons. Degeneration and loss of motor neurons cause progressive weakness and neurogenic muscular atrophy. The neuropathologic findings are largely identical between human and bovine SMA. They include motor neuron swelling, chromatolysis and loss of neurons in the ventral horn, axonal swelling in the spinal cord, and accumulation of neurofilaments in affected neurons (1). However, the single structural alterations are not specific for SMA but are shared with other unrelated disorders of the nervous system, so that they might be general reactions to various damaging events. Despite the remarkable similarity in the neuropathologic appearance of bovine SMA and the human forms of SMA (1) and also partially of amyotrophic lateral sclerosis (2, 3), genetic mapping (4, 5) could exclude involvement of bovine orthologues of the human genes for both diseases. Because the disease mechanism in SMA and amyotrophic lateral sclerosis was not understood until recently, the bovine form, with its apparently distinct causative gene, could serve as an animal model to understand general aspects of motor neuron degeneration. The high incidence of SMA calves, which die a few weeks after birth, in both European and American Brown cattle populations also stresses the need for a reliable gene diagnosis for animal welfare and for economic fitness of the Brown Swiss cattle breed within the cattle breeding industry.
The fine-mapping of the bovine SMA locus suggested three positional candidates, all of which might be functionally linked to survival/degeneration of motor neurons (5). The most obvious candidate based on its similarity to the human disease gene was the antiapoptotic protein BCL2. It is partly similar in function to the human SMA gene SMN and was reported to form functionally interacting and synergistic complexes with the latter gene product (6). However, we excluded most of the BCL2 sequence as a potential causative gene (5). The immediate neighbor of BCL2 on bovine chromosome 24 (BTA24) is the 3-ketodihydrosphingosine (KDS) reductase FVT1. FVT1 is part of the glycosphingolipid metabolism and catalyzes the second step in the biosynthesis of the central precursors of this pathway, sphingosine and ceramide. Several other enzymes of the glycosphingolipid pathways are known to be responsible for neurodegenerative disorders. Moreover, many intermediates of this pathway are highly bioactive and display toxicity and induce apoptosis at elevated levels (7). The third candidate gene is VPS4B, which is part of the endosomal protein sorting complex ESCRT-III and is responsible for its disassembly at the end of its functional cycle (8). Other constituents of this multiprotein complex have been shown to cause neurodegenerative diseases, with all of them showing motor neuron degeneration. Most closely related to SMA is the wobbler mutation in the mouse VPS54 gene that causes SMA and defective spermiogenesis (9).
Here we mapped the functional mutation causing bovine SMA to the gene FVT1. FVT1 codes for KDS reductase (10), which plays an important role in the biosynthesis of glycosphingolipids. Thus, bovine SMA and a similar human disease with identical name and neuropathological characteristics are caused by mutations in completely different genes. Our findings support the recently evolving model that neurodegenerative diseases can be caused by a very general defect in metabolism or other housekeeping functions that motor neurons (more than any other cell type) are very sensitive to. Furthermore, we provide an animal model to study human motor neuron diseases.
Results and Discussion
Marker Map.
In addition to the markers developed and used for mapping in our previous fine-mapping study (5) we developed four microsatellites and two SNP markers. These six markers lie within a chromosomal fragment of ≈0.21 cM length. Despite the large pedigree (n = 1,154) used for reconstruction of the marker map by the program CRI-MAP we were not able to estimate the most likely marker order because of lack of recombination. Therefore, we deduced the relative marker positions from the nucleotide sequence of a BAC clone covering the candidate region (GenBank accession no. AC164994) and the physical BAC map. The deduced marker positions and additional marker information are presented in supporting information (SI) Table 2 for microsatellites and SI Table 3 for SNPs.
Disease Mapping.
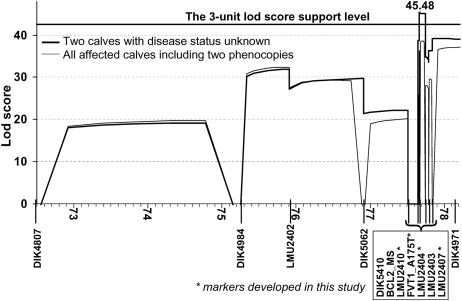
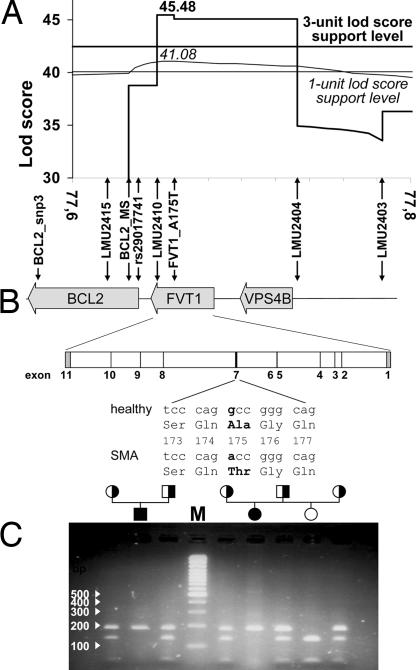
For the precise mapping of the disease-causing gene we built a complex pedigree of 421 animals including 32 inbreeding loops, 102 affected calves, 52 declared carriers (two and more affected calves with confirmed paternity), and 267 important relatives and available ancestors of affected animals. All 102 affected calves, 47 declared carriers, and 108 relatives were sampled and genotyped for a marker set of 17 microsatellites (SI Table 2) and partially for seven SNPs (SI Table 3). Only SNP FVT1_A175T, discovered and included in the map during candidate gene analysis, was genotyped for all sampled animals. Analyzing this complex pedigree with three-point analysis (the LINKMAP option of FASTLINK) we identified a significant linkage between SMA and the seven most distal markers on BTA24: BCL2_MS-LMU2410-FVT1_A175T-LMU2404-LMU2403-LMU2407-DIK4971 (Fig. 1). The most tightly linked marker interval is FVT1_A175T-LMU2404, with