Abstract
Colicin Ia is a 69 kDa protein that kills susceptible Escherichia coli cells by binding to a specific receptor in the outer membrane, colicin I receptor (70 kDa), and subsequently translocating its channel forming domain across the periplasmic space, where it inserts into the inner membrane and forms a voltage-dependent ion channel. We determined crystal structures of colicin I receptor alone and in complex with the receptor binding domain of colicin Ia. The receptor undergoes large and unusual conformational changes upon colicin binding, opening at the cell surface and positioning the receptor binding domain of colicin Ia directly above it. We modelled the interaction with full-length colicin Ia to show that the channel forming domain is initially positioned 150 Å above the cell surface. Functional data using full-length colicin Ia show that colicin I receptor is necessary for cell surface binding, and suggest that the receptor participates in translocation of colicin Ia across the outer membrane.
Keywords: active transport, colicin, outer membrane protein, protein import, TonB
Introduction
In addition to transporting iron complexes and vitamin B12, many TonB-dependent transporters are misappropriated by large proteins, called bacteriocins, to penetrate and kill susceptible Escherichia coli cells. Colicins (E. coli-specific bacteriocins) are toxic molecules secreted by certain bacterial strains to kill other bacteria in times of stress. It has been estimated that just one colicin molecule is required to kill a bacterial cell (Nomura, 1967). Colicins fall into two groups: group A colicins are translocated across the outer membrane with the help of the Tol system, consisting of proteins TolA, TolB, TolQ, TolR, and Pal (Lazdunski et al, 1998). Group B colicins require the Ton system, consisting of proteins TonB, ExbB, and ExbD (Braun et al, 2002). Both groups of colicins can bind to TonB-dependent transporters at the cell surface, but they appear to use different entry mechanisms for translocation.
How do large proteins penetrate the outer membrane? Many group A (Tol dependent) colicins bind to BtuB, a TonB-dependent transporter for cobalamin (Kadner, 1990; Supplementary Table I). These colicins appear to use BtuB as a specific cell surface receptor requiring a cotransporter for entry into the cell. The crystal structure of BtuB bound to the receptor binding domain of colicin E3 revealed a 1:1 complex, with colicin E3 protruding from the extracellular side of BtuB at an angle of approximately 45° to the membrane bilayer (Kurisu et al, 2003). Because colicins are elongated molecules, this orientation was suggested to aid recruitment of OmpF, the cotransporter for most colicins targeting BtuB (Kurisu et al, 2003; Law et al, 2003). The recent isolation of a BtuB–OmpF–colicin E9 complex confirmed recruitment of OmpF by colicin E9 and suggested that the N-terminal part of this colicin crosses the outer membrane through OmpF (Housden et al, 2005).
Group B colicins also target TonB-dependent transporters, but no requirement for a cotransporter has been identified (Supplementary Table I) and the entry mechanism is less well understood. One of the best characterized group B colicins is colicin Ia (Wiener et al, 1997; Stroud et al, 1998; Jakes et al, 1999), which targets the TonB-dependent transporter Cir (Konisky, 1972; Konisky and Cowell, 1972). Colicin Ia is a 69 kDa protein that can be divided into three domains according to function and structure (Supplementary Figure 1): the N-terminal translocation (T) domain, which is absolutely required for penetration of the target cell, a receptor binding (R) domain, which allows the colicin to specifically bind to Cir, and a C-terminal channel forming (C) domain that inserts into the inner membrane of the targeted cell, forming a lethal voltage-gated ion channel. The unusual architecture of colicin Ia makes it difficult to imagine how this protein is translocated across the outer membrane: the T- and C-domains are separated from the R-domain by a pair of alpha helices ∼160 Å long (Wiener et al, 1997).
Although no cotransporters have been identified for group B colicins, it is currently unclear whether TonB-dependent transporters can be used directly for cell permeation. In their ground state, TonB-dependent transporters consist of a 22-stranded transmembrane beta barrel with a luminal ‘plug' domain positioned inside (Ferguson and Deisenhofer, 2004; Wiener, 2005). TonB-dependent transport is normally initiated by binding a metal chelate at the extracellular surface of the transporter. Passage of the small molecule across the outer membrane requires energy derived from the protonmotive force and interaction with an integral inner membrane protein complex TonB–ExbB–ExbD (Postle and Kadner, 2003). Although many of the details are not understood, TonB appears to span or cross the periplasm and physically interact with the transporter; the transporter–TonB interaction occurs in a region of five residues near the transporter's N-terminus, called the TonB box. Mutations in the TonB box that change the conformation of the polypeptide, such as to proline or glycine, abolish the interaction with TonB, preventing energy coupling and subsequent transport of the metal complex (Kadner, 1990). The TonB–transporter interaction is illustrated by recent crystal structures of two TonB-dependent transporters bound to a periplasmic portion of TonB (Pawelek et al, 2006; Shultis et al, 2006). Both structures show that about half of the periplasmic surface of the plug domain is shielded by association with TonB; however, the plug domain remains inside the barrel in its ground-state conformation. Presumably, energy derived from the protonmotive force is required to produce a conformational change in the plug domain or pull it out, opening a transmembrane passage for the metal chelate. If colicins use TonB-dependent transporters in the same way that small molecules do, a larger transmembrane passage would be required.
Colicin I receptor is a TonB-dependent transporter expressed by many E. coli strains to bind and transport Fe3+ complexed to linear catecholates, such as dihydroxybenzoyl serine, across the outer membrane (Hantke, 1990). Cir was also implicated in the uptake of catechol-substituted cephalosporins (Curtis et al, 1988; Nikaido and Rosenberg, 1990; Critchley et al, 1991; Tatsumi et al, 1995). Colicin Ia misappropriates Cir to penetrate susceptible cells. In order to learn more about group B colicin translocation across membranes, we solved crystal structures of Cir alone and in complex with the R-domain of colicin Ia (residues 282–385). Our structures show that when colicin Ia binds, it stabilizes an unusual conformation of Cir that is open and exposed on the extracellular side, with the colicin Ia R-domain positioned directly above it. We then modelled the interaction between full-length colicin Ia and Cir, showing that the C-domain (conferring toxicity) is initially positioned approximately 150 Å above the cell surface. In order to penetrate the cell either through Cir or by an alternative mechanism, large conformational changes would be needed. We also asked whether Cir is required for killing by colicin Ia, and whether the translocation process involves additional outer membrane proteins. Finally, we investigated whether colicin Ia translocation requires a functional interaction between Cir and TonB, as would be required for small molecule transport.
Results
Cir consists of a 22-stranded beta barrel with a plug inside
We crystallized Cir in the presence of the detergent 2-hydroxyethyloctylsulfoxide (CHESO) (Locher et al, 1998) and solved the structure using multiwavelength anomalous diffraction on a selenomethionine-derivatized crystal, where we had substituted four hydrophobic residues with methionine (Chimento et al, 2003). We separately expressed and purified Cir and the R-domain of colicin Ia, and then isolated a 1:1 complex using size-exclusion chromatography. Crystals of the Cir–colicin R-domain complex grew from a detergent mixture containing n-dodecyl-N,N-dimethylamine-N-oxide (LDAO) and octyl tetraethylene glycol ether (C8E4). We solved the complex structure by molecular replacement using the structures of Cir and the R-domain of colicin Ia (PDB accession codes 2HDF and 1CII). Details of the structure determinations are provided in Table I.
Table 1.
