Abstract
The process by which fibronectin (FN), a soluble multidomain protein found in tissue fluids, forms insoluble fibrillar networks in the extracellular matrix is poorly understood. Cryptic sites found in FN type III domains have been hypothesized to function as nucleation points, thereby initiating fibrillogenesis. Exposure of these sites could occur upon tension-mediated mechanical rearrangement of type III domains. Here, we present the solution structures of the second type III domain of human FN (2FNIII), and that of an interaction complex between the first two type III domains (1−2FNIII). The two domains are connected through a long linker, flexible in solution. A weak but specific interdomain interaction maintains 1−2FNIII in a closed conformation that associates weakly with the FN N-terminal 30 kDa fragment (FN30 kDa). Disruption of the interdomain interaction by amino-acid substitutions dramatically enhances association with FN30 kDa. Truncation analysis of 1−2FNIII reveals that the interdomain linker is necessary for robust 1−2FNIII–FN30 kDa interaction. We speculate on the importance of this interaction for FN function and present a possible mechanism by which tension could initiate fibrillogenesis.
Keywords: cryptic site, fibrillogenesis, fibronectin, protein structure, self-association
Introduction
Fibronectin (FN) is an extracellular matrix (ECM) protein essential for normal cell adhesion and mobility in vertebrates (Mao and Schwarzbauer, 2005). The disulfide crosslinked FN homodimer is largely composed of three types of repeating domains, FNI, FNII and FNIII. These domain types are ubiquitous in modular proteins and structural comparisons show a high degree of homology even in cases of low sequence identity (Campbell and Spitzfaden, 1994). FN is soluble in blood and tissue fluids, but forms insoluble fibrillar networks in the ECM via a tightly regulated cellular process that depends on the presence of integrins and other cell surface receptors. The mechanism of initiation and regulation of fibrillogenesis is poorly understood, although initiation is believed to depend on cell-generated tension, which uncovers cryptic sites for self-association in the FN molecule (Geiger et al, 2001). Several cryptic sites are known to exist in FN type III domains (Ingham et al, 1997), and it has been hypothesized that they could serve as possible nucleation sites for FN polymerization and fibril assembly. The evidence for tension-induced exposure of interaction sites and the remarkable elasticity observed in FN fibrils (Erickson, 2002; Abu-Lail et al, 2006) led to the hypothesis that FNIII domain unfolding plays an important role in fibrillogenesis (Mao and Schwarzbauer, 2005). A number of recent studies have thus focused on the mechanical unfolding properties of FN type III domains (Oberhauser et al, 2002; Gao et al, 2003; Abu-Lail et al, 2006).
The first two type III domains of FN, 1FNIII and 2FNIII, make important contributions in the fibrillogenesis process. Denatured 1FNIII intermediates have been implicated in interactions with the FN N-terminal type I domains (Hocking et al, 1994), and peptide fragments from the same domain (Morla and Ruoslahti, 1992), or anti-1FNIII antibodies (Chernousov et al, 1991) can inhibit matrix assembly. Anastellin, a C-terminal fragment of 1FNIII (Briknarova et al, 2003), promotes FN polymerization to an insoluble, matrix-like form (Morla et al, 1994) through direct association with a number of type III domains (Ohashi and Erickson, 2005). Nevertheless, recombinant FN lacking 1FNIII was reported to assemble in a matrix form highly similar to that of native FN (Sechler et al, 2001). In contrast, 2FNIII is essential for robust matrix accumulation and fibril growth, and it contains at least one FN self-association site (Aguirre et al, 1994; Sechler et al, 2001). These results suggest that the first two type III domains of fibronectin (1−2FNIII) act together as a functional unit to regulate fibril assembly and promote elongation. Mechanical unfolding by atomic force microscopy (AFM) of 1FNIII or 1−2FNIII in poly-protein constructs showed that 1FNIII is stabilized in the 1−2FNIII context, and this was interpreted as evidence for a direct 1FNIII–2FNIII interaction (Oberhauser et al, 2002). Interestingly, 1FNIII and 2FNIII are connected in fibronectin through a relatively long, 21-amino-acid linker, whereas almost all other type III domains are connected by short, 2- to 4-amino-acid linkers.
Here, we present structural and biochemical evidence for the existence of a cryptic site in 1−2FNIII that interacts with the N-terminal FN 30 kDa fragment (FN30 kDa). In contrast to canonical type III domains, the first β-strand of 2FNIII is disordered in solution, although the remaining 2FNIII structure is highly similar to other type III protein domains. The linker connecting 1FNIII with 2FNIII is flexible and includes the disordered 2FNIII region. The presence of a direct 1FNIII–2FNIII interaction was confirmed using isolated domains and intact 1−2FNIII, and a restrained model of the 1−2FNIII structure was calculated using the 1FNIII structure (Gao et al, 2003) and the new 2FNIII structure. Wild-type 1−2FNIII exhibits only a weak association interaction with FN30 kDa. In contrast, a 1−2FNIII variant that disrupts the 1FNIII–2FNIII interaction shows significantly stronger association with the same fragment. A series of protein constructs of both 1−2FNIII and 2FNIII, with variable lengths of the interdomain linker, show that the strong 1−2FNIII–FN30 kDa interaction depends on the presence of that linker. This leads directly to a model, where disruption of the 1FNIII–2FNIII complex by cell-generated tension creates an open 1FNIII–2FNIII conformation that initiates fibrillogenesis by interaction with FN30 kDa.
Results
2FNIII structure
Over 21 000 FNIII domain entries exist currently in the SMART database (http://smart.embl.de), whereas over 80 distinct entries in the RCSB protein structure database contain one or more FNIII domains. The canonical structure of this domain type, a three-stranded and a four-stranded β-sheet in β-sandwich arrangement, is well understood. Unexpectedly, backbone dynamics studies (Supplementary Figure 1B and C) of the canonical 2FNIII domain indicated that approximately 20 amino acids of the 2FNIII N-terminus are largely flexible and unstructured. This N-terminal amino-acid region corresponds to the first β-strand (β-strand A) of the canonical FNIII structure (Supplementary Figure 1A). 2FNIII is, to the best of our knowledge, unique among the structurally characterized members of this domain class in lacking a secondary structure element in its native form.