a logarithm of odds (LOD) score of 38.652 (Fig. 1, all affected calves). The mapping and haplotype analysis performed in our previously published results mapped the disease locus distal to microsatellite BCL2_MS. Careful consideration of the segregating haplotypes showed that the ancestral haplotype signature of the most telomeric 0.5 cM (from BCL2_MS to DIK4971) was disrupted by only one recombination event. This single recombinant haplotype was transmitted to 20 affected calves by one Swiss bull with a key position in the disease pedigree. By genotyping four markers (LMU2415, BCL2_MS, rs29017741, and LMU2410) in recombinant and nonrecombinant animals we localized the recombination in the interval BCL2_MS-rs29017741 with an approximate length of 12 kb. According to this and previous results (5) we can exclude the complete coding sequence (CDS) of the BCL2 gene as a possible causal gene for bovine SMA. Nearly all (119 of 122) of the affected individuals inherited the same ancestral haplotype from both parents and are homozygous for all seven markers distal to BCL2_MS. Only three calves were not homozygous for these markers and inherited different and for SMA unusual haplotypes from one or both parents. The pedigree analysis strongly indicates that these three calves are most probably phenocopies (5). They are descendants of two German and one Swiss Braunvieh bull that sired ≈26,000, 31,000, and 31,000 calves, respectively, and no additional SMA cases. Not all true SMA cases are recognized by breeders, and not all recognized cases are reported to the breeder's association. According to the expected and observed number of cases from risk matings (sire and maternal grandsire are declared carriers) we calculate a reporting rate of 0.07 in Germany, i.e., on average one of 14 true SMA cases is reported to the breeder's association. The Swiss Brown Cattle Breeder's Federation estimated a higher reporting rate for Switzerland (0.125). Considering the expected number of cases from known risk mating as well as the appropriate reporting rate we estimate high probability (P > 0.99) that these three bulls are not SMA carriers and that the above three calves are phenocopies. Multipoint mapping with incomplete penetrance and full likelihood analysis in a large complex pedigree with numerous inbreeding loops is very computing-intensive. Therefore, we were not able to perform this analysis with our available computing power. Instead, we used an alternative mapping analysis by setting the disease status of three probable phenocopies to unknown (Fig. 1). One of the three phenocopies was excluded from both analyses because it is only loosely related to the remaining core pedigree and the dam is not known. The three-point analysis without two probable phenocopies mapped SMA with the highest LOD score (45.48) accurately to the SNP FVT1_A175T. The three-unit LOD score support interval covered the segments between markers LMU2410 and LMU2404 (Fig. 1). According to a three-point analysis with FASTLINK (Figs. 1 and 2A), the mapping results include only two positional candidates as possible causal genes for bovine SMA, FVT1 and VPS4B. Both genes are functional candidates as well (7, 9). An 18-point analysis by SimWalk estimated the maximum LOD score (41.08) at the same position as FASTLINK but gave a rather flat peak (Fig. 2A). Using the more conservative three-unit LOD score support interval at 18-point analysis gives a very broad confidence interval (1.83 cM) extending beyond the 20-fold confirmed recombination between BCL2_MS and LMU2410. Applying a one-unit LOD score support interval at 18-point analysis (SimWalk) leads to approximately the same confidence interval as estimated by the FASTLINK three-point analysis (Fig. 2A).
Fig. 1.
Three-point LOD scores for two alternative analyses by FASTLINK. The thin line shows results for all sampled animals; the analysis represented by the bold line excludes two presumptive phenocopies by setting their disease status to unknown. Only the chromosomal segment that yielded LOD scores above 10 is shown.
Fig. 2.
Fine-mapping and positional candidate gene analysis for bovine SMA. (A) Detail of the fine-mapping results showing only the peak interval. The positions of the used markers are shown relative to the candidate genes, illustrating exclusion of BCL2. The result of FASTLINK three-point analysis with its corresponding three-unit support level is shown in bold. Results of SimWalk 18-point analysis with its corresponding one-unit support level is shown in thin lines and italic letters. (B) Structure of the bovine SMA locus and of the FVT1 gene. Vertical arrows show the position of the markers used for mapping. Relative position of introns and exons is shown for FVT1 with introns in white, coding exons in black, and the 5′ and 3′ UTRs in gray. For exon 7 the nucleotide exchange and the resulting codon change are shown. (C) PCR-RFLP with MvaI analyzed on a 2% agarose gel showing segregation of SMA and the FVT1 genotype. The PCR product for the Ala variant is cut by MvaI producing fragments of 130 and 50 bp, whereas the Thr variant causing SMA remains uncut at 180 bp. SMA carriers and affected and disease-free animals are represented by half-filled, filled, and open symbols, respectively. The white numbers and arrows on the left show the size of the DNA marker bands (M).
Candidate Gene Analysis.