Data collection, crystallographic refinement, and model statistics
| Inflection | Peak | Remote | Cir–colicin complex | |
|---|---|---|---|---|
| Protein | Cir | Cir | Cir | Complex |
| Cell parameters | ||||
| Space group | C2 | C2 | C2 | C2 |
| a (Å) | 90.61 | 90.76 | 90.86 | 132.64 |
| b (Å) | 84.35 | 84.52 | 84.65 | 130.49 |
| c (Å) | 99.47 | 99.69 | 99.88 | 56.27 |
| β (deg) | 109.2 | 109.2 | 109.3 | 101.2 |
| Data collection statisticsa | ||||
| Beamline | APS-22ID | APS-22ID | APS-22ID | APS-22ID |
| Wavelength (Å) | 0.97940 | 0.97924 | 0.97240 | 1.0000 |
| Resolution range (Å) | 30–2.65 | 30–2.85 | 30–3.0 | 30–2.5 |
| Total reflections | 149 402 | 117 928 | 103 336 | 131 426 |
| Unique reflections | 40 440 | 32 616 | 27 940 | 32 311 |
| Completeness (%) | 99.1 (91.8) | 99.9 (100) | 99.9 (100) | 99.9 (99.9) |
| Redundancy | 3.7 (2.0) | 3.8 (3.8) | 3.7 (3.7) | 4.1 (4.0) |
| Rmerge (%)b | 0.095 (0.645) | 0.087 (0.779) | 0.086 (0.656) | 0.103 (0.570) |
| I/σI | 16.4 (1.85) | 18.2 (3.1) | 19.3 (3.3) | 15.3 (2.73) |
| MAD phases | ||||
| Phasing power (acentrics)c | 0/2.33 | 0.04/2.00 | 0.28/1.44 | |
| Phasing power overall | 2.33 | 1.92 | 1.38 | |
| FOM overall | 0.42 | |||
| Molecular replacement search models | 2HDF (Cir) | |||
| 1CII (R-domain) | ||||
| Model statistics | ||||
| Resolution (Å) | 15–2.65 | 15–2.5 | ||
| Rd | 0.2414 | 0.1910 | ||
| Rfreee | 0.2911 | 0.2493 | ||
| Refined model | ||||
| Number of residues | 584 | 701 | ||
| Number of detergent molecules | 2 | 2 | ||
| Number of water molecules | 18 | 255 | ||
| R.m.s.deviation, bonds (Å) | 0.010 | 0.010 | ||
| R.m.s.deviation, angles (deg) | 1.303 | 1.303 | ||
| Ramachandran plot (%) | ||||
| Most favored regions | 84.7 | 89.0 | ||
| Additionally allowed | 12.1 | 9.7 | ||
| Generously allowed | 2.8 | 0.7 | ||
| Disallowed regions | 0.4 | 0.7 | ||
| Average B-factor (Å2) | 59.0 | 44.9 | ||
| Range of B-factors (Å2) | 22.9–136.9 | 12.7–105.3 | ||
| Values in parentheses are for the highest-resolution shell (Å): 2.74–2.65 for inflection, 2.95–2.85 for peak, 3.10–3.00 for remote, and 2.59–2.50 for complex. | ||||
| Rmerge=∑∑I(h)j −〈I(h)〉/∑I(h), where I(h)j is the jth measurement of diffraction intensity of reflection h and 〈I(h)〉 is the average intensity of reflection h for all j measurements. | ||||
| Phasing power=∣FH(calc)∣/lack-of-closure; values are given for isomorphous and anomalous data. | ||||
| R=∑(∣Fo∣−∣Fc∣)/∑∣Fo∣. | ||||
| Rfree is calculated using a test set of 5% of the reflections excluded from refinement. | ||||
Like other TonB-dependent transporters, Cir contains an N-terminal plug domain inserted inside a 22-stranded transmembrane beta barrel (Figure 1). The plug domain consists of residues 26–163. The TonB box is visible in both structures at the N-terminal end of the plug domain and comprises residues 31–35, which adopt an extended conformation. The transmembrane beta barrel, consisting of residues 164–663, contains 22 beta strands connected by 11 long extracellular loops and 10 short periplasmic loops. The barrel has a height of about 40 Å and the cross-section of the barrel has an ellipsoid shape, which can be described by major and minor axes of approximately 50 and 40 Å, respectively. Corresponding dimensions for the pore size are 34 and 24 Å in the absence of the plug domain. Extracellular loops 7 and 8 (connecting strands 13 and 14, 15 and 16) are very long, containing 30 and 42 residues, respectively. In the Cir structure, these loops fold over the top of the plug domain and restrict access to the ligand binding pocket from the extracellular side (Figure 1A).
Figure 1.
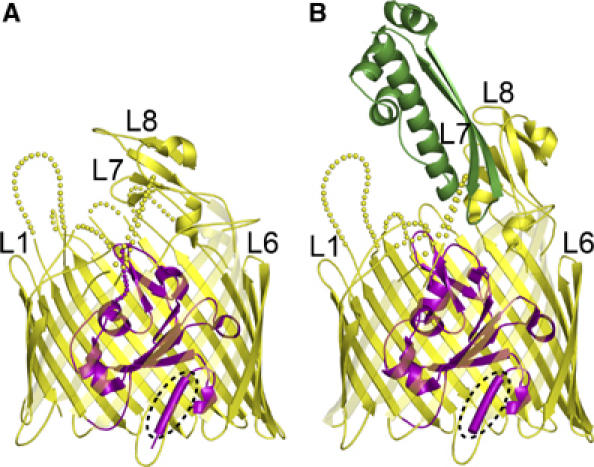
Ribbon diagrams of colicin I receptor (A) and the Cir–colicin Ia complex (B). The extracellular space is located at the top of the figure and the periplasmic space is at the bottom. Cir spans the outer membrane using a 22-stranded beta barrel (yellow) and contains a globular plug domain (magenta) within the barrel lumen. Residues comprising the TonB box (E31–T32–M33–V34–V35) are shown as a tube and encircled by a dotted line. Some extracellular loops are labelled for reference; disordered parts of the loops are indicated by dotted lines. In the Cir–colicin Ia complex, the R-domain of colicin Ia (green) interacts extensively with extracellular loops 7 and 8 of Cir. The fold of the colicin Ia R-domain is identical to that observed in the crystal structure of full-length colicin Ia (Wiener et al, 1997). All figures were prepared with PYMOL (DeLano).
The plug domain interacts extensively with the interior of the beta barrel: in the Cir–colicin structure, there are two salt bridges and 39 protein–protein hydrogen bonds coordinating the two domains. In addition, a large number of bound water molecules are found to solvate the plug–barrel interface: there are 23 bridging waters linking the two domains and approximately 70 non-bridging waters binding to either the barrel or plug in this interface (Supplementary Figure 2). The extensive solvation of the plug–barrel interface, seen in all TonB-dependent transporters, may facilitate rearrangement or removal of the plug domain during the transport process (Chimento et al, 2005).
Binding of colicin Ia is specific, tight, and mimics metal chelate trapping
Figure 1B shows the complex formed between the colicin Ia R-domain and Cir. The R-domain consists of a two-stranded beta sheet that folds around an alpha helix (Wiener et al, 1997). There are two regions on colicin Ia, where residues contribute to extensive H-bonding with Cir (Figure 2A). The first region includes a loop that forms a turn between the antiparallel beta strands and extends into the beta strand that immediately follows this loop (four residues in region 313–319). The second region covers the N-terminal end of the alpha helix and a part of the loop that immediately precedes it (seven residues in region 350–369). Both of these regions are at the tip of the colicin Ia R-domain and insert into the ligand binding pocket of Cir, penetrating approximately as deeply into the binding pocket as siderophores do in related systems (as deduced from a superposition of the structure of ferric citrate transporter (FecA) in complex with diferric dicitrate (Yue et al, 2003) with Cir–colicin Ia R-domain; shown in Figure 2B) and burying 989.5 Å2 of interaction surface upon complex formation. The corresponding H-bonding partner regions on Cir are mainly confined to residues 488–491 from loop 8 that adopt an imperfect beta strand conformation to pair in a parallel manner with the two-stranded beta sheet of colicin Ia; R490, within this strand, coordinates four additional colicin Ia residues through five hydrogen bonds. An arginine in loop 7, R436, coordinates four residues in colicin Ia through six hydrogen bonds, two of which form a salt bridge with colicin Ia residue D350. Additionally a single residue from loop 5 and one residue from an apical loop of the plug domain contribute to formation of hydrogen bonds (Supplementary Table II).
