Abstract
Expression profiling, ChiP–CHIP and phenotypic analysis were used to investigate the functional relationships of class III NAD+-dependent HDACs (Sirtuins) in fission yeast. We detected significant histone acetylation increases in Sirtuin mutants at their specific genomic binding targets and were thus able to identify an in vivo substrate preference for each Sirtuin. At heterochromatic loci, we demonstrate that although Hst2 is mainly cytoplasmic, a nuclear pool of Hst2 colocalizes with the other Sirtuins at silent regions (cen, mat, tel, rDNA), and that like the other Sirtuins, Hst2 is required for rDNA and centromeric silencing. Interestingly we found specific functions for the fission yeast Sirtuins Hst2 and Hst4 in gene regulation. Hst2 directly represses genes involved in transport and membrane function, whereas Hst4 represses amino-acid biosynthesis genes and Tf2 retrotransposons. A specific role for Hst4 in Tf2 5′ mRNA processing was revealed. Thus, Sirtuins share functions at many genomic targets, but Hst2 and Hst4 have also evolved unique functions in gene regulation.
Keywords: chromatin, fission yeast, HDAC, retrotransposon, Sirtuin
Introduction
The basic structural unit of chromatin is the nucleosome, which consists of two copies of each of the four core histones H2A, H2B, H3 and H4, around which 146 base pairs of DNA are wrapped. Each histone can be covalently modified in several ways including acetylation, methylation, phosphorylation, ADP ribosylation and ubiquitinylation. Histone acetylation is regulated by the activity of histone acetyltransferases (HATs) and histone deacetylases (HDACs). HDACs are highly conserved enzymes, which (with some exceptions) generally act as corepressors in gene repression mechanisms, or regulators of other chromatin-dependent processes such as DNA repair and DNA replication (Kurdistani and Grunstein, 2003; Verdin et al, 2003). HDACs are classified according to the structure of their active sites. The class I and class II HDACs share a conserved catalytic core, with some characteristic differences in the surrounding amino-acid sequences; however, the class III HDACs (Sirtuins) show no amino-acid sequence similarity to the other classes, and have an NAD+-dependent deacetylase activity (Blander and Guarente, 2004; North and Verdin, 2004). Sirtuins have been implicated in various biological processes such as lifespan extension by calorie restriction (Lamming et al, 2005; Vijg and Suh, 2006), regulation of non-histone proteins involved in cell survival that is, p53, p300, tubulin (North and Verdin, 2004) and histone deacetylation of transcriptionally silent chromosomal regions (Brachmann et al, 1995; Freeman-Cook et al, 1999, 2005; Perrod et al, 2001). The Sirtuins are represented in a wide range of organisms from Archaea to humans (Blander and Guarente, 2004). The mammalian Sir2 family comprises seven identified members; the budding yeast Saccharomyces cerevisiae has five members, whereas the fission yeast Schizosaccharomyces pombe encodes only three Sir2-like proteins Sir2, Hst4 and Hst2.
In budding yeast, ScSir2 has been shown to be required for silencing of all heterochromatic regions by preferentially deacetylating H4K16Ac in vivo (Robyr et al, 2002; Suka et al, 2002). The orthologue SpSir2 also acts at silent regions of S. pombe and affects H3K9Ac and H4K16Ac in vitro (Shankaranarayana et al, 2003). The other Sirtuins, ScHst4 and ScHst3 in S. cerevisiae and the Hst4 orthologue in S. pombe are all implicated in cell cycle regulation and heterochromatin silencing (Brachmann et al, 1995; Freeman-Cook et al, 1999). Recent studies showed that ScHst3, aided by ScHst4, regulates H3K56Ac levels during the cell cycle, and that this modification is required for genome integrity (Celic et al, 2006; Maas et al, 2006). The Hst2 ortholog in budding yeast, ScHst2, is mainly cytoplasmic (Perrod et al, 2001), but can directly silence the FLO10 gene involved in regulation of stress and flocculation (Halme et al, 2004). ScHst2 can also directly affect the rDNA locus under conditions of calorie restriction (Lamming et al, 2005, 2006). The Hst2 mouse ortholog SirT2 was recently found to be localized in the nucleus during early prophase and mitosis (Vaquero et al, 2006). In vitro SirT2 primarily deacetylates H4K16Ac followed by H3K9Ac (Vaquero et al, 2006).
Investigation of enzymatic specificities, expression profiles and binding locations clearly demonstrated the different cellular roles of Clr6, Clr3, Sir2 and Hos2 HDACs in S. pombe (Wiren et al, 2005). Some cellular functions were also found to be shared by these different HDACs. In particular, Clr3 and Sir2 were found to act cooperatively in the silent heterochromatin regions, as well as in other genomic locations, where they contribute to H3K14 and H3K9 deacetylation, respectively (Bjerling et al, 2002; Wiren et al, 2005). Clr6 was shown to act in promoter-mediated gene repression of stress and meiosis-induced genes, whereas Hos2 was required to boost high gene expression primarily by deacetylating H4K16Ac in coding regions (Wiren et al, 2005; Sinha et al, 2006). The latter finding was consistent with the observations in S. cerevisiae that ScHos2 binds highly expressed genes, ScHos2 is required for deacetylation of H4K16Ac and ScHos2 is required for full activation of GAL genes (Wang et al, 2002). To date, genome-wide studies have been performed on several HDACs in budding and fission yeast (reviewed in Ekwall, 2005; Millar and Grunstein, 2006); however, no such study has included the Sirtuins Hst2 and Hst4. The Sirtuin family is relatively small in fission yeast as compared with other species, with only three members, Sir2, Hst2 and Hst4. In this study, we investigated the cellular functions of all three Sirtuins in fission yeast, with particular focus on Hst2 and Hst4. We asked whether Hst2 and Hst4 are involved in regulation of genes involved in distinct cellular processes and whether this would be reflected in distinct phenotypes. Our approach was to combine microarray techniques, cDNA expression profiling, histone acetylation mapping by ChIP–CHIP and Sirtuin binding mapping by ChIP–CHIP, with phenotypic analysis. Interestingly, our data suggest that although all three Sirtuins share some functions and genomic targets, including the silent regions, each Sirtuin also has unique genomic targets and specialized roles in regulation of gene expression. We found that Hst2 directly represses several genes involved in transport activity, whereas Hst4 represses genes involved in amino-acid metabolism. In addition, Hst4 is required for Tf2 retrotransposon silencing and 5′ mRNA processing.