The high-resolution solution structure of 2FNIII was determined using standard triple-resonance NMR methods (Figure 1A and B). The final ensemble of structures was calculated from 2827 distance, geometry, and global orientation restraints, with an average of approximately 36 restraints per residue of the structured core domain (Supplementary Table I). The resulting ensemble precision is 0.20±0.04 Å for all backbone and 0.43±0.09 Å for all heavy atoms of non-mobile residues ({1H–}15N NOE⩾0.6), with over 94% of those residues in the most favored region of the Ramachandran plot. The overall structure of 2FNIII, other than β-strand A, closely resembles that of canonical FNIII domains. Structure alignments show that the backbone of essentially all structured 2FNIII residues can be superimposed with equivalent residues of other FNIII domains with average r.m.s. deviations smaller than 2 Å (Figure 1C and Supplementary Table II). The largest deviations are observed in the middle of the strongly twisted β-strand G (Figure 1D), which, in canonical FNIII domains, forms close contacts with the missing β-strand A.
Figure 1.
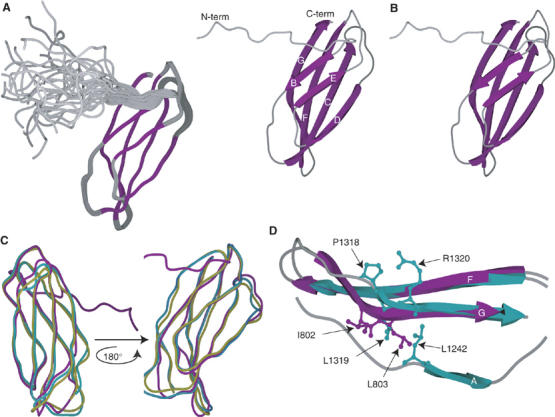
Solution structure of the 2FNIII domain and comparison with other type III domains. Shown here are (A) the 25-structure ensemble of 2FNIII structures. Purple-colored areas correspond to secondary structure elements. (B) Schematic stereo-representation of the minimized average structure of 2FNIII. β-strands are numbered according to canonical type III structures. (C) Superpositions of 2FNIII (purple) with human FN 8FNIII (turquoise) and avian tenascin 6FNIII (yellow). (D) The β-strands F and G of 2FNIII (purple) and human FN 8FNIII (turquoise) are shown here, as well as β-strand A of the latter. The side chains of the 2FNIII amino-acid residues likely responsible for the 2FNIII structural divergence (I802 and L803) are shown in purple. Also shown (turquoise) are the structurally equivalent 8FNIII residues (P1318-R1320) and L1242, which anchors the A and G β-strands.
Sequence alignment of the closest 2FNIII structural homologs and all identifiable 2FNIII domains from fibronectin orthologues (Figure 2) revealed a likely cause of the strand A disorder. In all 2FNIII domains, one of three hydrophobic residues that anchor β-strand A to β-strand G is substituted by a glutamate residue, E726. The remaining two hydrophobic residues are present (V728 and I731) but apparently insufficient to maintain an ordered structure. The volume occupied by the missing hydrophobic residue in canonical FNIII domains (L1242 in Figure 1D) is filled by two bulky hydrophobic residues of β-strand G in 2FNIII (I802 and L803 in the same figure); these do not follow the alternating side chain orientation rule of typical β-strands, and are thus responsible for the strong β-strand twist observed. As shown in Figures 1D and 2, other FNIII domains have one or no hydrophobic residues at the same position. We expect that, collectively, these substitutions cause disorder in the N-terminal β-strand of 2FNIII, but stabilize the remaining fold.
Figure 2.
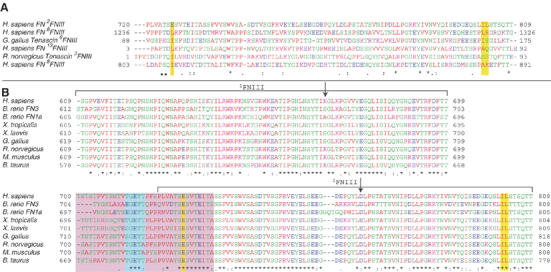
CLUSTAL-W sequence alignment of (A) 2FNIII and the five closest structural homologs of 2FNIII as identified by Dali (Holm and Sander, 1998), and (B) 1−2FNIII sequences from different FN proteins. The residue spans of 1FNIII and 2FNIII are denoted by brackets, and the positions of the two amino-acid substitutions engineered in this study, K669A and D767A, are indicated by arrows. Highlighted by yellow bars are the three 2FNIII residues (E726, I802 and L803) identified in our analysis as possibly responsible for the disordered 2FNIII N-terminus, and their sequence counterparts in other domains. Highlighted in a purple box are the residues of the disordered 1FNIII–2FNIII linker in all FN molecules. Shown in a cyan box are residues of the interdomain linker removal of which adversely affects the 1−2FNIII–FN30 kDa interaction.
1FNIII–2FNIII interaction
The flexible N-terminal region of 2FNIII increases the length of the 1FNIII–2FNIII linker to approximately 34 residues between structured domains. If flexible, this linker effectively uncouples the molecular motions of the two domains and allows the adoption of completely independent orientations in the absence of any interdomain interactions. We tested 1FNIII and 2FNIII for such interactions by monitoring their 1H–15N HSQC spectra for chemical shift perturbations upon addition of unenriched 2FNIII or 1FNIII, respectively.
Upon titration, several 1FNIII and 2FNIII resonances were perturbed significantly in a dosage-dependent manner (Supplementary Figure 2A–D), consistent with a weak interdomain interaction with dissociation rates that are fast on the NMR timescale. Fitting the chemical shift changes for different domain ratios and concentrations enabled us to estimate the Kd as approximately 100 μM for this interaction under the experimental conditions used (Supplementary Figure 3). The observed perturbations were subsequently mapped on the domain amino-acid sequences (Supplementary Figure 4A and B) and tertiary structures. The 1FNIII perturbations primarily localize along the β-strand connecting loops and turns at the C-terminal end of the molecule, while those of 2FNIII form a largely continuous surface on the exposed side of the four-stranded β-sheet (β-strands C, D, F and G).