Combining information from this and the previous study (5) BCL2 was excluded as positional candidate for bovine SMA. To find mutations in the two remaining candidate genes (Fig. 2A) we prepared cDNA covering the whole CDS of these genes. We obtained cDNA from different tissues of two animals with SMA and a healthy animal that was not a carrier. The cDNAs were identical in length between the different animals and tissues, so we excluded the presence of splice-site mutations in introns. Similar to BCL2 (5), we observed no differences in the CDS and in putative promoter or regulatory sequences of the candidate gene VPS4B. The intron sequences of both VPS4B and BCL2 contain several SNPs with low informativity for mapping because of high frequency (>0.9) of the allele in phase with the less common SMA allele (<0.05). Comparative sequencing of the cDNA from candidate genes FVT1 in affected and disease-free animals revealed a single difference in the CDS. The SMA-free control was identical to the database sequence (GenBank accession no. BC112615), but the two SMA calves both showed a single nucleotide exchange (nucleotide 562 in BC112615). The observed G-to-A transition in the FVT1 gene (Fig. 2B) changes an evolutionary highly conserved Ala in the vicinity of the enzyme's active site to a Thr residue (Fig. 3). The G/A polymorphism was assessed by PCR-RFLP with the restriction enzyme MvaI and showed Mendelian inheritance (Fig. 2C). We typed 122 affected animals with clinically and neuropathologically confirmed SMA diagnosis and found 119 of them were homozygous for the Thr variant (Thr-175). The three affected calves considered as phenocopies in this and the previous mapping study (5) were not homozygous for the Thr-175 variant. All available 94 parents of affected animals (phenocopies excluded) were heterozygous, as were the 30 additional declared SMA carriers that are included in our mapping pedigree. SMA in European Brown cattle shows a pronounced founder effect, relating all cases to the imported American Brown Swiss Bull “Meadow View Destiny.” The effectively small American Brown Swiss population is characterized by an initial genetic bottleneck, ≈100 years ago (≈20 cattle generations), followed by population expansion and strong selection (11). The accumulation of defective alleles is thought to originate from a combination of a small effective population size of American Brown Swiss population and an indirect selection due to linkage disequilibrium between the defective gene and the loci influencing the selected traits (12). In the majority of the cases a strong genetic bottleneck can eliminate some minor alleles in concerned subpopulation, but occasionally a bottleneck can amplify new mutations or minor alleles present in important founder animals. Therefore, the causal mutation for bovine SMA either was imported from Europe to America at the end of the 19th century or evolved from a new mutation during the genetic bottleneck. This rare disease allele was most likely amplified during a strong genetic bottleneck at the beginning of the 20th century and returned or immigrated into the effectively large European Brown population during the 1970s. To estimate the allele frequency of the Thr-175 variant in the modern German Braunvieh population, i.e., Braunvieh upgraded by American Brown Swiss, we genotyped 703 animals representing two different generations: 217 animals born between 1985 and 1990 and 486 animals born between 2000 and 2006 (Table 1). During approximately three cattle generations (15 years) the allele frequency of the Thr-175 variant increased from 1.84% to 4.42% in the current genetically active population. The observed allele frequency difference is significant (P = 0.019, SE = 0.0004; exact probability test) (13). For 48 randomly sampled modern German Braunvieh animals carrying the Thr-175 allele we analyzed the pedigrees and genotyped the 17 most distal markers on BTA24. The pedigree analysis showed that 77% of animals carrying the Thr-175 allele have well known SMA carriers as parents or grandparents. Seventeen percent have known SMA carriers in upper generations, and the remaining 6% have no known SMA carriers in their pedigree but have a large proportion of missing pedigree information. The haplotype analysis of these 48 animals showed that the Thr-175 allele is surrounded by the ancestral SMA haplotype at least for all markers distal to LMU2402 (≈2.23 cM).
Fig. 3.
Position of the evolutionarily conserved Ala that is changed to Thr in animals with SMA. The BLAST alignment with KDS reductases from other species shows the vicinity around the mutated position. The mutant position is in bold, and the catalytic motif with YXXK and the substrate binding Ser-13 residues upstream are underlined. In addition, the sequence of the S. cerevisiae homologue TSC10 is shown.
Table 1.
Sample size (n) and frequency of the Thr-175 allele in different cattle populations
| Sample | n | Frequency, % |
|---|---|---|
| Affected animals | 122 | 97.95* |
| Declared carriers | 95 | 50.00 |
| German Braunvieh born 1985–1990 | 217 | 1.84 |
| German Braunvieh born 2000–2006 | 486 | 4.42 |
| Original Braunvieh | 123 | 0.41† |
| Tyrolian Gray | 48 | 0.00 |
| Murnau-Werdenfelser | 53 | 0.00 |
| Murbodner | 48 | 0.00 |
*Only three animals considered as phenocopies are not homozygous for the Thr-175 variant.
†One Original Braunvieh animal shows the genotype and haplotype of typical SMA carriers.
Genotyping of FVT1_A175T in 123 “Original Braunvieh” animals, i.e., animals without any American Brown Swiss ancestors in their entire known pedigree, detected one heterozygous animal. This is equivalent to an allele frequency of 0.41%, which is significantly different (P = 0.0008, SE = 0.00005; Fisher exact test) (13) from the frequency in the modern German Braunvieh population. To the best of our knowledge, this is the only observation of SMA in Original Braunvieh. Haplotype analysis with available ancestors suggests an American Brown Swiss origin of the Thr-175 allele (i.e., false paternity) rather than a standing variation in the Original Braunvieh.