Results
Expression profiles reveal different functions for Sirtuins in regulation of gene expression
An important goal of this study was to define the roles of the different Sirtuins in gene regulation. Therefore, expression profiling of hst2Δ was carried out and the data compared with previously published data for the other HDACs (Wiren et al, 2005; Fagerstrom-Billai et al, 2007). Briefly, hst2Δ cells were grown to mid-logarithmic phase (5 × 106 cells/ml) in rich medium and RNA was extracted. Then, RNA was reverse transcribed into labelled cDNA, which was hybridized onto microarrays as described in Xue et al (2004). First, we carried out analyses of the general expression pattern of Sirtuins and compared them with class I and II HDAC mutants in S. pombe. We calculated the median expression ratio mutant/wild type (wt) and plotted the moving average of this ratio as a function of the wt gene expression levels (see Materials and methods). In this way, the global tendencies of gene expression in each HDAC mutant were compared. We observed different ‘global expression profiles' for each Sirtuin mutant. The ‘global expression profile' for the sir2Δ mutant showed a ratio of 1 for moderately expressed and highly expressed genes, indicating that the Sir2 activity was not required for regulating this range of gene expression levels (Figure 1A). In contrast, the silent genes and genes with low expression in wt showed a very high median expression ratio, indicating that Sir2 is mainly required in the silencing of non-expressed genes under these conditions in S. pombe. In the case of hst2Δ, the global expression was similar to that of sir2Δ, although less pronounced, suggesting that Hst2 also primarily represses expression of a non-expressed mRNAs. Surprisingly, the global expression profile of hst4Δ was in sharp contrast to those of sir2Δ and hst2Δ. Non-expressed genes in wt showed a weak increase of the median expression ratio, but remarkably, the hst4Δ mutant showed a general increase of median expression ratio for moderately to highly expressed genes (Figure 1A, green line). This indicated that whereas Sir2 mostly represses non-expressed genes, Hst4 acts more globally to tune down gene expression. This finding suggested that other HDAC classes may also play similarly specialized functions on transcription not detected by previous studies. To this end, we generated global expression profile plots for the class I and II HDAC mutants clr6-1, hos2Δ and clr3Δ using data from Wiren et al (2005). The global expression profiles of clr6-1 and clr3Δ mutants showed a general ratio of about 1, but non-expressed genes or weakly expressed genes had an increased median ratio, suggesting that Clr6 and Clr3 are mainly involved in keeping repressed genes silent (Figure 1B). As expected, a strikingly different expression profile was seen in the hos2Δ mutant. Highly expressed genes showed a markedly reduced median expression ratio, whereas weakly or non-expressed genes showed an increased median expression. This indicated that Hos2 is bifunctional in gene expression, consistent with the extensive upregulation (485 genes) and downregulation (455 genes) in hos2Δ (Wiren et al, 2005). Thus, the global expression profiles clearly revealed general roles of Sirtuins in repression of gene expression. Unlike Hos2, Sirtuins did not appear to be generally required for activation of gene expression.
Figure 1.
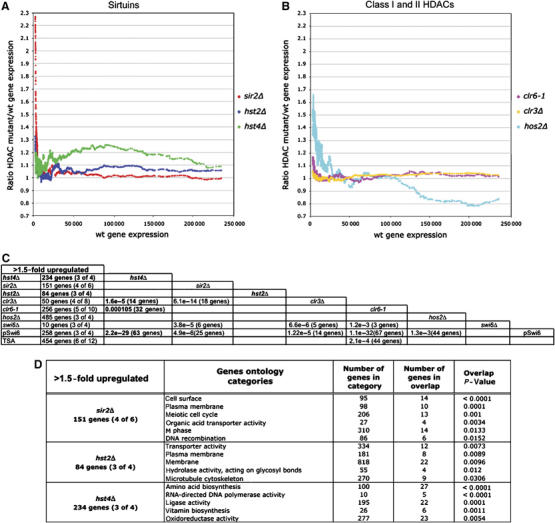
Expression profiling of HDAC mutants. (A, B) Moving average plots (window size=150; step size 1) of the median gene expression ratio mutant/wt plotted as a function of average transcription levels of 11 wt measurements of cultures in mid-logarithmic growth phase (using an arbitrary scale 0–250 000; data from Wiren et al (2005)). (C) The number of genes 1.5-fold upregulated in the different HDAC mutants are shown. The numbers within parentheses indicate the number of measurements, which fulfill the threshold limits, that is (4 of 6), four of six data points were above the 1.5-fold cut-off. New data on Sirtuins as part of this study are shown in bold letters. The other data are from Wiren et al (2005). The significance of overlaps between the gene lists is indicated by the hypergeometric probabilities, and the numbers of shared genes between the data sets are cross-tabulated. (D) The result of ‘GoMiner' analysis of genes derepressed in Sirtuin mutants (as indicated). The most significant gene ontology categories, the number of genes in each category, the number of genes in overlap and the P-values (two-sided Fisher's exact test) indicating the significance of the overlap are indicated.
Next, we employed a 1.5-fold cutoff value to define lists of hst4Δ and hst2Δ affected genes. Two hundred and thirty-eight and 132 genes, respectively, were downregulated in hst4Δ and hst2Δ mutants and no significant similarity was found to lists of downregulated genes in other HDAC mutants by hypergeometric distribution probability tests (data not shown). Regarding the Sirtuin-repressed genes that is,. those upregulated in hst4Δ and hst2Δ, mutants we found that in hst4Δ 234 genes were upregulated, of which a significant proportion were also upregulated in clr3Δ (14 genes; P=1.6e-5) and clr6-1 (32 genes; P=1.02e-4) (Figure 1C). In hst2Δ, we found 84 upregulated genes; however, the comparison did not reveal any significant similarities with lists of genes upregulated in other mutants. To investigate if genes involved in different cellular processes were affected by Sirtuin mutants, the sir2Δ-, hst4Δ- and hst2Δ-upregulated genes were subjected to a ‘GoMiner' search (Zeeberg et al, 2003). The most significant gene ontology categories and the P-values are shown in Figure 1D. Importantly, genes involved in distinct processes were represses by Hst2 and Hst4. For example, genes involved in amino-acid biosynthesis were upregulated in hst4Δ (P<0.0001), whereas gene involved in transport were upregulated in hst2Δ (P=0.0073). Thus, it was clear that different cellular functions were affected by the different Sirtuin mutants.
All Sirtuin mutants show nucleosome redistribution and their hyperacetylation patterns correlate with the global expression profiles
Our previous study showed that sir2Δ, hos2Δ but not clr3Δ and clr6-1 mutants had lower average nucleosome content and altered distribution frequency compared with wt cells (Wiren et al, 2005). We wished to know whether hst4Δ and hst2Δ mutants also affected genomic nucleosome density. To address this, we performed ChIP–CHIP experiments using Eurogentec SA-combined ORF+IGR S. pombe microarrays (see Materials and methods). wt, hst4Δ and hst2Δ cells were subjected to ChIP–CHIP using an antibody against histone H3 carboxy-terminal region ‘H3cter'. The nucleosome density was represented by histograms showing the ratio distribution of each mutant versus wt (Figure 2A). In the IGR and ORF regions, hst4Δ showed a marked reduction in distribution of histone density values, with a median value below 1.0, indicating that hst4Δ reduced the overall nucleosome content in comparison with wt. Immunoprecipitated fragments of hst2Δ revealed a weaker but similar tendency to reduce nucleosome density both in IGR and ORF regions. Reduced nucleosome density did not correlate with increased transcription in hst4Δ and hst2Δ mutants, indicating that it is not merely a consequence of increased transcription (data not shown). Taken together with similar findings on sir2Δ (Wiren et al, 2005), these data indicate that all Sirtuins in S. pombe are required to maintain a normal nucleosome density and distribution in the genome.
Figure 2.