Figure 2.
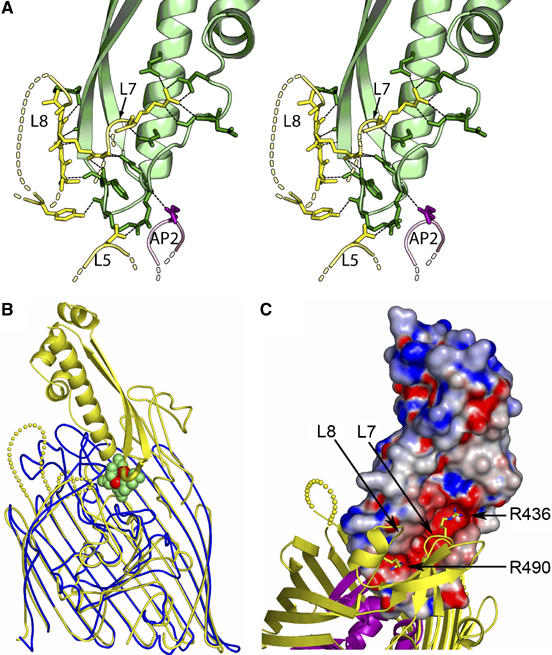
Recognition of Cir by colicin Ia. (A) Stereo illustration of the colicin Ia binding site, contributed by Cir residues from extracellular loops 5, 7, and 8, and from plug apical loop 2. Cir is colored yellow (barrel) and magenta (plug), and the R-domain of colicin Ia is colored green. H-bonds are shown as black dashed lines. Residues and distances are described in detail in the Supplementary data. (B) Superposition of the Cir–colicin complex (yellow) with E. coli ferric citrate receptor (blue) in complex with diferric dicitrate (green and red) (PDB ID 1PO3). The front strands of the beta barrels for both receptors have been removed for clarity. The loop (yellow tube) connecting the two beta strands of the colicin R-domain sits at approximately the same depth in the binding pocket as diferric dicitrate (green (citrate) and red (iron) spheres), mimicking siderophore binding. (C) Charge complementarity between the R-domain of colicin Ia and extracellular loops 7 and 8 of Cir. The R-domain of colicin Ia is shown in surface representation, colored by electrostatic potential using values of −10 to +10 kt from GRASP (Nicholls et al, 1991). Cir is drawn and colored as in Figure 1. Two arginine residues on Cir insert into the negatively charged groove on colicin Ia; R436 from loop 7 and R490 from loop 8 form multiple interactions with colicin Ia, and are drawn in stick representation. Other Cir–colicin Ia interactions are described in the text and in Supplementary Table II.
In total, 18 hydrogen bonds (16 of which are contributed from loops 7 and 8) stabilize the interaction between Cir and colicin Ia, which exhibits a dissociation constant of approximately 1 × 10−10 M in vivo (Konisky and Cowell, 1972). This can be compared to formation of a vitamin B12 receptor (BtuB)–colicin E3 complex, where BtuB was shown to use 10 residues distributed among several extracellular loops to bind colicin E3 (burying 1533 Å2 of interaction surface upon complex formation; Kurisu et al, 2003). Grasp electrostatic analysis reveals that R436 and R490 of Cir bind in a cleft of strong negative charge on colicin Ia (Figure 2C). Residues Asp350, Asp358, Asp362, and Glu369 on colicin Ia are primarily responsible for forming this negative patch. In contrast, a charged patch is not observed in the BtuB–colicin E3 complex structure (Kurisu et al, 2003). The relatively small buried surface for Cir–colicin Ia (Lo Conte et al, 1999) yields a surprisingly tight binding constant for complex formation, due to the very large number of hydrogen bonds found in the interaction area, including one salt bridge and a beta strand pairing.
The binding of colicin Ia to Cir can be summarized as follows: the tip of the R-domain mimics siderophore binding by sitting in the binding pocket at the approximate depth found for metal chelates, and both colicin Ia and small molecules form hydrogen bonds to the plug domain (a single H-bond in the case of colicin Ia). Whereas small molecules tend to have binding sites composed of residues from several beta strands and/or extracellular loops distributed around the beta barrel of Cir, colicin Ia uses two arginine residues in loops L7 and L8 almost exclusively for binding.
Cir undergoes large and unusual conformational changes upon binding colicin Ia
Binding of colicin Ia triggers a number of conformational changes in Cir, particularly at the extracellular side of the transporter (Figure 3; Supplementary Movie 1). The largest and most unusual movement is seen in extracellular loops 7 and 8: in the complex structure, these loops move as a rigid body to open outward by 37° as compared with the same loops in the uncomplexed Cir structure. The loops are also translated by 4 Å along a screw axis determined from a rotation and a translation vector relating the two pairs of loops (Gerstein et al, 1993). The combined translation and rotation result in a 17 Å displacement for residue 437 in loop 7 and a 21 Å displacement for residue 511 in loop 8. These loop movements are significantly larger than movements observed for metal chelate binding: in E. coli ferric citrate transporter FecA, residues in loops 7 and 8 move up to 11 and 14 Å, respectively, toward the bound substrate, closing the binding pocket at the extracellular surface and sequestering the ferric chelate (Ferguson et al, 2002; Yue et al, 2003). In contrast, the same extracellular loops of Cir move upon colicin Ia binding, but in the opposite direction: they are stabilized in an open conformation, exposing the plug domain to the extracellular milieu. This is the only example of substrate-induced ‘opening' of a TonB-dependent transporter, and by far the largest loop movements seen to date (conformational changes in BtuB upon colicin E3 binding are very small, perhaps reflecting BtuB's role as cell surface receptor but not transporter; Kurisu et al, 2003).
Figure 3.
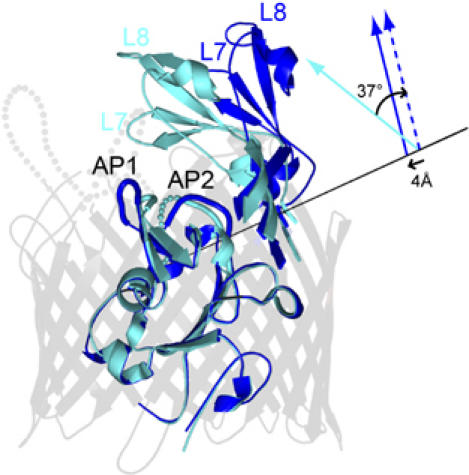
Conformational changes in Cir upon binding colicin Ia. A superimposition of uncomplexed Cir and colicin-bound Cir shows that the largest changes occur in the apical loops of the plug domain and extracellular loops 7 and 8 of the beta barrel. Significant movements are observed in apical loops 1 and 2 of the plug domain, as described in the text. The largest movements occur in extracellular loops 7 and 8, which undergo a rigid body movement around a screw axis (represented by a black line). The position/direction of the screw axis was determined as described in the Supplementary data. The solid cyan arrow schematically represents L7 and L8 of uncomplexed Cir and the solid blue arrow represents L7 and L8 of colicin-bound Cir. A rotation of 37° followed by a 4 Å translation along the same screw axis is required to superimpose L7 and L8 of uncomplexed Cir onto L7 and L8 of colicin-bound Cir. This movement effectively opens the extracellular side of the barrel, exposing the plug domain to the environment and potentially to colicin Ia. The beta barrel is colored light gray, uncomplexed Cir is colored cyan, and colicin-bound Cir is shown in blue.