In addition to the Original Braunvieh population, we sampled and genotyped three close neighboring cattle populations: Tyrolean Gray Cattle (n = 48) (Table 1), Murnau-Werdenfelser Cattle (n = 53), and Austrian Murbodner Cattle (n = 48). These three SMA-free breeds were homozygous for the Ala-175 variant.
The above findings are in accordance with the initial frequency estimate of the disease allele (4), demonstrate a complete linkage disequilibrium between Thr-175 and the SMA allele, and strongly suggest the Thr-175 variant as the causative mutation for the disease. Because of the absence of clearly observed recombination in the interval rs29017741 to DIK4971 and reduced informativity of LMU2404 and LMU2403 (most common marker allele in phase with SMA allele) there still exists the possibility that the Thr-175 mutation is not causative but in perfect linkage disequilibrium with the causative allele. However, the comparative cattle/human map (5) and analysis of bovine genome sequencing data strongly suggest the absence of additional functional candidates in the chromosomal fragment from VPS4B to the telomere just after the last available marker, DIK4971 (5).
Analysis of FVT1 and Its Mutant fvt1-A175T.
The gene FVT1 codes for a KDS reductase (10) that plays an important role in the biosynthesis of glycosphingolipids (KEGG pathway 00600, www.genome.jp/kegg). It catalyzes the second step in the de novo synthesis of the central metabolites ceramide and sphingosine. The bovine FVT1 sequence is highly similar to the human orthologue, with 87% identity on the nucleotide level and 92% identity and 97% homology for the respective proteins. There are no hints for the existence of isoforms of the bovine protein. FVT1 belongs to the short-chain dehydrogenase/reductase (SDR) family. It shows the SDR active-site motif YXXXK, which is preceded by a conserved Ser that is 13 residues upstream (14) as well as a nucleotide binding domain with a so-called Rossman fold and the nucleotide binding motif GXXXGXG. A topology model based on hydrophobicity and protease accessibility proposed that the enzyme has three hydrophobic transmembrane domains with a large hydrophilic domain in between, which faces the cytosol and contains the active site (10). The Ala residue that is mutated in the SMA animals lies in the cytosolic domain close to the conserved substrate-binding Ser and is conserved in FVT1 proteins of evolutionary distant species (Fig. 3). Although the substitution of Ala by Thr is usually regarded as neutral it can change the local conformation because of its bulkier (C-β branched) side chain. Close to the active site such small conformational changes are able to affect the enzymatic activity. Being an H-bond donor/acceptor like the neighboring substrate binding Ser-173, Thr-175 could also induce an alternative binding mode, affecting catalytic efficiency.
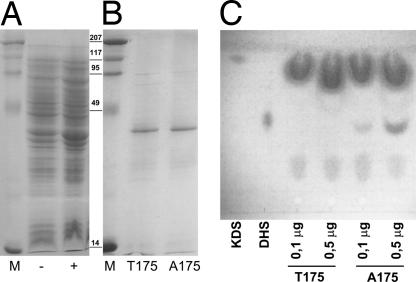
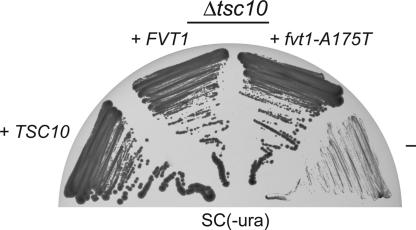
To characterize the protein variant found in SMA animals and its possible causality for the disease we constructed expression plasmids for both the SMA and the healthy variant. For efficient expression in Escherichia coli we omitted the first 25 aa that are very hydrophobic, putatively forming an α-helical membrane anchor, and abrogated expression of the human FVT1 protein in E. coli (10). The truncated mutant and normal proteins were efficiently expressed in E. coli (Fig. 4A) and purified to near homogeneity (Fig. 4B). The apparent molecular mass determined by SDS/PAGE is 34 kDa, matching the theoretical mass of 34.6 kDa. We then tested the enzymatic activity of both proteins in an in vitro assay. The FVT1 protein from an SMA-free calf showed KDS reductase activity. The reaction depended on both NADPH and the purified enzyme. In contrast, the same amount of the Thr-175 variant of FVT1, expressed and purified in parallel with the Ala-175 version, did not show any detectable enzymatic activity as analyzed by the appearance of the reaction product dihydrospingosine (Fig. 4C). Even a 5-fold higher amount of enzyme did not produce detectable amounts of dihydrospingosine. The fact that the observed variation not only cosegregates with the SMA phenotype but also leads to a loss of enzymatic activity in vitro is a very strong indication that the described Ala-to-Thr exchange is indeed causing bovine SMA. However, a complete disruption of the sphingolipid biosynthetic pathway by an inactive KDS reductase is likely to cause early embryonic lethality rather than postnatal progressive neurodegeneration. Therefore, we wanted to assess whether the observed loss-of-function mutation in FVT1 is also relevant in vivo. We thus expressed normal bovine FVT1 and the fvt1-A175T mutant in a yeast strain carrying a deletion of TSC10, the Saccharomyces cerevisiae homologue of FVT1. Expression of S.c. TSC10 rescues the slow-growth phenotype of a Δtsc10 strain in the absence of phytosphingosine (Fig. 5). Expression of FVT1 leads to an intermediate complementation showing that the bovine homologue does not fully complement the tsc10 knockout, which might be caused by the low homology (25%) of mammalian FVT1 to S.c. TSC10 (ref. 10 and Fig. 3). Δtsc10 cells expressing the SMA variant fvt1-A175T show the same growth phenotype as those complemented by normal FVT1 (Fig. 5). Thus, in vivo the A175T mutation in FVT1 does not lead to loss of KDS reductase activity. This could be explained by residual activity that was below the detection threshold of the in vitro enzymatic assay or by partial restoration of enzymatic function in vivo, e.g., by the action of heat-shock proteins, stabilization by intact membranes or by an unknown binding partner, or simply by higher expression levels from the recombinant plasmid. Thus, the Thr-175 variant is not completely inactive but can carry out its basic function in vivo. This could explain why the defect observed in vitro becomes manifest only after birth and selectively affects the motor neurons by a yet unknown mechanism.