A number of the larger perturbations involve charged 1FNIII and 2FNIII residues, suggesting that electrostatic forces contribute substantially to this interaction. 1FNIII has an overall positive charge under physiological conditions (pI 9.45), whereas 2FNIII is negatively charged (pI 3.79). To determine whether the interaction observed is specific between these two domains or simply a result of the opposite charges, we performed HSQC titration experiments using 1FNIII and the negatively charged 3FNIII under conditions identical to those used for the 1FNIII–2FNIII titrations. No observable changes could be detected in the HSQC spectra upon mixing 1FNIII/3FNIII (data not shown), thus indicating that the observed 1FNIII–2FNIII interaction is specific for this domain pair. In addition, significant chemical shift perturbations could be detected in the 1FNIII/2FNIII HSQC spectra at physiological ionic strength (data not shown).
Previous studies in FNIII domain pairs have shown a significant reduction of molecular motions in the second domain compared with isolated FNIII domains (Carr et al, 1997; Spitzfaden et al, 1997; Altroff et al, 2004). Heteronuclear {1H–}15N NOE and 15N T2 data collected for 1−2FNIII (Supplementary Figure 5) show that this is not the case for 2FNIII, since the N-terminal region of this protein, as well as the interdomain linker, remain flexible. Comparison of the backbone amide chemical shifts of 1FNIII and 2FNIII, isolated and in the 1−2FNIII context, shows that the interdomain interaction is present in this construct (Supplementary Figure 4C and D). The chemical shift perturbation patterns observed in 1−2FNIII best match those of 2FNIII in the presence of 1FNIII. 1FNIII perturbation patterns are relatively complicated, presumably due to the unstructured interdomain linker. The unstructured linker does not affect 2FNIII chemical shifts as its N-terminus is already flexible.
We performed simple calculations of the effective local 1FNIII and 2FNIII concentrations when connected by the linker; single 1FNIII and 2FNIII molecules were assumed to be present inside a sphere with a diameter that is the sum of the diameters of gyration of 1FNIII (26.6 Å) and 2FNIII (24.6 Å), plus the average end-to-end distance of a flexible 34-amino-acid chain. The latter was calculated as approximately 37.6 Å (Cα–Cα distance) from a large library of simulated linker chains. Local 1FNIII and 2FNIII concentrations were thus calculated to be approximately 4.5 mM, which, given an estimated Kd of 0.1 mM, implies that over 85% of 1FNIII molecules would be complexed with 2FNIII in the 1−2FNIII context. In practice, this estimate of the local concentration is expected to be low, since the linker will probably not be able to access all possible conformations. Note that we have only considered intrachain interactions in solution, because intermolecular interactions are considered to be unlikely under the conditions used. Analytical gel filtration of 1−2FNIII (Supplementary Figure 6) shows the presence of a single species; 15N T2 relaxation rates of 1−2FNIII (Supplementary Figure 5B) and analytical ultracentrifugation experiments (Supplementary Figure 7A) also indicate that this species is monomeric in solution.
1−2FNIII structure
The extent of chemical shift perturbations caused by the 1FNIII–2FNIII interaction is small, suggesting that the structures of the two domains remain highly similar to those calculated in the absence of this interaction. A possible approach toward determining the complex structure is then to use a rigid-body docking protocol to compute restraint models of intermolecular complexes (Clore and Schwieters, 2003; Dominguez et al, 2003). These protocols utilize chemical shift perturbations to delineate the interaction surfaces, and residual dipolar coupling (RDC) restraints measured for both complexed components to provide global orientation. Due to the ambiguous nature of such distance restraints, no specific information is entered into the calculation concerning interdomain interface packing. Despite the absence of specific packing information, the final model is often highly similar (backbone r.m.s. deviations ∼1 Å) to models derived from traditional methods (Clore and Schwieters, 2003).
This approach was used to calculate the structure of 1−2FNIII. Ambiguous NOE restraints were applied between residues of 1FNIII and 2FNIII that exhibit significant chemical shift perturbations and satisfy the solvent accessibility and surface continuity criteria described previously (Clore and Schwieters, 2003). Shift perturbations from both the 1FNIII/2FNIII titrations and 1−2FNIII (Supplementary Figure 4) were considered in order to reach a consensus. The final residues selected for distance restraints were S621, S625, K669, G674, V675, T698 and S701 for 1FNIII, and R751, E753, E755, D767, N789, Q799 and S800 for 2FNIII. In addition, 1DNH RDC restraints were collected in a strained polyacrylamide medium in the context of 1−2FNIII and used in the structure calculation. One hundred structures were calculated and the 50 lowest-energy models were retained for further analysis. The final family of structures consisted of two distinct population ensembles that fitted the experimental data equally well (Figure 3A and B). All ambiguous NOE restraints were satisfied in the vast majority of the calculated models, and the RDC restraint R-factors (Clore and Garrett, 1999) increased only slightly in the complex compared with those computed for isolated domains (1FNIII isolated R-factor ∼14.5%, complex R-factor ∼16.8%; 2FNIII isolated R-factor ∼9%, complex R-factor ∼9.8%).
Figure 3.
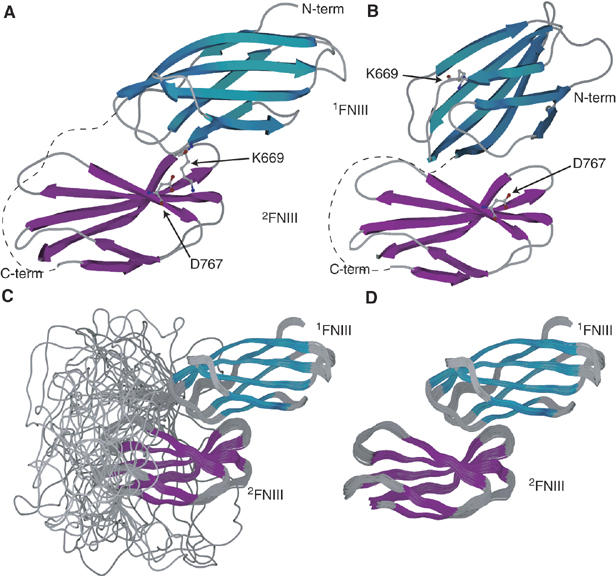
(A, B) Representative structures of the two different 1−2FNIII forms present in the structure calculation. Occupancy of form A in the final ensemble was 78%, whereas that of form B was 22%. Side chains for two residues, K669 and D767, involved in an electrostatic interaction in form A but not form B, are shown. D767 is also involved in electrostatic interactions with K672 in population A, and participates in an extended hydrogen bond and electrostatics network on the 1FNIII binding interface of 2FNIII. The interdomain linker is schematically represented as a dashed line. (C, D) 1−2FNIII structure. Shown here are: the final 39-structure ensemble of 1−2FNIII structures displaying (C) or omitting (D) the flexible 1FNIII–2FNIII linker. Secondary structure elements are colored cyan and purple for 1FNIII and 2FNIII, respectively.