The binding of the colicin Ia R-domain to Cir neither displaces the plug domain from the beta barrel nor does it open a smaller passage within the plug domain. This result was expected: in all TonB-dependent transporter structures solved to date (alone, with ligand, or with ligand and a periplasmic domain of TonB), the plug domain remains inside the barrel essentially in a ground-state conformation. Without energy from the protonmotive force, the plug domain continues to prevent entry of the substance bound at the extracellular side of the transporter. However, the binding of metal chelates produces significant conformational changes in the plug domain, and the same is found for the R-domain of colicin Ia. When comparing the plug domain of Cir alone and in complex with the colicin R-domain, movements of greater than 1.5 Å are observed in residue ranges 83–86 and 110–122 that belong to the two apical plug loops (Figure 3). The remaining regions of the plug domain are unaltered, displaying a low r.m.s.d. of only 0.518 Å in Cα positions. These include the four-stranded central beta sheet, the TonB box region, and the loop connecting the plug domain to the first beta strand of the barrel. If colicin Ia initiates the same events in Cir as small molecules do, we would expect the TonB box to become disordered upon binding substrate (Ferguson et al, 2002; Yue et al, 2003). Increased motion in this region has been suggested to signal TonB that a metal chelate is bound to the extracellular side of the transporter (Cadieux et al, 2003; Yue et al, 2003; Xu et al, 2006). Our structures show that the TonB box adopts the same conformation in the presence and absence of colicin Ia. However, the reagents used for crystallization have been shown to inhibit TonB box movement (Fanucci et al, 2003), and we address the issue of TonB signalling below.
Whereas conformational changes are observed in both domains of Cir, no changes are observed in the R-domain of colicin Ia when it binds to Cir. A superimposition of the colicin Ia R-domain from the original full-length structure (PDB ID 1CII) with the same domain from our Cir–colicin Ia complex structure shows no significant conformational changes, with an r.m.s.d. of 0.65 Å for Cα positions and 1.15 Å for all atoms (Supplementary Figure 3). This result has implications for initial stages of the translocation mechanism, because unfolding has been shown to facilitate the translocation of colicins A and E9 (Bénédetti et al, 1992; Duché et al, 1994; Penfold et al, 2004; Housden et al, 2005), and there is structural evidence for some unwinding of the coiled-coil at the ends of the R-domain of colicin E3 when it binds to BtuB (Kurisu et al, 2003). If unfolding occurs for colicin Ia, our structures suggest that the unfolding event either does not affect the R-domain or unfolding occurs sometime after binding to Cir.
A model for full-length colicin Ia bound to the cell surface
Using the knowledge that the R-domain of colicin Ia retains its native conformation upon binding to Cir, we modelled the interaction between full-length colicin Ia and Cir (Figure 4). The model assumes that the remainder of colicin Ia, including the long pair of alpha helices, the translocation domain, and the channel forming domain, retains its native structure upon binding. With these considerations, colicin Ia extends away from Cir, forming an angle of approximately 45° with the membrane bilayer and positioning the T- and C-domains about 150 Å above the bilayer, with an 80 Å lateral displacement from Cir. Our model likely represents the starting orientation of colicin Ia when bound to Cir on the surface of a bacterial cell, and shows the tremendous scale of the translocation event that eventually brings the C-domain into contact with the inner membrane.
Figure 4.
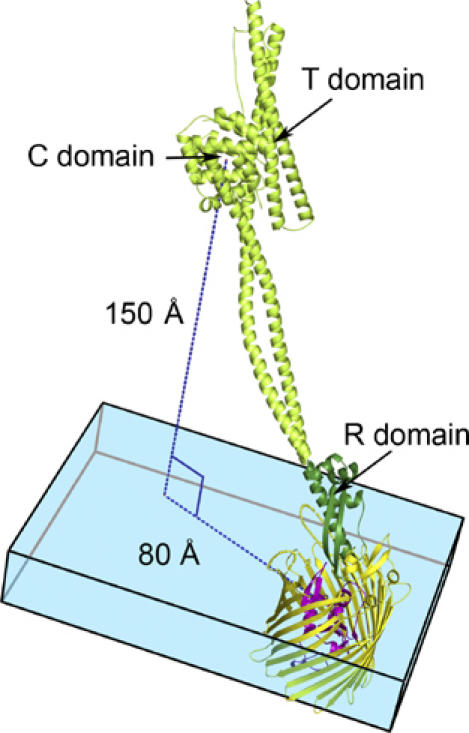
A model of full-length colicin Ia bound to Cir. Because no conformational changes occur in the R-domain of colicin Ia upon binding Cir (see text and Supplementary Figure 3), we assume that the remainder of colicin Ia does not change conformation upon binding. With this assumption, we have superimposed the R-domain of colicin Ia from the Cir–colicin complex structure with full-length colicin Ia (PDB ID 1CII). In the model, the colicin Ia R-domain is shown in dark green, with the rest of the colicin molecule (present only in the full-length colicin Ia structure) shown in light green. Cir is colored yellow and magenta, as in previous figures. The blue box denotes the thickness of the membrane bilayer, with Cir embedded in it. Colicin Ia binds to Cir with an angle of approximately 45° to the membrane bilayer, positioning the translocation (T) and channel-forming (C) domains of colicin Ia about 150 Å above the bilayer and translated by about 80 Å from Cir. Both the T- and C-domains are thought to enter the periplasm by a transport mechanism involving Cir and TonB–ExbB–ExbD.
The structural changes induced by complex formation can be summarized as follows. When the R-domain of colicin Ia binds to Cir, it stabilizes an open conformation by binding to extracellular loops L7 and L8 and holding them almost vertical, such that the colicin R-domain is positioned directly above the now exposed plug domain. The R-domain of colicin Ia does not unfold upon binding to Cir, implying that an unfolding event, if needed, occurs at a later stage in the translocation process. If colicin Ia truly mimics a metal chelate, then a transient pore in Cir would be created through which the colicin could be transported, but only after TonB interacts with the Cir–colicin complex and energy is applied. We note that 22-stranded beta barrels are the largest known transporters in the outer membrane (with the exception of multi-protein secretion systems), and if the plug were displaced, the resulting pore would be both large and unobstructed. However, the T- and C-domains of colicin Ia are located far above the membrane bilayer, and our structures do not suggest how they would access the pore. Also, the largest possible Cir pore (created by entirely removing the plug domain) would still require at least partial unfolding of the colicin T- and C-domains.
Cir is required for killing by colicin Ia
The large distances that domains of colicin Ia must traverse in order to use Cir as an import channel prompted us to look for other outer membrane proteins, which might be involved in colicin Ia translocation. First, we asked whether Cir is required for killing by colicin Ia. We found that wild-type E. coli strains BL21(DE3) and MC4100 are killed by colicin Ia over a concentration range of 30 μM to 30 pM (Table II). In contrast, strain ASH102, containing a cirA deletion, is completely resistant to killing under the conditions tested. We confirmed the requirement for Cir by making a chromosomal cirA deletion in BL21(DE3), which was also completely resistant to killing; sensitivity to colicin Ia was restored upon expressing cirA from a plasmid.
Table 2.