Fig. 4.
The A175T variant lacks KDS reductase activity. (A) Expression of FVT1 and fvt1-A175T in E. coli; SDS/PAGE of uninduced (−) and induced (+) whole-cell lysates. (B) Purification of the His6 fusion protein. Both the Thr-175 variant from an affected calf and the normal Ala-175 are expressed and purified in parallel under the same conditions with comparable results. (C) In vitro KDS reductase assay of the protein fractions shown in B.
Fig. 5.
The FVT1 and the fvt1-A175T mutation show an intermediate complementation of the S. cerevisiae Δtsc10 knockout. The Δtsc10 strain was transformed with plasmid-based S.c. TSC10, bovine FVT1, fvt1-A175T, and the empty expression plasmid (−). The functionality of the proteins was assessed by restreaking yeast transformants onto plates lacking phytosphingosine.
The sphingolipid metabolism is extensively linked to function and development of the nervous system, and several neurodegenerative diseases are described as consequences of its disruption. Mutation of the serine palmitoyl transferase, the enzyme that produces the substrate for FVT1, has been shown to cause hereditary sensory neuropathy (15), a neurodegenerative disorder that also affects motor neurons. In Tay-Sachs disease, a deficiency in hexosaminidase A (HexA), another enzyme of the mentioned metabolic network, leads to accumulation of the GM2 ganglioside and as a consequence to motor neuron degeneration and other symptoms. In rare cases, HexA deficiencies can also mimic amyotrophic lateral sclerosis or SMA (16). Deletion of glucosyl ceramide synthase (17) and disruption of ganglioside synthesis (18) produce severe neural defects after birth including motor neuron degeneration in experimental mouse models. These examples illustrate the importance of an intact sphingolipid metabolism for neuronal development and function. Experiments with fumonisin, a fungal toxin that inhibits ceramide synthase and leads to depletion of ceramide, showed that neuronal cells are vulnerable to decreased de novo synthesis of ceramide leading to inhibition of both axonal growth (19) and neuronal differentiation (20). We speculate that the nearly inactive FVT1 variant observed as the cause of SMA in cattle leads to limited availability of ceramide and derived metabolites. Recently, several genes responsible for motor neuron disease have been identified (21), and the emerging picture is that these genes do not belong to a motor neuron-specific pathway but that the common feature lies in the especially high vulnerability of motor neurons. Most of these genes have housekeeping functions like RNA processing, vesicle sorting, or biosynthesis and are ubiquitously expressed. Alterations in these functions that other cell types can cope with might thus selectively affect the motor neurons, which, because of their high metabolic activity and extensive transport processes along the axons, are most delicate. The FVT1 variant found to be responsible for bovine SMA fits this common theme. The activity of the enzyme seems to be largely reduced in vitro but nevertheless sufficient for vital functions as shown by the complementation of the yeast knockout strain and the viability of SMA-affected calves at birth. Biosynthesis of sphingolipids is a housekeeping function with special importance to neuronal cells. Sphingolipids are also involved in vesicle dynamics (22, 23), and a mutation in the protein VAPB that is involved in transport of ceramide to Golgi vesicles (24) has been reported to cause a form of familial SMA and amyotrophic lateral sclerosis (25).
The proposed causal mutation in bovine SMA is adding a component to the complex and still poorly understood pathogenicity of motor neuron diseases. We think that this study can contribute an important animal model for the understanding of human motor neuron diseases. It suggests a vital function for FVT1 and contributes to the understanding of the sphingolipid metabolism and the function of its compounds. Finally, perfect linkage disequilibrium with the bovine SMA will allow efficient gene/marker-assisted selection to eliminate disease carriers from breeding, thus avoiding future incidence of this recessively inherited disease. This will help to maintain livestock biodiversity as it withdraws an important economical threat from the environmentally adapted alpine cattle breed Braunvieh.
Materials and Methods
Animals and Phenotyping.
In total, 122 calves affected by SMA were sampled in the Austrian (one calf), German (37 calves), and Swiss (84 calves) Braunvieh population. Most of these (115 calves) have been reported by breeding organizations and sent alive to university hospitals (Innsbruck, Munich, and Zurich) for clinical and neurological examinations. Additionally, we sampled 96 parents of affected calves, 30 additional declared carriers, and 396 related animals. These 644 samples were used for fine-mapping, haplotyping, and risk analyses in complex pedigrees.