The two populations present in the final ensemble are related through an approximately 180° rotation and a small translation of the two domains with respect to each other. Due to the two-fold degeneracy present in the RDC restraints, the two models could not be discriminated by these initial data. Model A in Figure 3 represents 78% of the final family of structures (population A), whereas model B corresponds to the remaining 22% (population B). Two different approaches were used in order to determine which of the two population ensembles corresponds to the correct 1−2FNIII structure. First, we attempted to locate and disrupt interdomain interactions unique to one particular model, by designing specific amino-acid residue substitutions. Second, we collected additional RDC restraints for 1−2FNIII in a polyethylene glycol (PEG)/hexanol medium to break the two-fold symmetry inherent in the RDC data.
An electrostatic (salt-bridge) interaction between residues K669 of 1FNIII and D767 of 2FNIII is present only in population A (Figure 3A). The same residues in population B are exposed to solvent and are unlikely to contribute any stabilizing interactions. K669 experiences significant chemical shift perturbations in the 1FNIII–2FNIII interaction (Supplementary Figure 4), consistent with the structural interactions seen in population A. Domain variants with alanine substituting these two amino acids were created and tested for interdomain interactions in our HSQC chemical shift perturbation assay (Supplementary Figure 2E–H). Both of these variants appear to reduce the chemical shift change upon domain interaction for given protein concentrations and 1FNIII/2FNIII ratios. The dissociation constant for 1FNIII K669A and wild-type 2FNIII was estimated to be approximately 10-fold higher than that of the wild-type domains (Supplementary Figure 3), whereas shift perturbations for the interaction of 2FNIII D767A with wild-type 1FNIII were too small to allow an accurate Kd estimation. When 1FNIII K669A was added to 2FNIII D767A, no observable chemical shift perturbations were observed (data not shown), and analytical gel filtration experiments with a 1−2FNIII variant that contains both amino-acid substitutions show that the great majority of this 1−2FNIII construct is in a conformation with significantly larger hydrodynamic radius than wild-type 1−2FNIII (Supplementary Figure 6). NMR spectra of 1−2FNIII K669A/D767A (referred to henceforth as 1−2FNIII KADA) showed that this variant remains well folded and in a non-supramolecular aggregate state (data not shown); this was confirmed by analytical ultracentrifugation experiments (Supplementary Figure 7B). We expect that 1FNIII and 2FNIII completely dissociate in this variant, thus corresponding to a 1−2FNIII open conformation with an average hydrodynamic radius of approximately 110 Å, a two-fold increase over the 58 Å length of population A. In contrast, the K669A/D767A substitutions should not significantly affect the interactions in population B, thereby arguing against the validity of population B as a structural model of 1−2FNIII.
We performed singular value decomposition fits of the PEG/hexanol RDC restraints to the isolated 1FNIII and 2FNIII domains, as well as to structural models from populations A and B of our 1−2FNIII structure ensemble. The latter models were chosen to have the smallest backbone r.m.s.ds from the average of their respective populations. The structures of the individual domains are in good agreement with the new RDC restraints, having r.m.s.ds from those restraints of approximately 2.9 and 1.75 Hz for 1FNIII and 2FNIII, respectively (data not shown). The model of population A was also in good agreement, with an r.m.s.d. of approximately 3.1 Hz (3.9 Hz for the 1FNIII part and 2.3 Hz for the 2FNIII part) and a correlation R2 of experimental and calculated RDCs of approximately 97% (Supplementary Figure 8A). In contrast, the model of population B could not be fit well using the same procedure, having an r.m.s.d. to the data of over 10 Hz and a correlation R2 of only 67% (Supplementary Figure 8B). Therefore, both mutagenesis and NMR data indicate that population A is a better representation of the 1−2FNIII structure.
The 39-structure ensemble of population A is shown in Figure 3C and D, and a structural analysis is presented in Supplementary Table III. 1FNIII residues on the loops connecting β-strands A and B, and E and F interact with 2FNIII residues located on the strongly twisted four-stranded β-sheet, creating an overall appearance of 1FNIII being ‘wedged' along an N-terminal-to-C-terminal axis on the curved 2FNIII β-sheet. The interaction surface of the two domains is relatively small, approximately 540 Å2. Complex formation introduces only a relatively small change in the overall protein chain direction, as the N- and C-terminal direction vectors of 1−2FNIII differ by approximately 32°. This is comparable to the 10–40° angle formed by canonical type III domain pairs (Leahy et al, 1996; Spitzfaden et al, 1997; Altroff et al, 2004). The length of the interdomain linker is almost invariable in diverse species (Figure 2) and is considerably larger than is necessary to bridge the 1FNIII C-terminus and the 2FNIII N-terminus in this conformation. Indeed, calculations based on our model suggest that a linker length of about 15 residues would be sufficient to allow the observed 1FNIII–2FNIII interaction.