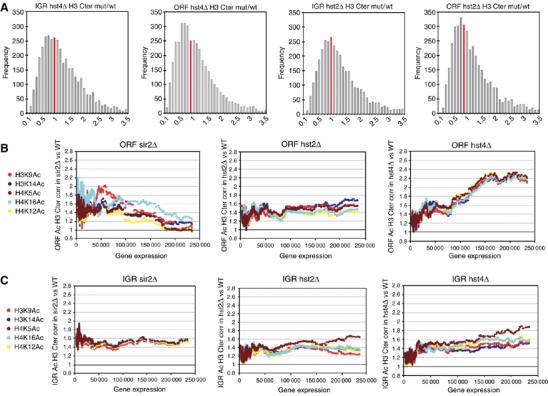
(A) Nucleosome occupancy in hst2 or hst4 HDAC mutants versus wt. The histograms represent the distribution of the ratios of H3cter fragments of IGR and ORF regions. The red bar indicates the frequency at a mutant/wt nucleosome ratio of 1. (B, C) Moving average plots (window size=150; step size 1) of different histone acetylation median ratio mutant/wt plotted in function of average transcription of 11 wt measurements of cultures in mid-logarithmic growth. (B) ORF histone acetylation and (C) IGR histone acetylation.
Histone acetylation neutralizes the positive charge of lysine and thereby weakens the interaction between histones and DNA, which facilitates mobilization of the nucleosome (Reinke and Horz, 2003). Therefore, the nucleosome density changes observed in hst4Δ and hst2Δ suggested a general increase in histone acetylation. To directly investigate the genome-wide effects on histone acetylation, we used a set of antibodies specific for five acetylated isoforms: H3K9Ac, H3K14Ac, H4K5Ac, H4K16Ac, H4K12Ac (Suka et al, 2001). ChIP was performed using wt, hst4Δ, hst2Δ and sir2Δ cells at mid-exponential growth phase in rich (YES) medium. The H3cter ChIP was used to correct acetylation levels for nucleosome density (see Materials and methods). To illustrate the genome-wide changes in histone acetylation, the density-corrected histone acetylation ratios (mutant/wt) were plotted against wt gene expression levels for each of the six different acetylation sites (Figure 2B and C). Interestingly, sir2Δ and hst4Δ mutants displayed opposite trends for acetylation changes in low and high ranges of gene expression, particularly in ORF regions (Figure 2B). sir2Δ showed increased histone acetylation of non-expressed and weakly expressed genes, while the ratio for highly expressed genes remained relatively unchanged. In contrast, hst4Δ histone acetylation of non-expressed and weakly expressed genes was relatively unchanged, whereas the global acetylation ratio was strongly increased for medium or highly expressed genes. In hst2Δ, global acetylation increases in ORFs seemed to be relatively independent of gene expression levels. The analysis of IGR regions indicated that all three Sirtuin mutants affect IGR acetylation (Figure 2C). In hst4Δ, we observed a similar but less pronounced trend in IGR as in ORF regions, that is, a preferential increase of highly expressed genes. The finding that ORF and IGR histone acetylation levels are generally increased in Sirtuin mutants suggested that all of the Sirtuins in fission yeast have a role in histone deacetylation. These data also indicated that one Sirtuin, Sir2, affects histone acetylation levels within the ORFs of genes whose expression is not required, whereas Hst4 affects changes within those ORFs whose expression is high under the growth conditions that we tested (Figure 2B). This trend was further strengthened by the analysis of mutant Sirtuin expression profiles. Hst4 dampens the expression of genes, whereas Sir2 keeps repressed genes silent (Figure 1A). Therefore, we infer that the histone acetylation changes catalyzed by Sir2 or Hst4 are key factors for achieving an appropriate balance in gene expression levels.
Hst2 is cytoplasmic and nuclear in fission yeast
Previous work showed that Sir2 and Hst4 are localized in the nucleus in fission yeast (Freeman-Cook et al, 1999; Freeman-Cook et al, 2005), but the cellular localization of Hst2 had previously not been determined in fission yeast. Therefore, to localize Hst2 in S. pombe cells, we generated an epitope-tagged strain in which the Hst2 protein was fused at the carboxyl terminus with c-myc epitopes, and expressed by the hst2+ promoter. As a nuclear envelope marker, we used a GFP fusion for the nuclear pore protein Pom152 (Ding et al, 2000; Bjerling et al, 2002). Immunofluorescence (IF) detection revealed that Hst2-myc was predominantly cytoplasmic; however, it was obvious from de-deconvolved IF images of interphase cells (Figure 3A) that some of the Hst2-myc protein (green) colocalized with the DAPI-stained (red) chromatin within the nuclear envelope delimited by Pom152-GFP (yellow). This finding is in good agreement with the observations on the mouse Hst2 ortholog SirT2, which can localize to the nucleus (Vaquero et al, 2006) and budding yeast ScHst2, which immunoprecipitates with the FLO10 promoter and the rDNA regions (Halme et al, 2004; Lamming et al, 2005, 2006). ScHst2 was recently reported to have a nuclear export sequence (NES) (Wilson et al, 2006). In the S. pombe Sirtuins, a homologous NES was only detected in Hst2 (data not shown). Therefore, unlike for the other two fission yeast Sirtuins, the NES may direct Hst2 to the cytoplasm, but a nuclear subpopulation of Hst2 is still able to interact with chromatin.
Figure 3.
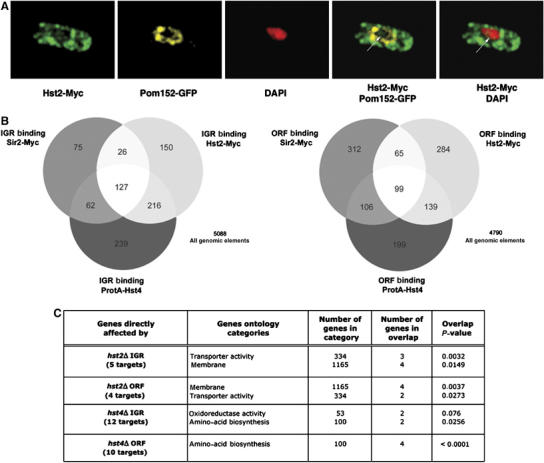
A nuclear pool of Hst2 shares chromatin binding targets with other Sirtuins in ORF and IGR regions (A) Deconvolved IF images of cells (Hu1627, Hst2-myc pom152-GFP) stained with DAPI (blue) anti-GFP (green) and anti-myc (red). Hst2-myc is mainly cytoplasmic, but a small fraction of Hst2-myc is localized at chromatin (arrow). (B) Venn diagrams illustrating the similarity of Sirtuin IGR and ORF binding data. (C) The result of ‘GoMiner' analysis of direct target gene. repressed by Hst2 and Hst4 (see main text for details). The most significant gene ontology categories, the number of genes in each category, the number of genes in overlap and the P-values (two-sided Fisher's exact test) indicating the significance for the overlap are indicated (see Materials and methods).
Hst2 and Hst4 have specific gene targets in fission yeast
Both phenotypic and genome-wide ChIP–CHIP data suggested that Sir2 and Clr3 act cooperatively at silent heterochromatin regions (Bjerling et al, 2002; Wiren et al, 2005). Hst4 has also been implicated in heterochromatin formation (Freeman-Cook et al, 1999); however, genome-wide binding data, before this study, were not available for Hst4 and Hst2. Therefore, we performed ChIP–CHIP to allow comparison of the global binding patterns of all three S. pombe Sirtuins. We used a strain with the endogenous hst4+ promoter driving expression of the protA-Hst4 fusion protein (Freeman-Cook et al, 1999), and another strain expressing Hst2-myc under the control of the endogenous hst2+ promoter. ChIP–CHIP was performed using a specific antibody against the c-myc epitope or IgG beads, which bind protA-Hst4. A ‘median percentile rank method' (Buck and Lieb, 2004) was used to determine statistically reproducible chromosomal binding maps of Hst2-myc and protA-Hst4 (Supplementary data S2), and these were compared with the Sir2-myc binding map (Wiren et al, 2005) (see lists of IGR and ORF binding targets in Supplementary data S3). A comparison of ORF and IGR binding of the three Sirtuins is represented as a Venn diagram in Figure 3B. Overlaps between Sir2, Hst4 and Hst2 binding were all highly significant with hypergeometric P-values, ranging from 8.8e-31 to 1.8e-165. In IGR regions, we found that the three Sirtuin enzymes shared 127 common targets, including many promoter regions and microarray probe fragments, representing all of the silent heterochromatin regions, that is, mat, rDNA, cen-dg, cen-dh and tel. Similarly, we found a significant number of shared targets in ORF regions.
