Abstract
Agrobacterium tumefaciens translocates T-DNA through a polar VirB/D4 type IV secretion (T4S) system. VirC1, a factor required for efficient T-DNA transfer, bears a deviant Walker A and other sequence motifs characteristic of ParA and MinD ATPases. Here, we show that VirC1 promotes conjugative T-DNA transfer by stimulating generation of multiple copies per cell of the T-DNA substrate (T-complex) through pairwise interactions with the processing factors VirD2 relaxase, VirC2, and VirD1. VirC1 also associates with the polar membrane and recruits T-complexes to cell poles, the site of VirB/D4 T4S machine assembly. VirC1 Walker A mutations abrogate T-complex generation and polar recruitment, whereas the native protein recruits T-complexes to cell poles independently of other polar processing factors (VirC2, VirD1) or T4S components (VirD4 substrate receptor, VirB channel subunits). We propose that A. tumefaciens has appropriated a progenitor ParA/MinD-like ATPase to promote conjugative DNA transfer by: (i) nucleating relaxosome assembly at oriT-like T-DNA border sequences and (ii) spatially positioning the transfer intermediate at the cell pole to coordinate substrate—T4S channel docking.
Keywords: ATPase, conjugation, partitioning, relaxosome, type IV secretion
Introduction
Bacterial conjugative DNA transfer contributes to genome plasticity and mediates the widespread transmission of antibiotic-resistance genes and other medically important virulence traits. The overall process of conjugation is divisible into three stages. First, the DNA transfer and replication (Dtr) proteins assemble as a relaxosome at a cognate origin-of-transfer (oriT) sequence (Pansegrau and Lanka, 1996). One relaxosome component, the relaxase, catalyzes strand-specific cleavage at the nic site and remains covalently bound to the 5′ end of the nicked strand destined for transfer (T-strand) (Pansegrau and Lanka, 1996). Next, in a reaction that is comparatively poorly understood at this time, a cognate membrane-bound substrate receptor or ‘coupling protein' (CP) (Schroder et al, 2002) recruits the relaxase-T-strand particle to a cognate type IV secretion (T4S) system. Finally, the T4S channel or ‘mating pore' translocates the DNA substrate across the cell envelope and into bacterial or eukaryotic target cells (Cascales and Christie, 2004b). In recent years, various structural and mechanistic features of conjugation have emerged (Christie et al, 2005 and references therein). However, fundamental questions remain concerning the molecular events underlying spatiotemporal coordination of the DNA substrate processing, recruitment and translocation reactions.
The Agrobacterium tumefaciens VirB/D4 T4S system localizes at cell poles (Lai et al, 2000; Kumar and Das, 2002; Judd et al, 2005), providing a convenient positional marker for characterizing early substrate recruitment reactions (Ding et al, 2002). The natural DNA substrate of the VirB/D4 T4S system is a ∼12-kb segment of transfer DNA (T-DNA), whose delivery to plant cells results in production of plant tumors (McCullen and Binns, 2006). The VirD2 relaxase cleaves oriT-like T-DNA border sequences in a relaxosomal complex (Yanofsky et al, 1986) thought to also include the ancillary subunits VirD1, VirC1, and VirC2 (Veluthambi et al, 1988; de Vos and Zambryski, 1989; Scheiffele et al, 1995). How these latter processing factors stimulate T-DNA border nicking is not known. Early studies showed that VirC1 bind a sequence termed overdrive located adjacent to the T-DNA right border repeat (Peralta et al, 1986; Toro et al, 1989), prompting a proposal that VirC1 bound to overdrive stimulates relaxosome assembly through recruitment of other processing factors. This activity or other possible VirC1 functions have not been characterized.
VirC1 belongs to a superfamily of ATPases containing a deviant Walker A nucleotide triphosphate-binding motif (KGGXXK[ST]) and other conserved A′ and B sequence motifs (Koonin, 1993). Members of this family include ParA required for accurate chromosomal and plasmid DNA partitioning (Bignell and Thomas, 2001; Ebersbach and Gerdes, 2005), MinD required for correct placement of septa during cell division (Shih and Rothfield, 2006), and Soj which plays a role in chromosome compaction required for nucleoid partition (Lee and Grossman, 2006). ParA proteins localize at or near cell centers or poles, exhibit oscillatory behavior between specific locations of a cell, or assemble dynamically as cytoskeletal structures (Ebersbach and Gerdes, 2004; Lim et al, 2005). These proteins function as spatial determinants for cognate DNA elements during segregation, mediating their positional effects through interactions with ParB proteins and a cis-acting ‘centromere-like' DNA element, parC (or parS). MinD and Soj also display distinct dynamic behaviors essential for division site selection and nucleoid compaction, respectively (Ebersbach and Gerdes, 2005; Shih and Rothfield, 2006).