The almost 10-fold decrease in affinity of our K669A substitution (Supplementary Figure 3) corresponds, in energetic terms, to a loss of approximately 1.3 kcal/mol in stabilization energy. This value is similar to that previously reported in proteins (Horovitz et al, 1990) and model peptides (Mayne et al, 1998) for disruption of single salt-bridge interactions. Nevertheless, the 1FNIII–2FNIII interaction is still present in this variant both for isolated domains (Supplementary Figure 2E and F) and 1−2FNIII K669A (data not shown). In contrast, the D767A substitution has an effect larger than that expected for a single salt-bridge disruption, effectively abolishing the interdomain interaction (Supplementary Figure 2G and H). Indeed, in addition to K669, D767 can also interact favorably with K672 and possibly K655 in our model. Further, D767 participates in an extended hydrogen bond and electrostatics network on the surface of the twisted β-sheet of 2FNIII; R751 and Q799, two other 2FNIII residues that show significant perturbations upon 1FNIII binding are also involved. Therefore, it is likely that D767 is a key residue for maintaining the 2FNIII surface in a conformation favorable for 1FNIII interaction. D767 is conserved or conservatively substituted in all FN proteins (Figure 2), whereas K669 is absent in some of them.
1−2FNIII interaction with FN30 kDa
Binding of 1FNIII and 2FNIII to the N-terminal FN fragment has been shown previously (Aguirre et al, 1994; Hocking et al, 1994; Sechler et al, 2001), although only the 2FNIII interaction has been demonstrated under native conditions. We performed surface plasmon resonance (SPR) experiments with immobilized FN30 kDa under physiological conditions to determine whether the observed 1FNIII–2FNIII interaction affects binding to FN30 kDa. Wild-type 1−2FNIII interacts with FN30 kDa weakly, with a Kd of approximately 85 μM, as estimated from equilibrium analysis (Figure 4A and Supplementary Figure 9A). In contrast, 1−2FNIII KADA, an open conformation of 1−2FNIII, binds to FN30 kDa strongly (Figure 4A and Supplementary Figure 9C). Kinetic analysis of binding by this variant fitted a simple 1:1 Langmuir model with a dissociation constant of approximately 1.4 nM (kon 4.04 × 104 M−1s−1, koff 5.56 × 10−5 s−1).
Figure 4.
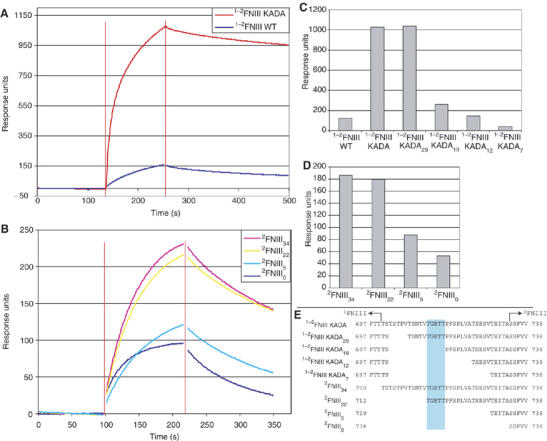
1−2FNIII–FN30 kDa interaction. Shown here are sample SPR sensogram traces of (A) the interaction of wild-type 1−2FNIII and 1−2FNIII KADA (5 μM protein concentration) and (B) the interaction of different 2FNIII linker length variants (20 μM protein concentration) with immobilized FN30 kDa. Data points close to injection start and end (denoted by red vertical lines) were removed for clarity. (C, D) Response levels at a time point 50 s after the end of the injection for the interaction of (C) different 1−2FNIII variants (5 μM protein concentration) and (D) different 2FNIII variants (20 μM protein concentration) with immobilized FN30 kDa. (E) The amino-acid sequences of the interdomain linker for the different 2FNIII and 1−2FNIII variants are shown here. The 1FNIII end and the start of the structured part of 2FNIII are denoted. Highlighted in cyan are linker residues, removal of which significantly affects FN30 kDa binding.
Further testing under the same conditions showed no appreciable binding of 1FNIII and the interdomain linker (data not shown) to immobilized FN30 kDa, whereas 2FNIII bound with intermediate kinetics and affinity (Supplementary Figure 9B). However, a series of 1−2FNIII KADA and 2FNIII constructs with variable lengths of the interdomain linker (Figure 4E) exhibited binding that decreased as the linker sequence was truncated from the N-terminus (Figure 4B–D). This trend is observed in both sets of constructs, suggesting that the effect is not due to the KADA substitutions but to the presence of the linker sequence. Removal of 1FNIII also affects binding adversely, as full-length 1−2FNIII KADA shows stronger binding affinity compared with a 2FNIII variant with the entire interdomain linker (Figure 4). Thus, it is likely that the high affinity of 1−2FNIII KADA for FN30 kDa is the result of cooperativity along multiple, independently weak binding sites. Interestingly, our variable linker constructs delineate a specific series of residues (Figure 4E), removal of which significantly decreases binding affinity to FN30 kDa. These residues form a generally conserved patch (Figure 2) in the middle of the linker, indicating that the interaction described here is of biological significance.
Discussion
An understanding of fibrillogenesis requires knowledge of the structural states and interactions of FN and FN components, both in solution and in fibrils. Although type III domains have been extensively studied both in FN (Leahy et al, 1996; Sharma et al, 1999; Gao et al, 2003) and in other systems (Leahy et al, 1992; Bisig et al, 1999), 2FNIII constitutes a unique case. It represents a well folded and stable (Litvinovich and Ingham, 1995) type III domain but with a flexible β-strand A in the native form. In spite of this flexibility, we have demonstrated that a 1FNIII–2FNIII interdomain interaction orients the two domains in FN in a way that maintains the chain direction. Recognition of residues responsible for abnormal chain deviations could be important for understanding the role of type III domains that form structural blocks in large proteins (Campbell and Spitzfaden, 1994).
The interaction between 1FNIII and 2FNIII presented here has been previously alluded to in mechanical unfolding (Oberhauser et al, 2002) and in vitro functional experiments (Chernousov et al, 1991; Morla and Ruoslahti, 1992; Sechler et al, 2001). Oberhauser et al (2002) showed, using AFM, that 1FNIII in the 1−2FNIII context exhibits greater mechanical stability than 1FNIII attached to the titin immunoglobulin domain I27. If the time constant for 1FNIII–2FNIII dissociation is smaller than the time constant in the mechanical unfolding experiment (∼40 ms), then 2FNIII could protect the C-terminal end of 1FNIII from unfolding. Under the conditions used in our study, the lifetime of the interaction between isolated 1FNIII and 2FNIII domains is relatively short (sub-millisecond); however, it is possible that, in the context of 1−2FNIII, and under the conditions used for the AFM experiments, the interaction is slow enough to allow such stabilization.