In addition to the shared targets, it was clear that each Sirtuin also had a large number of Sirtuin exclusive IGR and ORF binding sites not shared by other Sirtuins, indicating that they are able to associate independently with some loci. We used the exclusive binding targets to define lists of genes directly affected by Hst2 and Hst4 (Figure 3C; Supplementary data S4). It was clear from the gene ontology analysis of such genes that Hst4 directly represses genes involved in amino-acid biosynthesis (P<0.0001) and oxidoreductase activity, whereas Hst2 directly represses gene involved in membrane function and transport. In contrast, no statistically significant gene ontology category was overrepresented for Sir2-bound and Sir2-repressed genes. Thus, Hst2 and Hst4 directly repress genes involved in distinct cellular functions.
Distinct in vivo specificities of fission yeast Sirtuins
To investigate if the three different S. pombe Sirtuins have different enzymatic in vivo specificities, we systematically compared lists of IGR and ORF regions, where histone acetylation was changed in Sirtuin mutants to lists of IGR and ORF bound by Sirtuins (see Supplementary data S6). As expected, this comparison revealed that Sir2-myc-bound fragments were hyperacetylated in sir2Δ, most significantly at H3K9Ac, in both IGR (P=1.32e-21) and ORF regions (P=1.90e-23). This observation independently confirmed our previously published data for sir2Δ, and supported the notion that Sir2 acts on H3K9Ac in vivo (Wiren et al, 2005). However, when the exclusive binding targets were used (as opposed to the total number of targets), Sir2 binding matched best with hyperacetylation of H4K16Ac in sir2Δ at ORF (P=1.25e-25) and H4K12Ac in IGR regions (P=1.28e-06). Thus, this suggested that in the absence the other two Sirtuins, Sir2 would favor these two sites in vivo. Regarding Hst4, hyperacetylation of H3K14Ac in hst4Δ was most significant in Hst4-bound IGR regions. This was true both in the total number of Hst4-bound IGR regions (P=7.8e-26) and in Hst4-exclusive IGR regions, that is, without Hst2 or Sir2 (P=5.5e-07). A difference was observed in ORF regions, where in the case of the total number of Hst4 sites, we observed a slightly higher preference for hyperacetylation of H3K9Ac than of H4K12Ac in hst4Δ (P=1.30e-17 versus P=1.59e-16), whereas at Hst4-unique, ORF regions there was preferential hyperacetylation of H4K12Ac in hst4Δ (P=0.0175). The total number of fragments bound by Hst2-myc matched best with H3K14Ac hyperacetylation in hst2Δ both for IGR (P=3.06e-27), and in ORF regions (P=1.50e-21). Also, the exclusive Hst2 ORF targets correlated best with hyperacetylation of H3K9Ac in hst2Δ (P=0.00047). Thus, without the other two Sirtuins, Hst2 prefers H3K9Ac in vivo. Hence, comparison of enzyme binding with changes in histone acetylation in the respective Sirtuin mutants suggested that the Sirtuin mutants displayed characteristic in vivo specificities (See Discussion).
Hst4 is required for retrotransposon silencing and RNA processing
The expression and acetylation profiling of hst4Δ (Figures 1A and 2C) suggested that Hst4 is normally required to set appropriate levels of expression for medium to highly expressed genes primarily by interactions in the ORF regions. To define the highly expressed genes that were normally tuned down by Hst4, the lists were compared with lists of highly expressed genes in wt, upregulated genes in hst4 and binding of Hst4. The resulting intersection contained 20 direct Hst4 target genes and was dominated by Tf2 retrotransposons or pseudoretrotransposons (Figure 4A). Since our ChIP–CHIP analysis had revealed that all three Sirtuins bind to Tf2 elements to some extent, we analyzed Sirtuin binding at solo-LTR elements. In the S. pombe genome, all 13 Tf2 elements are flanked by LTR elements, but there are 186 solo-LTR elements thought to be the consequence of LTR–LTR recombination of previously existing LTR retroelements and a majority of these are within 1 kb of transcription start sites (Bowen et al, 2003). Thus, we generated a list of genes having a solo-LTR near or within the promoter region (177 genes). A total of 33.1% of these solo-LTR containing promoter elements showed Hst4 binding (59 of 644 Hst4 IGR targets were solo-LTR; P=9.68e-15). The other Sirtuins were less significantly bound to solo-LTR elements (Sir2 P=2.69e-06, Hst2 P=0.0202). Hence, solo-LTR element can target Sirtuins, in particular, Hst4, to Tf2 elements and many promoters.
Figure 4.
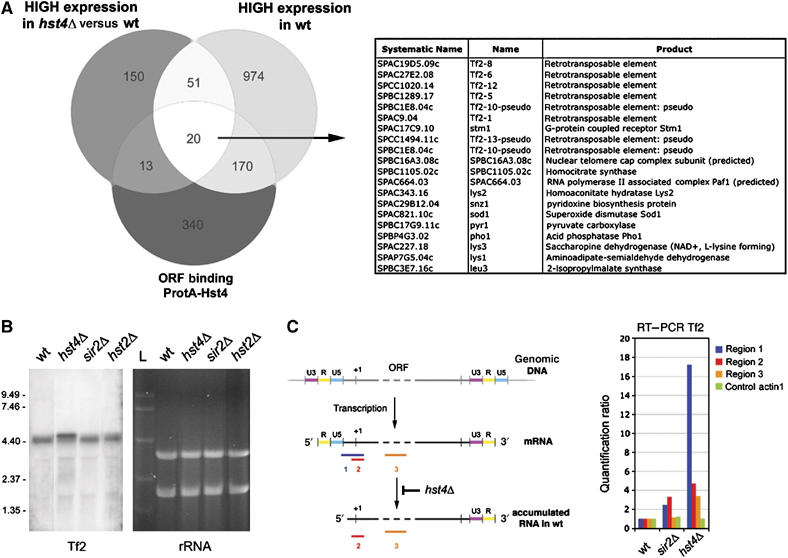
Tf2 retrotransposable elements are silenced and 5′ mRNA processed by Hst4. (A) A Venn diagram representation of genes highly expressed in wt, which are further upregulated in hst4 and ORF regions bound by Hst4. A list of the 20 genes in the intersect and a description of each gene product is provided (right). (B) Northern blot showing RNA levels of Tf2 elements in wt and Sirtuin mutants (as indicated). The EtBr-stained rRNA signals were used as loading controls (right). (C) A bar diagram showing quantitative RT–PCR analysis of Tf2 elements in Sirtuin mutants (right). A schematic representation of the structure of the Tf2 RNA is shown (left) and the processing step deficient in hst4Δ is indicated. Region 1 primers specifically amplify the unprocessed Tf2 mRNA, whereas regions 2 and 3 primers amplify the total Tf2 mRNA (unprocessed and processed forms).