Blood and sperm samples of 975 animals were sampled to analyze allele frequencies of candidate gene in Braunvieh and in three close neighbor populations: 703 samples of modern German Braunvieh, 123 of Original Braunvieh without introgression of American Brown Swiss, 48 Tyrolean Gray Cattle, 53 Murnau-Werdenfelser Cattle, and 48 Murbodner Cattle (Table 1). To obtain the sample of modern German Braunvieh we randomly took animals from all available samples that were sent to paternity testing in the respective year. To avoid bias we took no more than five animals with the same declared parent.
Marker Development.
In addition to SNP markers in LMAN1 and BCL2 genes and four microsatellite markers (BCL2_MS, LMU2401, LMU2402, and LMU2403) developed in our previous mapping study we added four microsatellite (LMU2404, LMU2407, LMU2410, and LMU2415) and two SNP markers to the candidate region investigated. Microsatellite markers were obtained by data-mining in combining information from BAC fingerprinting contigs, BAC end sequences, raw whole-genome shotgun sequences, and BLAST searches. Sequences obtained were searched for dinucleotide repeats and tested for informativity. The SNP markers were obtained by sequencing of two affected and one SMA-free animal. All markers considered for mapping and haplotyping in this study are shown in SI Table 2 for microsatellites and SI Table 3 for SNPs.
Markers were mapped by program CRI-MAP (26) in a complex pedigree comprising 1,154 animals by using a framework of well characterized markers from the U.S. Department of Agriculture bovine linkage map (www.marc.usda.gov/genome/genome.html). Where markers were closely spaced, so that no recombination was observed, the distance was deduced from the relative marker positions in the nucleotide sequence of a BAC clone covering the candidate region and the physical BAC map (www.bcgsc.ca/platform/mapping/data).
Genotyping.
Genomic DNA was prepared from semen, peripheral blood, hair roots, and paraffin-embedded tissue samples by using standard methods. For microsatellite markers (n = 17) (SI Table 2) the PCR products were analyzed on ABI377 and ABI310 DNA sequencers. Genotypes were assigned by using GENESCAN and GENOTYPER programs. Seven SNPs (SI Table 3) were scored by using PCR-RFLP analyzed on agarose gels and primer extension with MALDI-TOF analysis.
Using the programs CRI-MAP and SimWalk2 (27) we tested for paternity and assured quality of genotypes.
Pedigrees and Genetic Analysis.
The initial linkage analysis and testing of data consistency were performed by the CRI-MAP program (version 2.4) and used complex pedigree involving 1,154 individuals. For the recessive disease locus, only affected and confirmed carrier animals were scored. Thus, CRI-MAP ignores some of the available information and does not use allele frequencies. Therefore, we preferred the conventional linkage analysis with full likelihood analysis, as provided by LINKAGE or FASTLINK (28, 29), to fine-map the disease locus. Because of computer memory and time constraints, we performed the full likelihood analysis in a smaller pedigree. We reduced the maximal pedigree (n = 1,154) by removing 733 animals, mostly founders without genotypes and most full and half siblings of affected animals. To avoid a large proportion of missing data and thus to reduce the maximum-likelihood parameter space we also removed 20 affected calves and their ancestors. The removed affected calves are uninformative for disease mapping, i.e., homozygous for all markers genotyped in the entire most telomere region, and are very loosely related to the remaining part of the pedigree, and samples of one or both parents are not available. To avoid mating loops, we preferentially ignored maternal relationships of nonaffected animals and some relationships of nonsampled parents or grandparents of affected calves. The resulting pedigree comprised 421 animals. This pedigree was still characterized by 32 inbreeding loops that were broken at genotyped animals with well inferable haplotypes. For linkage analyses with FASTLINK we fixed the disease allele frequency to 0.04 and assumed a purely recessive mode of inheritance, with a penetrance of 1.0 for the disease genotype. The marker allele frequencies used were obtained by simple counting of both alleles of 22 unrelated founders and maternal alleles of 148 additional animals with unrelated mothers. We used only safely deduced maternal alleles. Using MLINK, ILINK, and LINKMAP options of the FASTLINK programs we performed two-point and three-point analyses of the candidate region. To map the disease locus against a fixed map of markers we proceeded by three-point analysis as described (30).
The most likely set of maternal and paternal haplotypes was estimated with the SimWalk2 program. The haplotype analysis was performed on the basis of the maximal pedigree with 1,154 individuals. Because of computer memory and time constraints, we split this pedigree into appropriate overlapping parts. The same core pedigree with 421 animals, allele frequencies, marker map, and inheritance mode was used for a multipoint analysis by the program SimWalk2.
Candidate Gene Analysis.
Whole RNA was prepared from frozen liver, kidney, and brain tissue from two killed calves with clinically and neuropathologically confirmed SMA diagnoses and a SMA-free control by the TRIzol method. From the quantified RNA cDNA was synthesized with a random hexamer primer and murine leukemia virus reverse transcriptase. A total of 0.5 μl of the cDNA was used as template for specific PCR amplification of the respective candidate genes. Primer pairs vps4b_for1 ctagcaaagcagcccagga/vps4b_rev1 aaaggaacgctgaggtgaga and vps4b_start ggtctccgaacaggttcca/vps4b_int1 ctcccagaaccaaaattcca were used to generate two overlapping fragment covering the whole CDS of the VPS4B gene. PCR products were purified with microcon PCR centrifugal filter devices (Millipore, Bedford, MA) and sequenced on both strands by using the amplification primers as well as internal primers.