Functional studies suggest that 1FNIII and 2FNIII interact with FN30 kDa (Aguirre et al, 1994; Hocking et al, 1994; Sechler et al, 2001), an FN fragment essential for fibrillogenesis (Schwarzbauer, 1991). 2FNIII is also essential for robust fibril accumulation, whereas recombinant FN lacking 1FNIII was reported to form normal fibrils (Sechler et al, 2001). Our results indicate that 1−2FNIII and isolated 2FNIII bind FN30 kDa relatively weakly in their native forms. In contrast, the open 1−2FNIII conformation represented by our KADA variant binds FN30 kDa strongly and almost irreversibly. A truncation analysis of this open conformation showed that all three components of this construct (1FNIII, interdomain linker and 2FNIII) are necessary to achieve tight binding, although it is possible that in vivo some of these requirements, notably that of 1FNIII, could be relaxed due to the presence of additional interacting domains. We have established a strong correlation in vitro between the presence of the interdomain linker and FN30 kDa binding. Of particular interest is a series of generally conserved residues in the middle of this linker (Figures 2 and 4E) that cannot be explained in structural terms as the entire linker is flexible. Removal of these residues decreases FN30 kDa binding in vitro.
A possible model of fibril formation and elongation emerges from our results and previously published data (Figure 5). 1−2FNIII exists in the closed conformation in the solution state of FN and the weak interaction between 1−2FNIII and FN30 kDa might help maintain the FN globular structure (Figure 5A). During fibril formation, FN binds to cell surface receptors and is stretched by cell-generated tension (Figure 5B). This disrupts the globular structure and causes 1FNIII and 2FNIII to dissociate; subsequently, the 1−2FNIII open conformation interacts strongly with the FN30 kDa region of other FN molecules, thereby creating a FN homodimer along the FN N-terminus (Figure 5C). As FN is disulfide crosslinked at the C-terminus, this FN self-association interaction could potentially create a FN fibril that, upon elongation, is rendered insoluble. Thus, our proposed mechanism places a regulatory role on the 1FNIII–2FNIII interaction at the initiation step of fibrillogenesis. The presence of this interaction would ensure that fibril formation does not occur spontaneously, but that, through the requirement for tension-induced dissociation, it is under firm cellular control. We believe that this model of weakly interacting domains, dissociating to uncover cryptic sites, may well also be applicable to other stress-sensing molecules.
Figure 5.
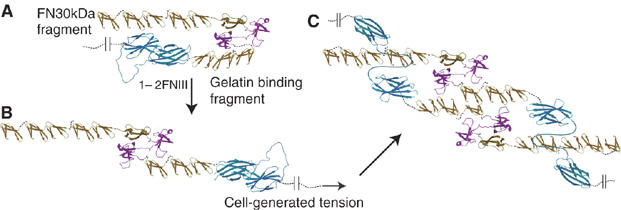
Possible fibrillogenesis mechanism: FN molecules exist in solution, with the 1−2FNIII domain pair in closed conformation, likely interacting with the FN N-terminus as part of a larger globular structure (A). Under tension, the FN globular structure and the 1FNIII–2FNIII interaction are disrupted (B, C). This allows the 1−2FNIII open conformation to strongly associate with the N-terminus of other FN molecules (C) and, along with the disulfide crosslinks at the FN C-terminus, create FN fibrils. The different domain types of FN are shown in gold (FNI), purple (FNII) or cyan (FNIII). The different FN fragments shown correspond to: FN30 kDa, 1−5FNI; gelatin binding domain, 6FNI-1−2FNII-7−9FNI.
Materials and methods
Protein preparation and analysis
Gene fragments encoding human FN residues 608–701 (1FNIII), 721–809 (2FNIII), 700–809 (2FNIII34), 712–809 (2FNIII22), 729–809 (2FNIII5), 734–809 (2FNIII0), 810–900 (3FNIII) or 608–809 (1−2FNIII) were cloned as GST fusions using a pGEX-6P-2 expression vector (GE Biosciences). Modified genes with amino-acid substitutions or internal truncations were created using a PCR-based method and cloned in a modified pET-16 expression vector (Novagen) incorporating a 3C protease cleavage site. These genes include the 1−2FNIII K669A/D767A double mutant (1−2FNIII KADA), as well as variants of this double mutant with linker truncations, 1−2FNIII KADA29 (FN residues 608–701 and 707–809), 1−2FNIII KADA19 (residues 608–701 and 717–809), 1−2FNIII KADA12 (residues 608–701 and 724–809) and 1−2FNIII KADA7 (residues 608–701 and 729–733). Details of the protein expression and purification protocols can be found in the supporting information. As prepared, 2FNIII includes six N-terminal non-FN residues (cloning artifacts, amino-acid sequence GPLGSH), whereas all other constructs include five N-terminal residues (GPLGS). The proteolytic N-terminal FN 30 kDa fragment (FN30 kDa) was purchased from Sigma.
Gel filtration chromatography was performed using a calibrated Superdex-75 analytical gel filtration column (GE Biosciences) equilibrated with PBS buffer. Analytical ultracentrifugation equilibrium experiments were performed on samples in PBS buffer using a Beckman Optima XL-A analytical ultracentrifuge. UV absorbance was monitored at 280 nm. The duration of the run was 48 h at 20 000 r.p.m. and it was conducted at 25°C. The data were fit to an ideal monodisperse model using the program Origin (OriginLab).
NMR experiments
All experiments were performed at 30°C using 11.7 T, 14.1 T, 17.6 T and 22.3 T spectrometers equipped with triple-resonance, triple-axis gradient probeheads (Soffe et al, 1995). NMR sample buffers corresponded to 20 mM NaCl, 20 mm NaPi pH 7.0 for 1FNIII, 2FNIII, 3FNIII wild-type and variants, and 150 mM NaCl, 20 mM NaPi pH 7.0 for 1−2FNIII wild type and variants. Details of the NMR experiment performed are provided in the supporting information. All assignments have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession numbers 7127 and 7128 for wild-type 2FNIII and 1−2FNIII, respectively.