Next, we tested all three Sirtuin mutants for Tf2 expression by northern blots (Figure 4B). Clearly, Tf2 RNA was upregulated only in the hst4 mutant and the Tf2 RNA also showed a decreased mobility in hst4Δ, indicating a larger molecular size. During the first steps of reverse transcription, the closely related Tf1 retrotransposon in S. pombe uses a self-priming mechanism, which trims down the 5′ part of the LTR mRNA to generate a template (Lin and Levin, 1998). Therefore, we reasoned that the Tf2 RNA species detected by northern blots in wt, hst2Δ and sir2Δ had been processed by this molecular event, and a 5′ processing defect may be occurring in the hst4 mutant. To test this possibility, we carried out 5′RACE mapping of Tf2 RNA transcripts in hst4 and wt. The 5′ cDNA sequences in wt were consistent with the processed form of Tf2 RNA; however, the 5′ sequences in hst4Δ (see Supplementary data S7) matched perfectly with the intact Tf2 mRNA, indicating that Hst4 is required for the specific cleavage in the 5′ part of Tf2 RNA. To further investigate this phenomenon, we performed quantitative RT–PCR analysis of Tf2 transcripts in wt, sir2 and hst4 mutants (Figure 4C). The unprocessed Tf2 transcript specifically detected by region 1 RT–PCR was about 16-fold more abundant in hst4Δ as compared with wt. This was strikingly different from RT–PCR of regions 2 and 3, which detected both processed and unprocessed forms of Tf2 RNA and showed 3- to 5-fold increased levels in hst4Δ. Thus, Hst4 has a role in Tf2 silencing and 5′ mRNA processing.
A role for Hst2 in heterochromatin silencing
Extensive studies have been performed in fission yeast on sir2Δ and hst4Δ mutants showing severe silencing defects in heterochromatin regions (Freeman-Cook et al, 1999, 2005; Shankaranarayana et al, 2003). We wished to know whether Hst2 also regulates heterochromatin silencing. Our ChIP–CHIP binding data revealed that Hst2 as well as Sir2 and Hst4 directly bind to the silent regions, suggesting that Hst2 may also be required for heterochromatin formation. We therefore generated hst2Δ strains with ura4 marker genes integrated into the various heterochromatin regions in S. pombe: the mating-type region, the telomere region, the rDNA region, the outer repetitive domains of centromere 1, the imr1 region and the central core region of centromere 2. Briefly, silencing of the S. pombe ura4 reporter can be detected by reduced growth on medium lacking uracil (−ura) or by increased growth on media containing 5-fluoro-orotic acid (FOA). Loss of silencing is manifested as increased growth on −ura medium and decreased growth on 5-FOA. All Sirtuin mutant strains with an inserted ura4+ reporter within the rDNA locus showed plating efficiencies on +ura media similar to the wt strain harboring the same reporter, but on −ura plates, sir2Δ and hst2Δ strains showed plating efficiencies about 5-fold higher than wt (Figure 5A). On FOA media, sir2Δ and hst2Δ strains both showed reduced plating efficiencies as compared with wt, and this was most apparent for sir2Δ, indicating that the rDNA silencing defect of hst2Δ is somewhat weaker than that of sir2Δ. In the case of the centromere 1 imr repeats (imr1), both sir2Δ and hst4Δ were previously reported to be required for silencing. We found that the hst2Δ mutant also alleviated the ura4 silencing at imr1 slightly, indicating that Hst2 activity is normally required to silence imr1 together with Hst4 and Sir2 activity (Figure 5B). Our analyses of the other silent regions did not reveal any detectable alleviation of silencing by the hst2 mutation (Supplementary Figure S5). Therefore, we concluded from these silencing assays that Hst2 in S. pombe plays a similar role as the other Sirtuins in transcriptional silencing of the rDNA regions, as well as the imr1 regions at the centromere. A complete overview of Sirtuin silencing function, from our analysis and others laboratories, is presented in Figure 5C. Each Sirtuin mutant displays a characteristic spectrum of silencing defects in the various heterochromatin regions. The genome-wide binding maps (Supplementary Table S3) revealed that Sir2, Hst2 and Hst4 bind all of the silent regions. Hence, it is likely that Sirtuins have partly redundant roles in heterochromatin formation.
Figure 5.
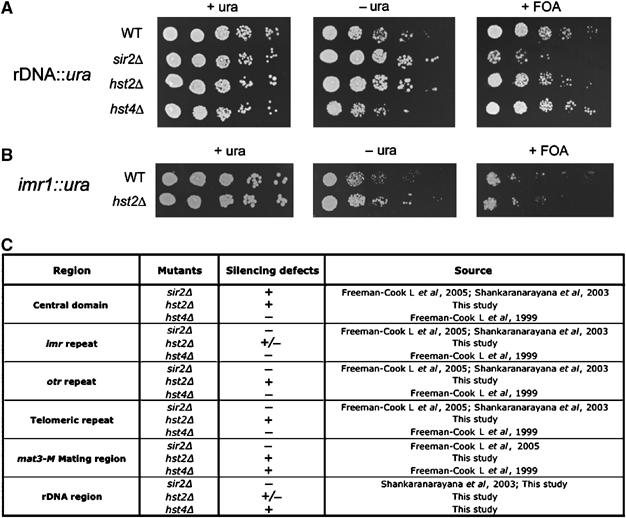
Hst2 is required for rDNA and imr1 silencing in S. pombe (A) hst2Δ but not hst4Δ mutants derepress the rDNA locus. Fivefold dilutions of cells were plated onto EMM plates as a growth control (+ura), onto EMM plates lacking uracil (−ura) and onto 5-FOA plates (+FOA) to assay silencing of the ura4+ reporter gene inserted in the rDNA locus. Strains used were (wt) Hu393, (sir2Δ) Hu1408, (hst2Δ) Hu1409 and (hst4Δ) Hu1430. (B) hst2Δ derepresses the imr1 locus. Fivefold dilutions were plated onto EMM plates as a growth control (+ura), onto EMM plates lacking uracil (−ura) and onto 5-FOA plates (+FOA) to assay silencing of the ura4+ reporter gene inserted in the imr1 locus. Strains used were (wt) FY0498 and (hst2Δ) Hu1587. (C) A table summarizing the current and previous studies of Sirtuin mutant silencing phenotypes (−, defect; ±, partial or weak defect; +, no defect , that is, like wt).
Hst2 and Hst4 are involved in microtubule function and UV DNA damage response
To further understand the functional relationships of Sirtuins members, we analyzed the phenotypes of single and double mutant combinations of hst2Δ, sir2Δ and hst4Δ. Various growth conditions and drug treatments were used to compare the different Sirtuin mutants (Figure 6A). As expected, we reproduced a number of previously observed results for hst4Δ cells (Freeman-Cook et al, 1999), that is, growth defects, sensitivity to hydroxyurea, temperature, the microtubule destabilizing drug thiabendazole (TBZ) and DNA damage sensitivities (phleomycin and UV). Moreover, in good agreement with an earlier study (Freeman-Cook et al, 2005), our analysis of sir2Δ mutant did not reveal any detectable phenotypes under the same conditions. Also in the case of hst2Δ, we were not able to detect a particular sensitivity to the various growth conditions and drug treatments. To check for synthetic interactions between Sirtuins, we generated strains carrying double deletions. Growth of the double mutants was assayed using the different conditions: low temperature (25°C), treatments with TBZ and UV (Figure 6B). At normal incubation temperature, that is, 30°C, all mutants except hst4Δ grew at wt rates. hst4Δ and doubles with hst4Δ had a slightly reduced growth rate. At 25°C, hst4Δ and doubles with hst4Δ grew very poorly, and in addition, we observed a slightly reduced growth in sir2Δhst2Δ. Interestingly, UV irradiation experiments revealed that sir2Δ hst4Δ and hst2Δ hst4Δ double mutants were more sensitive to UV than hst4Δ. This suggested that Sir2 and Hst2 partially compensate for the absence of Hst4 in the hst4Δ cells after UV-induced DNA damage. Treatment with the microtubule destabilizing drug TBZ revealed another synthetic interaction between hst2Δ and hst4Δ, since this double mutant grew less well than the single mutants. In addition, sir2Δ hst2Δ showed increased TBZ sensitivity as compared with the single mutants. This suggested that Hst2 has roles in microtubule function, in parallel with Hst4 and Sir2, and that Sir2 has roles in UV damage response and cell, together with Hst2 and Hst4.