For FVT1 the primer pairs fvt_for1 gcctgcgccgagctg/fvt_rev1 gcttgcagccaggtttctga and fvt_for2 cgcacgtagtggttacagga/fvt_rev2 cataccccacggcaatgtgt were used to amplify and sequence the complete CDS.
The whole CDS of BCL2, intron 1, and parts of promoter regions and intron 2 were analyzed in our previous study (5).
PCR-RFLP Test for A175T Mutation.
A fragment bearing a single MvaI restriction site was amplified from genomic DNA by using the primers fvt_A175T_f atcgccaccatgaaggaac and fvt_A175T_r ctgggctgaaaggaatcaat. The Ala variant resulted in MvaI cleavage with fragments of 130 and 50 bp, respectively. The digestion products were analyzed on 2% agarose gels.
Expression and Purification of FVT1 Variants.
The CDSs for FVT1 from an animal with SMA and a disease-free control were prepared for expression in E. coli by amplification of the cDNA with the primer pair Bam_FVT1_f1 ggatccaagcccctcgccctgccc/Hind_FVT1_r1 aagcttctgtgtcccacttagcagca. The resulting product coded for an N-terminally truncated form of the FVT1 protein starting with amino acid 26. The amplified fragment was digested with the restriction enzymes BamHI and HindIII, gel-purified, and ligated to an equally treated pQE30 expression vector (Qiagen, Hilden, Germany), yielding a His6-tagged construct of 318 aa. E. coli cells bearing the respective plasmid were grown in LB medium at 37°C to an OD600 = 0.6, induced with 1 mM IPTG, harvested 5 h after induction, and frozen. The thawed cells were resuspended in lysis buffer (25 mM Tris·HCl, pH 7.5/300 mM NaCl/10% glycerol/1% Triton X-100/10 mM 2-mercaptoethanol/10 mM imidazole/1 mM PMSF/1× protease inhibitor mixture). After addition of 1 mg/ml lysozyme cells were kept on ice for 30 min, sonicated, and centrifuged for 20 min at 4,000 × g to clear the lysate. One milliliter of Ni-NTA agarose beads was added to the cleared lysate and incubated for 30 min with agitation. Ni-NTA agarose was packed on a mini column and successively washed with lysis buffer containing 20 and 40 mM imidazole. Elution with 350 mM imidazole yielded the recombinant, almost pure FVT1 and fvt1-A175T.
In Vitro KDS Reductase Assay.
A reaction mixture containing 25 mM Tris·HCl (pH 7.5), 150 mM NaCl, 10% glycerol, 0.2% Triton X-100, 10 mM 2-mercaptoethanol, 1 mM PMSF, 100 μM NADPH, 100 μM KDS, and 0.1 μg of purified FVT1 protein was incubated at 37°C for 1 h. Lipids were extracted from the reaction mixture with chloroform/methanol 1:1, and the organic phases were pooled and dried in a Speedvac. Lipids were resolved on Silica 60 HPTLC plates (Nanosil 20; Macherey-Nagel, Düren, Germany) with chloroform/methanol/NH4OH 40:10:1. Sphingoid bases were detected by spraying the developed TLC plates with 0.2% ninhydrin in ethanol and heating at 180°C for 5 min.
In Vivo Yeast Complementation Assay.
The plasmids pAK80-W0671 (FVT1) and pAK80-SMA031 (fvt1-A175T) were obtained by amplification of the FVT1 cDNA from a control (W0671) and an affected animal (SMA031) and ligation into pGEM-T easy. From the respective pGEM-T easy plasmids the NotI fragment was cloned to pAK80 (13) (gift of A. Kihara, Hokkaido University, Sapporo, Japan) to yield pAK80-W0671 and pAK80-SMA031. As positive control for complementation the TSC10 gene was amplified and ligated to pGEM-T easy, and the NotI fragment was cloned to pAK80 to yield pAK80-TSC10. Plasmids pAK80-W0671(FVT1), pAK80-SMA031(fvt1-A175T), pAK80-TSC10, and pAK80 were transformed into the Δtsc10 strain (KHY625). Transformants were selected on synthetic complete plates lacking Ura plus 5 μM phytosphingosine and 0.0015% Nonidet P-40 as a dispersant. To assess the functionality of these constructs, colonies were restreaked onto synthetic complete plates lacking Ura and incubated for 4 days at 30°C, and growth of colonies was analyzed.
Supplementary Material
Acknowledgments
We thank Tierzuchtforschung (Grub, Germany), the Breeding Organisations for Braunvieh in Germany and Switzerland, and the Breeders of Murnau-Werdenfelser, Austrian Murbodner, and Tyrolean Gray cattle for providing samples for the population surveys. The yeast strain KHY625 and the expression plasmid pAK80 were gifts of A. Kihara, whom we want to thank for his cooperativity and technical advice. This work was funded by Deutsche Forschungsgemeinschaft Grants KR 2866/1 (to S.K.) and SFB646 (to K.S.).
Abbreviations
- SMA
spinal muscular atrophy
- KDS
3-ketodihydrosphingosine
- CDS
coding sequence
- LOD
logarithm of odds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607721104/DC1.