Killing by colicin Ia requires Cir, but does not require other abundant outer membrane proteins
| Bacterial strain | Colicin Ia |
|---|---|
| BL21(DE3) (Cir and all porins present) | Sensitive |
| ASH102 (Cir-minus) | Resistant |
| MC4100 (Cir and all porins present) | Sensitive |
| RAM725 (MC4100, OmpF-minus [ompF::lacZ]) | Sensitive |
| RAM726 (MC4100, OmpC-minus [ompC::lacZ]) | Sensitive |
| RAM105 (MC4100, OmpF-minus, LamB-minus, OmpC-plus) | Sensitive |
| RAM123 (RAM105, OmpF-minus, LamB-minus, OmpC-plus, OmpG-plus) | Sensitive |
| B2045 (RAM105, OmpF-minus, LamB-minus, OmpC-minus, OmpG-plus) | Sensitive |
| RAM1129 (MC4100, TolC-minus [tolC::Kmr] | Sensitive |
| A586 (TolC point mutant) | Sensitivea |
| EW1b (TolC partial deletion) | Sensitivea |
| BL21(DE3) (Cir-minus)b | Resistant |
| BL21(DE3) (Cir-minus)+pET20b-cirA | Sensitive |
| BL21(DE3) (Cir M33C/V35P) | Resistant |
| Some tolC mutants are sensitive to colicin Ia but are killed less efficiently than wild type. | |
| The chromosomal cirA deletion in BL21(DE3) is described in Supplementary data. | |
We then asked whether other E. coli outer membrane proteins modulate sensitivity to colicin Ia. We obtained deletion strains lacking OmpF, OmpC, LamB, or TolC (Table II). We observed killing at the same levels as in wild-type cells for deletion strains of OmpF, OmpC, LamB, and TolC, demonstrating that these outer membrane proteins are not required for binding and/or translocating colicin Ia across the outer membrane. However, we observed a 10-fold lower susceptibility to colicin Ia for a strain expressing a TolC point mutant, and a 100-fold lower susceptibility for a strain harboring a partial TolC deletion. These effects are small when compared with a control experiment testing the TolC partial deletion strain for sensitivity to colicin E1 (requiring both BtuB and TolC for sensitivity), which was completely resistant over a micromolar to picomolar range (not shown). To assess subtle differences in colicin sensitivity of TolC deletion strains, we carried out killing assays in liquid culture (Supplementary Figure 4). Strains MC4100 (wild type), A586 (TolC point mutant), RAM1129 (TolC complete deletion), and EW1b (TolC partial deletion) were monitored for growth upon addition of colicin Ia. All strains were killed upon addition of colicin Ia, although EW1b showed increased resistance in both plate and solution assays. Notably, the complete TolC deletion strain (RAM1129) was killed as effectively as wild type. Thus, TolC is also unlikely to serve as a secondary transporter for colicin Ia. Nevertheless, the possibility exists that an unidentified outer membrane protein acts as a cotransporter, or that other outer membrane proteins might substitute for the missing outer membrane proteins in deletion strains.
The TonB boxes of Cir and colicin Ia are both required for killing
The experiments described above establish Cir as the primary cell surface receptor for colicin Ia, but they do not indicate whether Cir also functions as a transporter in this process. TonB-dependent transporters require a functional TonB box sequence, allowing a productive interaction with TonB protein to provide the energy necessary for active transport. Cir contains a TonB box sequence near its N-terminus; mutations in TonB boxes of other metal chelate transporters have been shown to abolish the interaction with TonB protein, preventing small molecule transport (Postle and Kadner, 2003). We made a M33C/V35P mutation in the TonB box of Cir (Supplementary Table III) and expressed the mutant in the BL21(DE3) cirA deletion strain described above. Unlike cells expressing wild-type cirA from a plasmid (which are sensitive to colicin Ia), the cirA TonB box mutant is fully resistant to killing (Table II), suggesting that Cir's natural transport function is required by colicin Ia. Our result agrees with earlier data showing that another TonB box mutation (V35G; called V10G in the original paper) yields a Cir mutant that binds colicin Ia with wild-type affinity, but is not killed. The mutant was rescued (restoring the ability of colicin Ia to kill) by the TonB suppressor mutant Q160L (Bell et al, 1990), providing genetic evidence that Cir and TonB must interact in order for colicin Ia to kill cells. These experiments confirm the requirement for a functional TonB box sequence in Cir, and the requirement for an interaction between Cir and TonB protein for killing by colicin Ia.
Group B (Ton dependent) colicins, such as colicin Ia, also contain a TonB box sequence near the N-terminus (Supplementary Figure 1 and Supplementary Table III). Mutations in this region were shown to abolish killing by the related Ton-dependent colicins B (Mende and Braun, 1990) and M (Pilsl et al, 1993). We made a site-directed M25P mutation in the TonB box of colicin Ia and found that cells susceptible to killing by wild-type colicin Ia are not killed by the mutant (data not shown). To confirm that the M25P mutant retains toxicity, that is, the ability to form voltage-gated channels, we assayed its activity in planar lipid bilayers. The colicin Ia TonB box mutant makes normal channels, displaying good channel forming activity, normal voltage-dependent gating, and normal single channel conductance (Supplementary Figure 5).
We asked whether the failure of our colicin Ia and Cir mutants to kill/be killed was due to an inability to bind their respective wild-type partners. We used analytical ultracentrifugation to characterize wild-type Cir, colicin Ia, and complexes of the wild-type proteins, and both wild-type/mutant complexes. Cir and colicin Ia were observed to be monodisperse monomers, and they formed a 1:1 stoichiometric complex with high affinity (Figure 5; Supplementary Figures 6–8 and Supplementary Table IV). Complex formation was confirmed for wild-type Cir interacting with the colicin Ia M25P mutant, and for the Cir M33C/V35P mutant interacting with wild-type colicin Ia (Supplementary Table IV). The equilibrium sedimentation experiments are consistent with high-affinity complex formation. Based on the protein concentrations used, they give estimates of binding affinities much tighter than 0.1 μM for wild-type and mutant proteins. Colicin Ia is thought to bind Cir with a Kd of 10−10 M (Konisky and Cowell, 1972), and the AUC experiments would not have detected binding changes in the picomolar to nanomolar range. However, a Kd of greater than 10−7 M (minimal estimate) is probably still tight enough for killing to occur. Also, our structures suggest that the TonB box mutations in either Cir or colicin Ia are separated by large distances from the binding site, and are thus unlikely to influence binding substantially.
Figure 5.
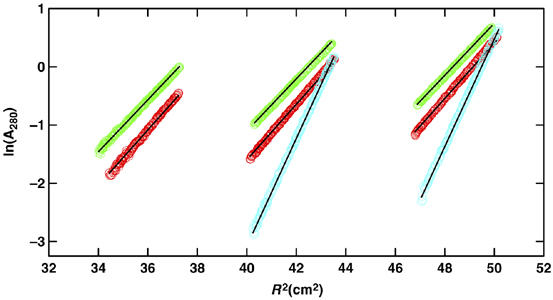
Colicin I receptor and colicin Ia form a 1:1 complex. Sedimentation equilibrium profiles shown in terms of lnA280 versus the radius squared for data collected at 10k r.p.m. and 4.0°C for the colicin I receptor (red), full-length colicin Ia (green), and a 1:1 mixture of the two (cyan). Data for the colicin I receptor and colicin Ia were collected at loading A280 of approximately 0.4 (left), 0.8 (center), and 1.2 (right), whereas data for the 1:1 mixture were collected at loading A280 of 0.65 (center) and 1.2 (left). The solid lines represent data expected for species having molecular masses corresponding to protein monomers in their 1:1 complex.
Taken together, the killing, binding, and functional assays show that Cir is necessary for colicin binding to the cell surface, because deleting it confers resistance to killing, and that Cir probably has an active role in translocation of colicin Ia across the outer membrane, because disrupting the Cir TonB box (abolishing energy coupling to TonB–ExbB–ExbD) permits colicin binding but prevents killing. The colicin Ia TonB box mutant cannot kill, and yet binds to Cir and retains its ability to form voltage-gated channels. Thus, both mutants appear to be defective in translocation. Our data suggest that colicin Ia transport across the outer membrane requires two distinct interactions with TonB protein, as postulated for colicins B and M (Braun et al, 2002).