Figure 6.
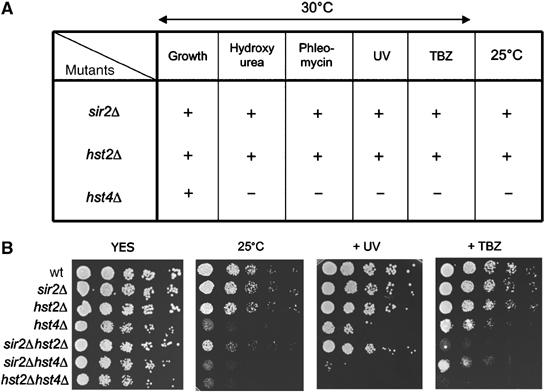
Genetic interactions between Sirtuin mutants in S. pombe (A) A table summarizing the different single mutant phenotypes observed (plating data are not shown). Fivefold dilutions of mid-exponential phase cells were plated onto (YES) rich media. Plating assays were performed at 30°C for hydroxyurea (1 and 2 mM), phleomycin (0.25, 1.0, 1.5 μg/ml), UV (60, 80, 100 μJ cm−2) and thiaminobendazole (TBZ) (5, 10, 15 μg/ml). Growth defects are represented by + (no observed effect) and − (defect). (B) Plating assays of single and double mutants, which reveal synthetic genetic interactions. Fivefold dilutions of mid-exponential phase cells were plated onto YES rich media at 30°C. Cells on the two left plates were not treated, the middle plate was exposed with 80 μJ cm−2 and the right plate contained 10 μg/ml of TBZ.
Discussion
Genome-wide functions of the Hst2 and Hst4 enzymes
Two fission yeast HDAC genes, clr3+ and clr6+, were discovered by forward genetic screens for factors required for heterochromatin silencing (Ekwall and Ruusala, 1994; Grewal et al, 1998). Subsequently, the hst4+ and sir2+ genes were shown to be required for heterochromatic silencing (Freeman-Cook et al, 1999; Shankaranarayana et al, 2003), and hos2 was shown to enhance heterochromatin silencing (Olsson et al, 1998). However, it was clear that HDACs would have additional cellular roles apart from their functions in heterochromatin. In total, there are six HDACs in fission yeast, four of which we have described previously in terms of their genome-wide functions: Clr6, Hos2, Clr3 and Sir2 (Wiren et al, 2005). Here, we have attempted to dissect the genome-wide functions of the Hst2 and Hst4 enzymes, thus completing the genome-wide analysis of HDACs in fission yeast. We wished to know what the general roles are for Hst2 and Hst4 in regulation of gene expression—are they activators or repressors? We asked whether they are specifically involved in regulation of genes involved in certain cellular processes, and if so, whether their different roles in gene regulation would be reflected in distinct phenotypes of hst2 and hst4 mutants. We also asked if compared with Sir2, Hst2 and Hst4 show different substrate specificity at in vivo targets? Finally, we characterized one interesting Hst4-specific process, namely retrotransposon silencing and RNA processing.
General roles for Hst2 and Hst4 in regulation of gene expression—are they activators or repressors?
The paradigm role of histone acetylation in gene regulation is that it is required for gene activation (Allfrey et al, 1964; Brownell et al, 1996). HDACs are therefore generally thought of as corepressors of transcription. However, both in fission and budding yeast, evidence has been presented supporting a role for Hos2 in promoting gene expression (Wang et al, 2002; Wiren et al, 2005). In fission yeast, ORF regions generally accumulate histone hyperacetylation in a hos2Δ mutant and this correlates with reduced expression of genes, which are highly expressed in wt cells (Wiren et al, 2005). Surprisingly, this study showed a similar hyperacetylation of histones in ORF regions in hst4Δ mutant; however, in contrast to hos2Δ, expressed genes were globally upregulated in hst4Δ. Thus, a significant fraction of the hst4Δ upregulated genes were already relatively highly expressed in wt cells and become further derepressed in hst4Δ (Figure 4A). Hence, it seems as if Hos2 and Hst4 in S. pombe both affect histone acetylation levels primarily within expressed ORFs; however, they appear to have completely opposing functions in regulation of gene expression. To test if they affected the same set of genes but in opposite ways, we compared the gene lists of genes downregulated in hos2Δ and upregulated in hst4Δ and found no significant similarity (data not shown). This suggested that the two enzymes mainly act upon distinct sets of highly expressed genes. It is possible that the down-tuning of expression by SpHst4 is similar to the role of Rpd3 in budding yeast, which reduces histone acetylation and basal transcription globally in large chromosomal regions (Vogelauer et al, 2000). In some chromosomal regions near Tf2 elements, acetylation changes could be observed to extend past the regions directly bound by SpHst4. The ‘global expression profile' for hst2 was similar to that of sir2, with a less pronounced tendency for derepression of genes, which show low expression in wt (Figure 1A). Thus, in contrast to Hos2, both Hst4 and Hst2 primarily function in repression of gene expression.
The Sirtuins show different substrate specificities in vivo
Genome-wide binding and genotypic analysis of Clr3 and Sir2 in fission yeast had revealed shared functions to control genes and heterochromatin regions (Bjerling et al, 2002; Freeman-Cook et al, 2005; Wiren et al, 2005). The genome-wide binding and phenotypic analysis presented here revealed that Hst2 colocalizes with Sir2 and Hst4 in heterochromatic regions and at many genes. Probably because of the extensive co-occupancy between Sirtuins, the comparison of Sirtuin binding maps with histone acetylation changes in Sirtuin mutants did not reveal any striking differences between the mutants (Supplementary Figure S6A and B). However, when analyzing acetylation changes in Sirtuin mutants at binding sites unique to each Sirtuin, there were clear differences (Supplementary Figure S6C and D). For example, the only significant hyperacetylation detected in hst2 at Hst2 exclusive ORF regions was of H3K9Ac, whereas significant hyperacetylation of H4K12Ac and H4K16Ac was detected in hst4 at Hst4 exclusive ORF regions. When considering all Sir2 binding sites, H3K9Ac was the most significant hyperacetylated modification in sir2 both in IGR and ORF regions, confirming our previous genome-wide study (Wiren et al, 2005). However, for Sir2 exclusive ORF regions, we found increased H4K16Ac as the most significant hyperacetylation in sir2. In this context, it is interesting to note that the Sir2 enzyme was reported to act on both H4K16Ac and H3K9Ac in vitro (Shankaranarayana et al, 2003). We found that at Sir2 exclusive IGR regions, H4K12 hyperacetylation in sir2 was significant. In conclusion, our experiments suggest that the Sirtuins have different substrate specificities in vivo. One of these in vivo specificities agrees with previous in vitro experiments, that is, H4K16Ac for SpSir2.