NMR structure calculations
Structure calculations were performed using the XPLOR-NIH software package (Schwieters et al, 2003). 2FNIII calculations were based on established simulated annealing protocols. In contrast, 1−2FNIII calculations employed a rigid-body docking protocol described previously (Clore and Schwieters, 2003), appropriately modified for two structured domains and a flexible linker in a single polypeptide chain. XPLOR-NIH was also used for short simulations of possible linker conformations under conditions that generated random coil conformations with favorable Ramachandran statistics. Details of these different types of calculations can be found in the supporting information. The structures and structure calculation restraints for 2FNIII have been deposited in the RCSB Protein Databank under accession numbers 2H41 and 2H45 for the average minimized structure and the 25-structure ensemble, respectively; the population A structure ensemble of 1−2FNIII is deposited under accession number 2HA1, along with the calculation restraints.
Surface plasmon resonance
SPR experiments were performed on a BIAcore 2000 instrument (Biacore AB, Uppsala, Sweden). The FN30 kDa was immobilized on the dextran matrix of the sensor chip by amine coupling (Biacore, 1994) in 10 mM sodium acetate buffer at pH 5.0. All experiments were carried out at 25°C in HBS running buffer (20 mM HEPES at pH 7.4, 150 mM NaCl and 0.005% (v/v) Tween 20), with flow rates of 50 and 1 μl/min for the kinetic and equilibrium experiments, respectively. Regeneration of the sensor surface was achieved with 30-s exposure to 50 mM HCl. The equilibrium binding constant was measured for the binding of wild-type 1−2FNIII by injecting 100 μl of this construct (at concentrations of 17–270 μM) over immobilized FN30 kDa at 1 μl/min until equilibrium was reached. Responses at equilibrium, corrected for background from control flow cells, were recorded as Req for individual injections. Scatchard analysis was used to evaluate the binding constants from triplicate data sets using linear regression, which gives –1/Kd as slope and Rmax as intercept on the x-axis. The on- and off-rates (kon and koff) of 2FNIII and the 1−2FNIII KADA variant binding to 30 kDa FN30 kDa were measured by injecting a series of sample dilutions (2FNIII: 20–80 μM and 1−2FNIII variant 31–250 nM) over the immobilized fibronectin fragment. To reduce mass transport effects, the immobilization level was kept low (1000 RU) and the flow rate was kept high (50 μl/min) (Kortt et al, 1999). Due to the slow koff for 1−2FNIII KADA (5.6 × 10−5 s−1), the dissociation rate was measured over 15 min to obtain a reliable value. The kon and koff were obtained by fitting the 1:1 Langmuir binding and the heterogeneous ligand models to components of the sensorgrams. Experiments of similar type at a specific protein concentration level were conducted for the different 2FNIII (20 μM protein concentration) and 1−2FNIII KADA (5 μM protein concentration) variants with variable linker lengths, over FN30 kDa immobilized at 5000 RU to increase the sensitivity of the sensor. The sensogram response levels at a fixed point 50 s after the end of the injection, corrected for background from the control flow cell, were used as a measure of the relative binding affinities of the variants. The data were analyzed using the BIAevaluation software (Biacore, 1997) provided with the instrument.
Figures and notes
Amino-acid numbering for the different proteins corresponds to that in their respective entries; the accession numbers for the different FN proteins are as follows: Homo sapiens UniProt P02751, Brachydanio rerio FN3 Q6JAN2, B. rerio FN1a O93405, Xenopus tropicalis Q501R6, X. laevis Q91740, Bos taurus P07589, Rattus norvegicus P04937 and Mus musculus P11276. The FN protein of Gallus gallus used here corresponds to translation of the ensemble ENSGALT00000005654. The 2FNIII structural homologs identified correspond to the RCSB entries 1FNF (H. sapiens FN 8FNIII), 1QR4-B (G. gallus Tenascin 6FNIII), 1FNH (H. sapiens FN 13FNIII), 1TDQ-A (R. norvegicus Tenascin 3FNIII) and 1TEN (H. sapiens Tenascin 3FNIII). The subscript in the variable linker length 1−2FNIII KADA and 2FNIII constructs denotes the length or the linker in amino acids. Using the same nomenclature, our original constructs would correspond to 1−2FNIII KADA34 and 2FNIII13. The exact amino-acid sequences of all variants are described in the methods and are also shown in Figure 4E.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr Jonathan Boyd and Mr Nick Soffe for technical assistance with the NMR instrumentation, Dr Sophie Ribaud for the 2FNIII pGEX construct and Professor Jean E Schwarzbauer (Princeton University) for helpful discussions and comments related to the manuscript. Financial support was provided by the Wellcome Trust and BBSRC. Funding for the 22.3 T (950 MHz 1H frequency) superconducting magnet was provided by the Wellcome Trust, and for modifications to the NMR laboratory by the Edward Penley Abraham Fund.