Discussion
Tol- and Ton-dependent colicins have different requirements for translocation across the outer membrane
We now have two structural models for TonB-dependent transporters interacting with two different types of colicin: BtuB interacting with the group A (Tol dependent) colicin E3 (Kurisu et al, 2003) and Cir interacting with the group B (Ton dependent) colicin Ia (this work). The two structures are similar in the positioning of each colicin at the cell surface, since both E3 and Ia bind at an angle of approximately 45° to the membrane. Such an orientation positions the T- and C-domains far above the membrane, and far from the cell surface receptor (about 80 Å away for the Cir–colicin Ia R-domain structure). The initial colicin E3 orientation on BtuB led to the proposal that the 45° angle facilitates recruitment of OmpF, which is a required cotransporter for this system (Kurisu et al, 2003). Recruitment of OmpF was later demonstrated for the related colicin E9 by the isolation of a BtuB–OmpF–colicin E9 complex from cells (Housden et al, 2005). It was further shown that the T-domain of colicin E9 was responsible for recruiting OmpF, implying that the T-domain may enter the cell through OmpF, where it can then interact with TolB (Lazdunski et al, 1998; Loftus et al, 2006).
Although colicin Ia binds at a similar angle to Cir, to date no cotransporter has been identified and the entry mechanism remains unclear. The 160 Å long pair of alpha helices separating the T- and C- domains from the R-domain was suggested to allow the colicin to span the cell envelope, with the R-domain remaining attached to the cell surface, while the C-domain penetrated the inner membrane (Stroud et al, 1998). Consistent with this hypothesis, another channel forming colicin (Tol-dependent colicin A) was shown to remain bound to the outer membrane when its channel forming domain inserted into the inner membrane (Bénédetti et al, 1992). An alternative explanation for the large separation of the R-domain from the T- and C-domains mirrors the findings for BtuB: once colicin Ia is bound to Cir, the 45° angle that it makes with the membrane might allow colicin Ia to search the surrounding regions for a cotransporter or other entry passage into the cell. However, the crystal structure of Ton-dependent colicin B argues against both hypotheses (Hilsenbeck et al, 2004). In this structure, the R- and T-domains are intertwined and separated from the C-domain by a single helix less than half as long as observed for colicin Ia. This colicin is neither long enough to span the cell envelope, nor would it have as much flexibility to search for a possible cotransporter.
Several other differences to the BtuB–colicin E3 (or E9) system are apparent. First, binding of colicin Ia stabilizes large and unusual conformational changes in two extracellular loops of Cir, which open outward, exposing the interior of the beta barrel to the extracellular environment and to the colicin. This is the first example of substrate-induced opening in a TonB-dependent transporter, showing the largest loop movements ever observed. If Cir functions to transport colicin Ia across the outer membrane, this is the type of conformational change that would be required prior to penetration by the colicin. In contrast, BtuB does not exhibit significant conformational changes when colicin E3 binds, perhaps reflecting its sole function as cell surface receptor (Kurisu et al, 2003).
Second, while cirA deletions consistently rendered cells insensitive to killing by full-length colicin Ia, the two outer membrane proteins (OmpF and TolC) known to act as cotransporters for Tol-dependent colicins are not required for colicin Ia translocation. Like colicin Ia, colicins Ib, B, D, and M target TonB-dependent transporters but have no known cotransporter (Braun et al, 2002; Cao and Klebba, 2002). It is notable that BtuB only binds Tol-dependent colicins (A, E1–E9), and in each case, a cotransporter is required. For Ton-dependent colicins, the possibility exists that an unidentified cotransporter is used (we did not screen for rare outer membrane proteins and we were not able to screen for essential outer membrane proteins, such as YaeT; Ruiz et al, 2006). Further possibilities are that Ton-dependent colicins are indiscriminate in their use of a cotransporter, with many different outer membrane proteins fulfilling this function, or that Ton-dependent colicins move down the outside (hydrophobic) wall of a beta-barrel protein, as proposed for colicin N (Bainbridge et al, 1998). These possibilities have not yet been tested.
Third, BtuB and Cir differ in their requirements for a functional TonB box sequence (and thus in their requirement for a productive interaction with TonB) for colicin translocation. BtuB appears to function primarily as a cell surface receptor during Tol-dependent colicin translocation, requiring neither a functional TonB box sequence nor interaction with TonB protein to effect colicin translocation (Gudmundsdottir et al, 1989; Cadieux et al, 2000). In contrast, Cir requires a functional TonB box sequence for killing by colicin Ia, implying that a productive Cir–TonB interaction is necessary for colicin uptake. Furthermore, our killing assays, sedimentation equilibrium experiments and electrophysiological measurements on wild-type and TonB box mutants in Cir and colicin Ia demonstrate that both proteins require functional TonB box sequences for colicin transport across the outer membrane. Thus, in addition to differing from Tol-dependent colicin uptake, Ton-dependent colicin translocation is more complicated than small molecule transport, which requires only one functional interaction between the transporter and TonB (Postle and Kadner, 2003).
A requirement for unfolding or large conformational changes in colicin Ia
Our model for full-length colicin Ia bound to Cir at the cell surface positions the T- and C-domains about 150 Å above the bilayer and 80 Å away from Cir. Whether colicin Ia is translocated by Cir or by some other mechanism, large conformational changes and/or unfolding would be necessary to bring the T- and C-domains into contact with the bilayer. Some colicins function more efficiently when unfolded: colicin A denatured in urea kills cells more quickly than natively folded colicin A (Bénédetti et al, 1992). Although some unwinding of the coiled-coil at the end of the R-domain was observed in the BtuB–colicin E3 R-domain crystal structure (Kurisu et al, 2003), no such unfolding was observed in a complex of BtuB with colicin E9 (Housden et al, 2005), or when the colicin Ia R-domain bound to the Cir receptor. However, the BtuB–colicin E3 R domain complex was formed in the absence of calcium, which has been shown to significantly enhance binding of the R-domain to BtuB (Mohanty et al, 2003). In contrast, Housden et al measured calorimetric data from in vivo complexes formed by BtuB, with whole colicin E9 or with a disulfide mutant of E9 engineered to prevent unwinding of the R-domain. The thermodynamics of both complexes were virtually identical, suggesting that no unwinding occurs when the wild-type colicin binds to BtuB. Since colicins E3 and E9 share >90% sequence identity in their receptor binding and translocation domains, the discrepancy between the findings of the two groups must be due to differences in the conditions under which the complexes were formed. Therefore, Tol- and Ton-dependent colicins appear to initiate translocation in the same manner: receptor binding does not induce unfolding of either type of colicin.
Exactly when and how colicins unfold remains to be investigated. However, the requirement for unfolding occurs in other systems as well: many bacterial and plant toxins targeting eukaryotic cells must unfold to kill, and unfolding is commonly achieved by a change in pH when the toxin encounters the acidic environment of the endosome (Falnes and Sandvig, 2000). With colicins entering bacterial cells from the extracellular milieu, it is unclear whether they would encounter sufficient pH differences to initiate unfolding.
Conclusions
The crystal structure of Cir bound to the R-domain of colicin Ia illustrates how colicin Ia initially attaches to the outer membrane of a target cell. Our data suggest that Tol- and Ton-dependent colicins (that target TonB-dependent transporters) are transported by different mechanisms. The structural changes we observed and the functional assays we carried out are consistent with a role for Cir in translocation of colicin Ia, in addition to Cir's demonstrated role as cell surface receptor. Other modes of entry are possible, but no data exist implicating other outer membrane protein(s), or other mechanisms, in transport of colicin Ia or any other Ton-dependent colicin. Translocation of colicin Ia appears to require functional interactions between Cir and TonB, and also between colicin Ia and TonB. Further experiments are required to decipher the energetics of transport and colicin Ia's pathway across the outer membrane.