Hst2 and Hst4 repress genes in distinct cellular processes and repression by Hst4 involves the Ssn6 corepressor
We found that all three Sirtuins were involved in repression of genes, which define characteristic sets of cellular functions (Figure 1D). However, when exclusive Sirtuin binding data (unique binding of one Sirtuin only) were overlayed with gene expression data to define directly affected genes, this revealed significant cellular functions only for Hst2 and Hst4 and not for Sir2 (Figure 3C; Supplementary Table S4). Therefore, apart from its role in silencing of heterochromatin regions, it is unclear if Sir2 has a specific role in gene repression in S. pombe, and it is likely that the 151 sir2 upregulated genes are indirect effects of the sir2 gene deletion. In contrast, the gene ontology searches revealed statistically significant gene regulatory roles for Hst2 in transport activity and membrane function, and for Hst4 in oxidoreductase activity and amino-acid biosynthesis. Interestingly, all of the Hst4-repressed ORF targets, which are involved in amino-acid biosynthesis (leu3, lys3, lys1, SPBC1105.02c and SPAC222.08c), are also direct targets for the Ssn6-Tup11/12 corepressor complex and are affected by Ssn6 overexpression (Fagerstrom-Billai et al, 2007), suggesting that Ssn6 could be involved in targeting of the class III HDAC Hst4 to repress this set of genes. In budding yeast, Ssn6/Tup1 physically interacts with both class I and II HDACs, and Ssn6/Tup1 has shared roles with such HDACs in gene repression (Watson et al, 2000; Wu et al, 2001; Robyr et al, 2002; Davie et al, 2003). Our findings in S. pombe show that Ssn6 is also able act in concert with a class III HDAC. Regarding the physiological role of Hst4, we assume that high expression of amino-acid biosynthesis genes is not needed under the experimental conditions tested, that is, in rapidly growing cells in rich medium supplemented with external amino acids.
Are the different roles of Hst2 and Hst4 in gene regulation reflected in distinct phenotypes?
The hst2Δ sir2Δ and the hst2Δ hst4Δ double mutants in S. pombe showed synthetic genetic interactions on TBZ plates (Figure 6). Since TBZ is a microtubule destabilizing drug, this indicated that the Sirtuin mutants cause compound perturbations in microtubule function. Interestingly, we also found that several genes involved in microtubule cytoskeleton function were misregulated in hst2Δ (Figure 1A). Therefore, it is conceivable that a double deletion of hst2Δ and hst4Δ, or sir2Δ, aggravates this gene regulation defect and causes an increased TBZ sensitivity. Alternatively, the increased TBZ sensitivity of the double mutants could be due to additive defects in chromosome segregation since all three mutants also affect centromeric heterochromatin function (Figure 5C). In general, mutations that affect centromeric heterochromatin structure in fission yeast are supersensitive to TBZ (Ekwall et al, 1999). In any case, our phenotypic analysis clearly demonstrated synthetic genetic interactions between Sirtuin mutants in S. pombe. Synthetic interactions between hst2 and sir2 were also reported in the silencing of budding yeast rDNA (Perrod et al, 2001; Lamming et al, 2005, 2006) and life-span regulation under calorie restriction (Lamming et al, 2005). Thus, in both yeasts, Hst2 is able to backup Sir2 functions, although this affects different cell physiology functions in the different yeasts.
A role for Hst4 in retrotransposon silencing
Our genome-wide data indicated that all three Sirtuins bind Tf2 retrotransposons to some extent. However, Hst4 showed the most extensive binding to Tf2s, and only one of the Sirtuin mutants, hst4Δ, displayed defective silencing of Tf2 elements. Additionally, only Hst4 showed more significant binding to promoter regions containing solo-LTR elements, which are derived from Tf2s, and only hst4Δ has a direct role in both silencing of Tf2s and a role in 5′ processing of the Tf2 mRNA. Transposable elements in plants are known to be silenced via epigenetic mechanisms involving small regulatory RNA, the RNAi machinery, a remodelling factor ddm1, DNA methylation and a class II HDAC sil1/Hda6 (Lippman et al, 2003; May et al, 2005). In budding yeast, Ty1 retrotransposons inserted in rDNA region are subject to silencing mediated by SIR2 (Bryk et al, 1997). However, to our knowledge, the Sir2 family of HDAC enzymes (class III) has not previously been implicated in mRNA processing of retrotransposons. Since all three Sirtuins are also involved in heterochromatin silencing of centromere repeats (Figure 5C), and that this process also involves the RNA processing via the RNAi machinery (Volpe et al, 2002), we were interested in determining if the RNAi machinery would also be involved in Tf2 silencing. Expression profiling of the RNAi machinery mutants, ago1, dcr1 and rdp1 revealed only slight defects in Tf2 silencing (Hansen et al, 2005). We found no role for Dicer in Tf2 mRNA processing and, unlike dcr1, the hst4 mutant did not accumulate unprocessed centromeric pre-siRNA (M Durand-Dubief and K Ekwall, unpublished observation). Thus, it is clear that the Tf2 silencing function of Hst4 is mechanistically distinct from its function in centromeric silencing.
Materials and methods
Microarrays
All the microarray data in this study have been submitted to Gene Expression Omnibus (GEO) at http://www.ncbi.nlm.nih.gov/geo/ with accession number GSE6114. The combined microarray strategy in this study was performed essentially as outlined in (Wiren et al, 2005). cDNA expression profiling of hst2Δ mutant was carried out according to (Xue et al, 2004).
We used the S. pombe ORF and combined IGR+ORF spotted microarrays (Eurogentec, Belgium). For histone acetylation maps, ChIP–CHIP method was essentially used using according to Robyr and Grunstein (2003). Antibodies against H3K9Ac, H3K14Ac, H4K5Ac, H4K16Ac and H4K12Ac (Suka et al, 2002) were used. The histone ‘H3cter' antibody was purchased from Upstate and used for ChIP according to manufacturer's recommendations. For the Hst2-myc binding map experiments, we used the ChIP–CHIP procedure described by Kurdistani et al (2002). For ProtA-Hst4 binding map experiments, immunoprecipitation was performed with rabbit IgG sepharose beads (Sigma). Corrections for nucleosome loss were performed as described in Wiren et al (2005). See Supplementary data S1 for further details.
Media and spotting assays
Standard fission yeast genetic techniques and media were used according to Moreno et al (1991). For microarray analysis, cells were grown in YES media. Silencing assays plating and serial dilution experiments were performed as described (Allshire et al, 1994). Fivefold dilutions were plated onto EMM plates as a growth control (+ura), onto EMM plates lacking uracil (−ura) and onto 5-FOA plates (+FOA). Thiabendazole (TBZ; Sigma) was dissolved in DMSO to give stock solution at 20 mg/ml.
Construction of strains
Haploidization of diploid heterozygote strains h+/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 hst2+/hst2Δ::kanMx4 and h+/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 hst4+/hst4Δ::kanMx4 (Bioneer Corporation) was performed in YES liquid media containing 10 μg/ml of TBZ during 2–3 days. The cells were then plated on YES (–ade) plates containing 150 μg/ml G418 (Sigma). G418-resistant haploid colonies with red or pink colony color were then selected, checked by PCR and backcrossed with the wt strain Hu303. 13 × Myc epitope tagging of Hst2 was performed according to Bahler et al (1998). All S. pombe strains used in this study are listed in Supplementary data S1.