References
- Abu-Lail NI, Ohashi T, Clark RL, Erickson HP, Zauscher S (2006) Understanding the elasticity of fibronectin fibrils: unfolding strengths of FN-III and GFP domains measured by single molecule force spectroscopy. Matrix Biol 25: 175–184 [DOI] [PubMed] [Google Scholar]
- Aguirre KM, McCormick RJ, Schwarzbauer JE (1994) Fibronectin self-association is mediated by complementary sites within the amino-terminal one-third of the molecule. J Biol Chem 269: 27863–27868 [PubMed] [Google Scholar]
- Altroff H, Schlinkert R, van der Walle CF, Bernini A, Campbell ID, Werner JM, Mardon HJ (2004) Interdomain tilt angle determines integrin-dependent function of the ninth and tenth FIII domains of human fibronectin. J Biol Chem 279: 55995–56003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacore (1994) BIAcore 2000 instrument handbook. Biacore AB: Uppsala, Sweden
- Biacore (1997) BIAevaluation 3.0 software handbook. Biacore AB: Uppsala, Sweden
- Bisig D, Weber P, Vaughan L, Winterhalter KH, Piontek K (1999) Purification, crystallization and preliminary crystallographic studies of a two fibronectin type-III domain segment from chicken tenascin encompassing the heparin- and contactin-binding regions. Acta Crystallogr D 55: 1069–1073 [DOI] [PubMed] [Google Scholar]
- Briknarova K, Akerman ME, Hoyt DW, Ruoslahti E, Ely KR (2003) Anastellin, an FN3 fragment with fibronectin polymerization activity, resembles amyloid fibril precursors. J Mol Biol 332: 205–215 [DOI] [PubMed] [Google Scholar]
- Campbell ID, Spitzfaden C (1994) Building proteins with fibronectin type III modules. Structure 2: 333–337 [DOI] [PubMed] [Google Scholar]
- Carr PA, Erickson HP, Palmer AG III (1997) Backbone dynamics of homologous fibronectin type III cell adhesion domains from fibronectin and tenascin. Structure 5: 949–959 [DOI] [PubMed] [Google Scholar]
- Chernousov MA, Fogerty FJ, Koteliansky VE, Mosher DF (1991) Role of the I-9 and III-1 modules of fibronectin in formation of an extracellular fibronectin matrix. J Biol Chem 266: 10851–10858 [PubMed] [Google Scholar]
- Clore GM, Garrett DS (1999) R-factor, free R, and complete cross-validation for dipolar coupling refinement of NMR structures. J Am Chem Soc 121: 9008–9012 [Google Scholar]
- Clore GM, Schwieters CD (2003) Docking of protein-protein complexes on the basis of highly ambiguous intermolecular distance restraints derived from 1H/15N chemical shift mapping and backbone 15N–1H residual dipolar couplings using conjoined rigid body/torsion angle dynamics. J Am Chem Soc 125: 2902–2912 [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM (2003) HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125: 1731–1737 [DOI] [PubMed] [Google Scholar]
- Erickson HP (2002) Stretching fibronectin. J Muscle Res Cell Motil 23: 575–580 [DOI] [PubMed] [Google Scholar]
- Gao M, Craig D, Lequin O, Campbell ID, Vogel V, Schulten K (2003) Structure and functional significance of mechanically unfolded fibronectin type III1 intermediates. Proc Natl Acad Sci USA 100: 14784–14789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Bershadsky A, Pankov R, Yamada KM (2001) Transmembrane extracellular matrix–cytoskeleton crosstalk. Nat Rev Mol Cell Biol 2: 793–805 [DOI] [PubMed] [Google Scholar]
- Hocking DC, Sottile J, McKeown-Longo PJ (1994) Fibronectin's III-1 module contains a conformation-dependent binding site for the amino-terminal region of fibronectin. J Biol Chem 269: 19183–19187 [PubMed] [Google Scholar]
- Holm L, Sander C (1998) Touring protein fold space with Dali/FSSP. Nucleic Acids Res 26: 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz A, Serrano L, Avron B, Bycroft M, Fersht AR (1990) Strength and co-operativity of contributions of surface salt bridges to protein stability. J Mol Biol 216: 1031–1044 [DOI] [PubMed] [Google Scholar]
- Ingham KC, Brew SA, Huff S, Litvinovich SV (1997) Cryptic self-association sites in type III modules of fibronectin. J Biol Chem 272: 1718–1724 [DOI] [PubMed] [Google Scholar]
- Kortt AA, Nice E, Gruen LC (1999) Analysis of the binding of the Fab fragment of monoclonal antibody NC10 to influenza virus N9 neuraminidase from tern and whale using the BIAcore biosensor: effect of immobilization level and flow rate on kinetic analysis. Anal Biochem 273: 133–141 [DOI] [PubMed] [Google Scholar]
- Leahy DJ, Aukhil I, Erickson HP (1996) 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 84: 155–164 [DOI] [PubMed] [Google Scholar]
- Leahy DJ, Hendrickson WA, Aukhil I, Erickson HP (1992) Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science 258: 987–991 [DOI] [PubMed] [Google Scholar]
- Litvinovich SV, Ingham KC (1995) Interactions between type III domains in the 110 kDa cell-binding fragment of fibronectin. J Mol Biol 248: 611–626 [DOI] [PubMed] [Google Scholar]
- Mao Y, Schwarzbauer JE (2005) Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol 24: 389–399 [DOI] [PubMed] [Google Scholar]
- Mayne L, Englander SW, Qiu R, Yang JX, Gong YX, Spek EJ, Kallenbach NR (1998) Stabilizing effect of a multiple salt bridge in a prenucleated peptide. J Am Chem Soc 120: 10643–10645 [Google Scholar]
- Morla A, Ruoslahti E (1992) A fibronectin self-assembly site involved in fibronectin matrix assembly: reconstruction in a synthetic peptide. J Cell Biol 118: 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morla A, Zhang Z, Ruoslahti E (1994) Superfibronectin is a functionally distinct form of fibronectin. Nature 367: 193–196 [DOI] [PubMed] [Google Scholar]
- Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM (2002) The mechanical hierarchies of fibronectin observed with single-molecule AFM. J Mol Biol 319: 433–447 [DOI] [PubMed] [Google Scholar]
- Ohashi T, Erickson HP (2005) Domain unfolding plays a role in superfibronectin formation. J Biol Chem 280: 39143–39151 [DOI] [PubMed] [Google Scholar]
- Schwarzbauer JE (1991) Identification of the fibronectin sequences required for assembly of a fibrillar matrix. J Cell Biol 113: 1463–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Marius Clore G (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160: 65–73 [DOI] [PubMed] [Google Scholar]
- Sechler JL, Rao H, Cumiskey AM, Vega-Colon I, Smith MS, Murata T, Schwarzbauer JE (2001) A novel fibronectin binding site required for fibronectin fibril growth during matrix assembly. J Cell Biol 154: 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Askari JA, Humphries MJ, Jones EY, Stuart DI (1999) Crystal structure of a heparin- and integrin-binding segment of human fibronectin. EMBO J 18: 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffe N, Boyd J, Leonard M (1995) The construction of a high-resolution 750 MHz probehead. J Magn Reson series A 116: 117–121 [Google Scholar]
- Spitzfaden C, Grant RP, Mardon HJ, Campbell ID (1997) Module–module interactions in the cell binding region of fibronectin: stability, flexibility and specificity. J Mol Biol 265: 565–579 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