Materials and methods
Crystallization, structure determination, and analysis
A Cir mutant containing 4 methionine substitutions (for phasing) was expressed using a modified pET20b vector in BL21(DE3) cells. Membranes were isolated and solubilized with Elugent (Calbiochem) and the solubilized protein was purified using nickel affinity, ion exchange, and size-exclusion chromatography columns. Detergent exchange was performed during the final chromatography step to either CHESO (Bachem) or LDAO (Fluka). The colicin Ia R-domain was purified from inclusion bodies expressed in BL21(DE3) cells and refolded in the presence of Cir. The Cir–colicin Ia complex was chromatographed on a size-exclusion column equilibrated with LDAO and C8E4 (Anatrace). Cir crystals grew in 32–40% PEG 2000 MME, 5% isopropanol, 20–50 mM SrCl2, and 100 mM Tris–HCl, pH 7.5. Cir–colicin complex crystals grew in 22% PEG 2000 MME, 200 mM NaCl, 10% glycerol, and 100 mM MES, pH 6.2.
Diffraction data were collected at the SER-CAT 22ID beamline of the Advanced Photon Source (Argonne, IL). All data were integrated and scaled using HKL2000 (Otwinowski and Minor, 1997). Cir crystallized in the monoclinic spacegroup C2 with unit cell dimensions a=90.61 Å, b=84.35 Å, c=99.47 Å, β=109.2°, having one molecule in the asymmetric unit. SHELXD (Schneider and Sheldrick, 2002) located nine selenium atoms, which were used in autoSHARP (Vonrhein et al, 2006) for phase determination. The structure was built using O (Jones et al, 1991) and Coot (Emsley and Cowtan, 2004), and was refined using REFMAC (Winn et al, 2003). The Cir–colicin complex crystallized in spacegroup C2 with unit cell dimensions a=132.64 Å, b=130.49 Å, c=56.27 Å, β=101.2°, having one (complex) molecule in the asymmetric unit. The structure was solved by molecular replacement using Phaser (McCoy et al, 2005), and the model was built and refined as described above.
The buried interaction surface for Cir–colicin Ia was calculated using AREAIMOL (part of the CCP4 program suite; CCP4, 1994) using a probe sphere (representing solvent) of 1.4 Å radius. The interaction surface was calculated as ((asa Cir+asa col)−asa cir/col)/2, where asa is the solvent accessible surface area, defined as the surface mapped out by the center of the spherical probe of radius 1.4 Å as if it were rolled around the van der Waals surface of the protein.
Functional analyses
Bacterial strains and plasmids, including recombinant DNA techniques, are described in the Supplementary data. Full-length colicin Ia (wild-type or the TonB box mutant) was expressed and purified as described (Mel and Stroud, 1993; Qiu et al, 1994). Susceptibility to killing by colicin Ia was determined by spotting 10-fold dilutions of purified full-length colicin Ia (in PBS, diluted with LB broth) onto top agar overlays of the strains to be tested on LB plates containing ampicillin (100 μg/ml) where appropriate. The degree of susceptibility was determined from the lowest colicin concentration at which no perceptible growth was observed in the application spot. The colicin Ia TonB box mutant (M25P) was evaluated for its ability to form voltage-gated channels using methods described previously (Kienker et al, 2000). Cir and colicin Ia (wild-type proteins and TonB box mutants for each protein) were evaluated individually and for complex formation by analytical ultracentrifugation using the detergent octyl-POE (Bachem). Sedimentation equilibrium data collected at different speeds and different loading concentrations were analyzed globally in terms of various species analysis models using SEDPHAT 4.1b (Schuck) to obtain the sample molecular mass. Solution densities (ρ) were measured at 20.0°C on a Mettler–Toledo DE51 density meter and corrected to values for ρ at 4°C. Values of the partial specific volume (v) were calculated based on the amino-acid composition using SEDNTERP (Philo).
Supplementary Material
Supplementary Figure
Supplementary Information
Acknowledgments
We thank Mark McIntosh, Rajeev Misra, and Margaret Riley for bacterial strains, and Cynthia Cornelissen, Harris Bernstein, and Wei Yang for critically reading the manuscript. This work was supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (SKB, PL, SG, RG, MA, TJB) and the National Cancer Institute (LE). KSJ and PKK acknowledge support from National Institutes of Health Grant GM31986. Diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html.
Accession numbers Coordinates and structure factors for Cir and the Cir–colicin R-domain complex have been deposited in the Protein Data Bank with accession codes 2HDF and 2HDI.
References
- Bainbridge G, Armstrong GA, Dover LG, Whelan KF, Lakey JH (1998) Displacement of OmpF loop 3 is not required for the membrane translocation of colicins N and A in vivo. FEBS Lett 432: 117–122 [DOI] [PubMed] [Google Scholar]
- Bell PE, Nau CD, Brown JT, Konisky J, Kadner RJ (1990) Genetic suppression demonstrates interaction of TonB protein with outer membrane transport proteins in Escherichia coli. J Bacteriol 172: 3826–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénédetti H, Lloubès R, Lazdunski CJ, Letellier L (1992) Colicin A unfolds during its translocation in Escherichia coli cells and spans the whole cell envelope when its pore has formed. EMBO J 11: 441–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Patzer SI, Hantke K (2002) Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84: 365–380 [DOI] [PubMed] [Google Scholar]
- Cadieux N, Bradbeer C, Kadner RJ (2000) Sequence changes in the ton box region of BtuB affect its transport activities and interaction with TonB protein. J Bacteriol 182: 5954–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieux N, Phan PG, Cafiso DS, Kadner RJ (2003) Differential substrate-induced signaling through the TonB-dependent transporter BtuB. Proc Natl Acad Sci USA 100: 10688–10693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Klebba PE (2002) Mechanisms of colicin binding and transport through outer membrane porins. Biochimie 84: 399–412 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chimento DP, Kadner RJ, Wiener MC (2005) Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins 59: 240–251 [DOI] [PubMed] [Google Scholar]
- Chimento DP, Mohanty AK, Kadner RJ, Wiener MC (2003) Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat Struct Biol 10: 394–401 [DOI] [PubMed] [Google Scholar]
- Critchley IA, Basker MJ, Edmondson RA, Knott SJ (1991) Uptake of a catecholic cephalosporin by the iron transport system of Escherichia coli. J Antimicrob Chemother 28: 377–388 [DOI] [PubMed] [Google Scholar]
- Curtis NA, Eisenstadt RL, East SJ, Cornford RJ, Walker LA, White AJ (1988) Iron-regulated outer membrane proteins of Escherichia coli K-12 and mechanism of action of catechol-substituted cephalosporins. Antimicrob Agents Chemother 32: 1879–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. www.pymol.org
- Duché D, Baty D, Chartier M, Letellier L (1994) Unfolding of colicin A during its translocation through the Escherichia coli envelope as demonstrated by disulfide bond engineering. J Biol Chem 269: 24820–24825 [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Falnes P, Sandvig K (2000) Penetration of protein toxins into cells. Curr Opin Cell Biol 12: 407–413 [DOI] [PubMed] [Google Scholar]
- Fanucci GE, Lee JY, Cafiso DS (2003) Spectroscopic evidence that osmolytes used in crystallization buffers inhibit a conformation change in a membrane protein. Biochemistry 42: 13106–13112 [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Deisenhofer J (2004) Metal import through microbial membranes. Cell 116: 15–24 [DOI] [PubMed] [Google Scholar]