IF microscopy
S. pombe cells were prepared for IF microscopy according to Bjerling et al (2002). See Supplementary data S1 for further details.
RNA analysis
For Northern blotting, total RNA of Hu0303 (wt), Hu1103 (hst4Δ), Hu1098 (sir2Δ) and Hu1251 (hst2Δ) was extracted according (Xue et al, 2004). See Supplementary data S1 for further details.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
KE is a Royal Swedish Academy of Sciences Research Fellow, supported by grants from Knut and Alice Wallenberg Foundation, Swedish Cancer Society, Swedish Research Council (VR) M Bergvalls stiftelse and EU ‘The Epigenome' NoE network. This work was initiated during a short sabbatical stay for KE at MG's laboratory at UCLA. AW is a VR Research Fellow, supported by grants from VR and the Swedish Cancer Society. We thank L Pillus for kindly providing strains, H Levin for kindly providing a list of solo-LTR regions and Rebecca Silverstein for critical reading of the manuscript.
References
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G (1994) Position effect variegation at fission yeast centromeres. Cell 76: 157–169 [DOI] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE (1964) Acetylation and methylation of histones and their possible role in the regulation of Rna synthesis. Proc Natl Acad Sci USA 51: 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bjerling P, Silverstein RA, Thon G, Caudy A, Grewal S, Ekwall K (2002) Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol Cell Biol 22: 2170–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Guarente L (2004) The Sir2 family of protein deacetylases. Annu Rev Biochem 73: 417–435 [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL (2003) Retrotransposons and their recognition of pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res 13: 1984–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev 9: 2888–2902 [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851 [DOI] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ (1997) Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev 11: 255–269 [DOI] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD (2004) ChIP–chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83: 349–360 [DOI] [PubMed] [Google Scholar]
- Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A (2006) The sirtuins hst3 and hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol 16: 1280–1289 [DOI] [PubMed] [Google Scholar]
- Davie JK, Edmondson DG, Coco CB, Dent SY (2003) Tup1–Ssn6 interacts with multiple class I histone deacetylases in vivo. J Biol Chem 278: 50158–50162 [DOI] [PubMed] [Google Scholar]
- Ding DQ, Tomita Y, Yamamoto A, Chikashige Y, Haraguchi T, Hiraoka Y (2000) Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells 5: 169–190 [DOI] [PubMed] [Google Scholar]
- Ekwall K (2005) Genome-wide analysis of HDAC function. Trends Genet 21: 608–615 [DOI] [PubMed] [Google Scholar]
- Ekwall K, Cranston G, Allshire RC (1999) Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics 153: 1153–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Ruusala T (1994) Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics 136: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerstrom-Billai F, Durand-Dubief M, Ekwall K, Wright AP (2007) Individual subunits of the ssn6-tup11/12 corepressor are selectively required for repression of different target genes. Mol Cell Biol 27: 1069–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman-Cook LL, Gomez EB, Spedale EJ, Marlett J, Forsburg SL, Pillus L, Laurenson P (2005) Conserved locus-specific silencing functions of Schizosaccharomyces pombe sir2+. Genetics 169: 1243–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman-Cook LL, Sherman JM, Brachmann CB, Allshire RC, Boeke JD, Pillus L (1999) The Schizosaccharomyces pombe hst4(+) gene is a SIR2 homologue with silencing and centromeric functions. Mol Biol Cell 10: 3171–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Bonaduce MJ, Klar AJ (1998) Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR (2004) Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415 [DOI] [PubMed] [Google Scholar]
- Hansen KR, Burns G, Mata J, Volpe TA, Martienssen RA, Bahler J, Thon G (2005) Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol Cell Biol 25: 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK, Grunstein M (2003) Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol 4: 276–284 [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Robyr D, Tavazoie S, Grunstein M (2002) Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet 31: 248–254 [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA (2005) HST2 mediates SIR2-independent life-span extension by calorie restriction. Science 309: 1861–1864 [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA (2006) Response to comment on ‘HST2 mediates SIR2-independent life-span extension by calorie restriction'. Science 312: 1312. [DOI] [PubMed] [Google Scholar]
- Lin JH, Levin HL (1998) Reverse transcription of a self-primed retrotransposon requires an RNA structure similar to the U5-IR stem–loop of retroviruses. Mol Cell Biol 18: 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R (2003) Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol 1: E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas NL, Miller KM, Defazio LG, Toczyski DP (2006) Cell cycle and checkpoint regulation of histone h3 k56 acetylation by hst3 and hst4. Mol Cell 23: 109–119 [DOI] [PubMed] [Google Scholar]
- May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA (2005) Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet 1: e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Grunstein M (2006) Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol 7: 657–666 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- North BJ, Verdin E (2004) Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol 5: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson TG, Ekwall K, Allshire RC, Sunnerhagen P, Partridge JF, Richardson WA (1998) Genetic characterisation of hda1+, a putative fission yeast histone deacetylase gene. Nucleic Acids Res 26: 3247–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrod S, Cockell MM, Laroche T, Renauld H, Ducrest AL, Bonnard C, Gasser SM (2001) A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J 20: 197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Horz W (2003) Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11: 1599–1607 [DOI] [PubMed] [Google Scholar]
- Robyr D, Grunstein M (2003) Genomewide histone acetylation microarrays. Methods 31: 83–89 [DOI] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M (2002) Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437–446 [DOI] [PubMed] [Google Scholar]
- Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI (2003) Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol 13: 1240–1246 [DOI] [PubMed] [Google Scholar]
- Sinha I, Wiren M, Ekwall K (2006) Genome-wide patterns of histone modifications in fission yeast. Chromosome Res 14: 95–105 [DOI] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M (2002) Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet 32: 378–383 [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D (2006) SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev 20: 1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Dequiedt F, Kasler HG (2003) Class II histone deacetylases: versatile regulators. Trends Genet 19: 286–293 [DOI] [PubMed] [Google Scholar]
- Vijg J, Suh Y (2006) Ageing: chromatin unbound. Nature 440: 874–875 [DOI] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M (2000) Global histone acetylation and deacetylation in yeast. Nature 408: 495–498 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M (2002) Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298: 1412–1414 [DOI] [PubMed] [Google Scholar]
- Watson AD, Edmondson DG, Bone JR, Mukai Y, Yu Y, Du W, Stillman DJ, Roth SY (2000) Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev 14: 2737–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JM, Le VQ, Zimmerman C, Marmorstein R, Pillus L (2006) Nuclear export modulates the cytoplasmic Sir2 homologue Hst2. EMBO Rep 7: 1247–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiren M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, Laurenson P, Pillus L, Robyr D, Grunstein M, Ekwall K (2005) Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J 24: 2906–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Suka N, Carlson M, Grunstein M (2001) TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol Cell 7: 117–126 [DOI] [PubMed] [Google Scholar]
- Xue Y, Haas SA, Brino L, Gusnanto A, Reimers M, Talibi D, Vingron M, Ekwall K, Wright AP (2004) A DNA microarray for fission yeast: minimal changes in global gene expression after temperature shift. Yeast 21: 25–39 [DOI] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, Riss J, Barrett JC, Weinstein JN (2003) GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol 4: R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data