References
- 1.el-Hamidi M, Leipold HW, Vestweber JG, Saperstein G. Zentralbl Veterinarmed A. 1989;36:731–738. doi: 10.1111/j.1439-0442.1989.tb00786.x. [DOI] [PubMed] [Google Scholar]
- 2.Siso S, Pumarola M, Ferrer I. J Comp Pathol. 2003;128:132–139. doi: 10.1053/jcpa.2002.0616. [DOI] [PubMed] [Google Scholar]
- 3.Troyer D, Leipold HW, Cash W, Vestweber J. J Comp Pathol. 1992;107:305–317. doi: 10.1016/0021-9975(92)90006-g. [DOI] [PubMed] [Google Scholar]
- 4.Medugorac I, Kemter J, Russ I, Pietrowski D, Nuske S, Reichenbach HD, Schmahl W, Forster M. Mamm Genome. 2003;14:383–391. doi: 10.1007/s00335-002-3024-3. [DOI] [PubMed] [Google Scholar]
- 5.Krebs S, Medugorac I, Russ I, Ossent P, Bleul U, Schmahl W, Forster M. Mamm Genome. 2006;17:67–76. doi: 10.1007/s00335-005-0102-3. [DOI] [PubMed] [Google Scholar]
- 6.Iwahashi H, Eguchi Y, Yasuhara N, Hanafusa T, Matsuzawa Y, Tsujimoto Y. Nature. 1997;390:413–417. doi: 10.1038/37144. [DOI] [PubMed] [Google Scholar]
- 7.Linn SC, Kim H, Keane E, Andras L, Wang E, Merrill A. Biochem Soc Trans. 2001;29:831–835. doi: 10.1042/0300-5127:0290831. [DOI] [PubMed] [Google Scholar]
- 8.Scott A, Chung HY, Gonciarz-Swiatek M, Hill GC, Whitby FG, Gaspar J, Holton JM, Viswanathan R, Ghaffarian S, Hill CP, et al. EMBO J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt-John T, Drepper C, Mussmann A, Hahn P, Kuhlmann M, Thiel C, Hafner M, Lengeling A, Heimann P, Jones, et al. Nat Genet. 2005;37:1213–1215. doi: 10.1038/ng1661. [DOI] [PubMed] [Google Scholar]
- 10.Kihara A, Igarashi Y. J Biol Chem. 2004;279:49243–49250. doi: 10.1074/jbc.M405915200. [DOI] [PubMed] [Google Scholar]
- 11.Yoder DM, Lush JL. J Hered. 1937;28:154–160. [Google Scholar]
- 12.Georges M, Dietz AB, Mishra A, Nielsen D, Sargeant LS, Sorensen A, Steele MR, Zhao X, Leipold H, Womack JE, et al. Proc Natl Acad Sci USA. 1993;90:1058–1062. doi: 10.1073/pnas.90.3.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raymond M, Rousset F. Evolution (Lawrence, Kans) 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 14.Duax WL, Griffin JF, Ghosh D. Curr Opin Struct Biol. 1996;6:813–823. doi: 10.1016/s0959-440x(96)80012-1. [DOI] [PubMed] [Google Scholar]
- 15.Bejaoui K, Wu C, Scheffler MD, Haan G, Ashby P, Wu L, de Jong P, Brown RH., Jr Nat Genet. 2001;27:261–262. doi: 10.1038/85817. [DOI] [PubMed] [Google Scholar]
- 16.Johnson WG, Wigger HJ, Karp HR, Glaubiger LM, Rowland LP. Ann Neurol. 1982;11:11–16. doi: 10.1002/ana.410110103. [DOI] [PubMed] [Google Scholar]
- 17.Jennemann R, Sandhoff R, Wang S, Kiss E, Gretz N, Zuliani C, Martin-Villalba A, Jager R, Schorle H, Kenzelmann M, et al. Proc Natl Acad Sci USA. 2005;102:12459–12464. doi: 10.1073/pnas.0500893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamashita T, Wu YP, Sandhoff R, Werth N, Mizukami H, Ellis JM, Dupree JL, Geyer R, Sandhoff K, Proia RL. Proc Natl Acad Sci USA. 2005;102:2725–2730. doi: 10.1073/pnas.0407785102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harel R, Futerman AH. J Biol Chem. 1993;268:14476–14481. [PubMed] [Google Scholar]
- 20.Riboni L, Prinetti A, Bassi R, Caminiti A, Tettamanti G. J Biol Chem. 1995;270:26868–26875. doi: 10.1074/jbc.270.45.26868. [DOI] [PubMed] [Google Scholar]
- 21.Talbot K, Ansorge O. Hum Mol Genet. 2006;15:182–187. doi: 10.1093/hmg/ddl202. [DOI] [PubMed] [Google Scholar]
- 22.Hoekstra D, Maier O, van der Wouden JM, Slimane TA, van Ijzendoorn SC. J Lipid Res. 2003;44:869–877. doi: 10.1194/jlr.R300003-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Rohrbough J, Rushton E, Palanker L, Woodruff E, Matthies HJ, Acharya U, Acharya JK, Broadie K. J Neurosci. 2004;24:7789–7803. doi: 10.1523/JNEUROSCI.1146-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry RJ, Ridgway ND. Mol Biol Cell. 2006;17:2604–2616. doi: 10.1091/mbc.E06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishimura A, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, et al. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lander ES, Green P. Proc Natl Acad Sci USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobel E, Lange K. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 28.Lathrop GM, Lalouel JM. Am J Hum Genet. 1984;36:460–465. [PMC free article] [PubMed] [Google Scholar]
- 29.Cottingham RW, Jr, Idury RM, Schaffer AA. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger JD, Ott J. Handbook of Human Genetic Linkage. Baltimore: Johns Hopkins Univ Press; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.