- Ferguson AD, Chakraborty R, Smith BS, Esser L, van der Helm D, Deisenhofer J (2002) Structural basis of gating by the outer membrane transporter FecA. Science 295: 1715–1719 [DOI] [PubMed] [Google Scholar]
- Gerstein M, Anderson BF, Norris GE, Baker EN, Lesk AM, Chothia C (1993) Domain closure in lactoferrin: two hinges produce a see–saw motion between alternative close-packed interfaces. J Mol Biol 234: 357–372 [DOI] [PubMed] [Google Scholar]
- Gudmundsdottir A, Bell PE, Lundrigan MD, Bradbeer C, Kadner RJ (1989) Point mutations in a conserved region (TonB box) of Escherichia coli outer membrane protein BtuB affect vitamin B12 transport. J Bacteriol 171: 6526–6533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K (1990) Dihydroxybenzoylserine—a siderophore for E. coli. FEMS Microbiol Lett 67: 5–8 [DOI] [PubMed] [Google Scholar]
- Hilsenbeck JL, Park H, Chen G, Youn B, Postle K, Kang C (2004) Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 Åresolution. Mol Microbiol 51: 711–720 [DOI] [PubMed] [Google Scholar]
- Housden NG, Loftus SR, Moore GR, James R, Kleanthous C (2005) Cell entry mechanism of enzymatic bacterial colicins: porin recruitment and the thermodynamics of receptor binding. Proc Natl Acad Sci USA 102: 13849–13854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakes KS, Kienker PK, Finkelstein A (1999) Channel-forming colicins: translocation (and other deviant behaviour) associated with colicin Ia channel gating. Q Rev Biophys 32: 189–205 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kadner RJ (1990) Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol Microbiol 4: 2027–2033 [DOI] [PubMed] [Google Scholar]
- Kienker PK, Jakes KS, Finkelstein A (2000) Protein translocation across planar bilayers by the colicin Ia channel-forming domain. Where will it end? J Gen Physiol 116: 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konisky J (1972) Characterization of colicin Ia and colicin Ib. Chemical studies of protein structure. J Biol Chem 247: 3750–3755 [PubMed] [Google Scholar]
- Konisky J, Cowell BS (1972) Interaction of colicin Ia with bacterial cells. Direct measurement of Ia–receptor interaction. J Biol Chem 247: 6524–6529 [PubMed] [Google Scholar]
- Kurisu G, Zakharov SD, Zhalnina MV, Bano S, Eroukova VY, Rokitskaya TI, Antonenko YN, Wiener MC, Cramer WA (2003) The structure of BtuB with bound colicin E3 R-domain implies a translocon. Nat Struct Biol 10: 948–954 [DOI] [PubMed] [Google Scholar]
- Law CJ, Penfold CN, Walker DC, Moore GR, James R, Kleanthous C (2003) OmpF enhances the ability of BtuB to protect susceptible Escherichia coli cells from colicin E9 cytotoxicity. FEBS Lett 545: 127–132 [DOI] [PubMed] [Google Scholar]
- Lazdunski CJ, Bouveret E, Rigal A, Journet L, Lloubès R, Bénédetti H (1998) Colicin import into Escherichia coli cells. J Bacteriol 180: 4993–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J (1999) The atomic structure of protein–protein recognition sites. J Mol Biol 285: 2177–2198 [DOI] [PubMed] [Google Scholar]
- Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch JP, Moras D (1998) Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95: 771–778 [DOI] [PubMed] [Google Scholar]
- Loftus SR, Walker D, Maté MJ, Bonsor DA, James R, Moore GR, Kleanthous C (2006) Competitive recruitment of the periplasmic translocation portal TolB by a natively disordered domain of colicin E9. Proc Natl Acad Sci USA 103: 12353–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D 61: 458–464 [DOI] [PubMed] [Google Scholar]
- Mel SF, Stroud RM (1993) Colicin Ia inserts into negatively charged membranes at low pH with a tertiary but little secondary structural change. Biochemistry 32: 2082–2089 [DOI] [PubMed] [Google Scholar]
- Mende J, Braun V (1990) Import-defective colicin B derivatives mutated in the TonB box. Mol Microbiol 4: 1523–1533 [DOI] [PubMed] [Google Scholar]
- Mohanty AK, Bishop CM, Bishop TC, Wimley WC, Wiener MC (2003) Enzymatic E-colicins bind to their target receptor BtuB by presentation of a small binding epitope on a coiled-coil scaffold. J Biol Chem 278: 40953–40958 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Nikaido H, Rosenberg EY (1990) Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: study with beta-lactam antibiotics containing catechol and analogous groups. J Bacteriol 172: 1361–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M (1967) Colicins and related bacteriocins. Annu Rev Microbiol 21: 257–284 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW (2006) Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312: 1399–1402 [DOI] [PubMed] [Google Scholar]
- Penfold CN, Healy B, Housden NG, Boetzel R, Vankemmelbeke M, Moore GR, Kleanthous C, James R (2004) Flexibility in the receptor-binding domain of the enzymatic colicin E9 is required for toxicity against Escherichia coli cells. J Bacteriol 186: 4520–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo J. http://www.jphilo.mailway.com/
- Pilsl H, Glaser C, Gross P, Killmann H, Olschlager T, Braun V (1993) Domains of colicin M involved in uptake and activity. Mol Gen Genet 240: 103–112 [DOI] [PubMed] [Google Scholar]
- Postle K, Kadner RJ (2003) Touch and go: tying TonB to transport. Mol Microbiol 49: 869–882 [DOI] [PubMed] [Google Scholar]
- Qiu XQ, Jakes KS, Finkelstein A, Slatin SL (1994) Site-specific biotinylation of colicin Ia. J Biol Chem 269: 7483–7488 [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ (2006) Advances in understanding bacterial outer membrane biogenesis. Nat Rev Microbiol 4: 57–66 [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM (2002) Substructure solution with SHELXD. Acta Crystallogr D 58: 1772–1779 [DOI] [PubMed] [Google Scholar]
- Schuck PS. http://www.analyticalultracentrifu gation.com/sedphat/sedphat.htm
- Shultis DD, Purdy MD, Banachs CN, Wiener MC (2006) Outer membrane active transport: structure of the BtuB:TonB complex. Science 312: 1396–1399 [DOI] [PubMed] [Google Scholar]
- Stroud RM, Reiling K, Wiener M, Freymann D (1998) Ion-channel-forming colicins. Curr Opin Struct Biol 8: 525–533 [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Maejima T, Mitsuhashi S (1995) Mechanism of tonB-dependent transport of KP-736, a 1,5-dihydroxy-4-pyridone-substituted cephalosporin, into Escherichia coli K-12 cells. Antimicrob Agents Chemother 39: 613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G (2006) Automated structure solution with autoSHARP. In: Doublie, S (ed) Crystallographic Methods. Totowa: Human Press [DOI] [PubMed] [Google Scholar]
- Wiener M, Freymann D, Ghosh P, Stroud RM (1997) Crystal structure of colicin Ia. Nature 385: 461–464 [DOI] [PubMed] [Google Scholar]
- Wiener MC (2005) TonB-dependent outer membrane transport: going for Baroque? Curr Opin Struc Biol 15: 394–400 [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ (2003) Macromolecular TLS refinement in REFMAC at moderate resolutions. Method Enzymol 374: 300–321 [DOI] [PubMed] [Google Scholar]
- Xu Q, Ellena JF, Kim M, Cafiso DS (2006) Substrate-dependent unfolding of the energy coupling motif of a membrane transport protein determined by double electron–electron resonance. Biochemistry 45: 10847–10854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WW, Grizot S, Buchanan SK (2003) Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J Mol Biol 332: 353–368 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure
Supplementary Information