Here, we present experimental evidence suggesting that A. tumefaciens appropriated a ParA/MinD-like ATPase, VirC1 to stimulate two early steps of conjugation. Together with its partner protein VirC2, VirC1 stimulates relaxosome assembly at T-DNA border sequences. Independently of VirC2, VirC1 functions as a spatial determinant for the processed T-complex by recruiting the DNA substrate to the polarly localized T4S channel. Both VirC1 functions require NTP energy as suggested by phenotypes of strains producing VirC1 Walker A mutant proteins. These VirC1 activities manifest in enhanced interkingdom T-DNA transfer and A. tumefaciens virulence. Our findings expand the repertoire of functions described to date for members of the ParA/MinD ATPase superfamily.
Results
VirC1 and VirC2 strongly stimulate T-strand generation
We first quantified the stimulatory effects of VirC1 and VirC2 on processing of the transfer intermediate from T-DNA on the Ti plasmid. In response to sensory perception of the plant phenolic, acetosyringone (AS), the VirA/VirG two-component system activates expression of the virulence (vir) regulon as well as the repABC operon controlling Ti plasmid replication and copy number (Cho and Winans, 2005). Consistent with these findings we showed that the Ti plasmid copy number increased in wild-type (WT) and Dtr mutant (virC, virD) strains from about 1 per circular chromosome in uninduced cells to 4–5 within 24 h of AS induction (Figure 1A). As expected, T-strand production also increased in response to AS induction with a kinetics profile generally matching profiles for vir gene transcription (Chen and Winans, 1991) and Vir protein accumulation, for example, VirC1, VirD2, and VirB9 (Supplementary Figure S1). T-strand levels began increasing exponentially within 3 h of AS induction, and reached an estimated 12–14 molecules per Ti plasmid by 24 h (Figure 1B). By contrast, mutants lacking either or both virC1 or virC2 produced T-strands at three- to fourfold lower levels, suggesting that both VirC proteins function together to stimulate T-DNA processing. Two VirC1 Walker A mutants, VirC1K15Q (Figure 1B) and VirC1K15E (data not shown) also did not support efficient T-strand production, suggesting that VirC1 stimulates T-DNA substrate processing by an NTP-dependent mechanism.
Figure 1.
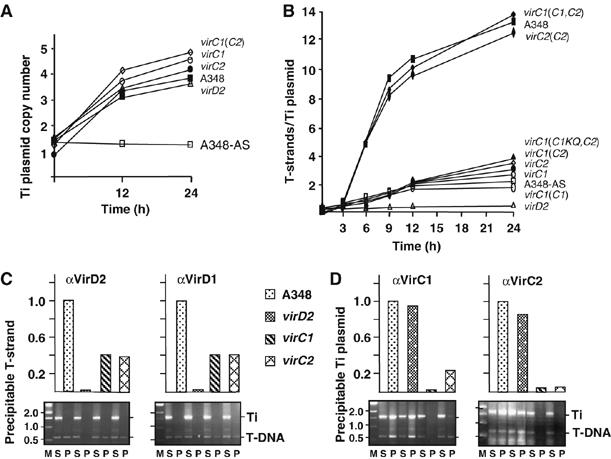
Quantitative effects of VirC1 and VirC2 on generation of the T-DNA transfer intermediate. (A) Increase in Ti plasmid copy number in response to AS induction. Strains: A348 WT strain; virD2, Mx311; virC1, Mx365 (this mutation is polar on downstream virC2); virC2, Mx364; virC1(C2), Mx365(pKA114) producing VirC2 from the IncP replicon. All strains except for A348-AS were induced with AS for the times indicated at 22°C. (B) Effects of VirC proteins on cellular levels of T-strand. Strains: same as (A) plus virC1(C1,C2), Mx365(pKAB188) producing VirC1 and VirC2 from an IncP replicon; virC1(C1KQ,C2), Mx365(pKAB190) producing VirC1K15Q, and VirC2 from an IncP replicon. T-strand levels were determined as described in the text. (C, D) Effects of VirC proteins on accumulation of the VirD2–T-strand intermediate. Strains listed at center were treated without (for VirD2) or with (for VirD1, VirC1, and VirC2) formaldehyde before cell lysis. Antibodies listed at the top of each histogram were used to immunoprecipitate the cognate Vir protein. Levels of co-precipitated T-strand or Ti plasmid were determined by quantitative (upper histogram) and nonquantitative (lower gel) ChIP (TrIP) assays as described previously (Cascales and Christie, 2004b). Upper histogram: T-strand or Ti plasmid levels in WT strain A348 are normalized to 1.0, and levels in vir mutant strains are depicted as a fraction of WT levels. Lower gel: agarose gels showing PCR amplification products generated with primers against a Ti plasmid gene fragment (Ti) and a T-DNA fragment (T-DNA). Lanes: M, molecular mass markers with sizes in kb listed at left; PCR products were generated using supernatant (nonprecipitated) (S) and immunoprecipitated (P) material.
From data in Figure 1A and B, it can be estimated that WT cells accumulate as many as 50 T-strand molecules within a 24-h induction period, whereas the virC mutants accumulate only 12–15 T-strands during this period. The accumulated T-strands likely exist as covalent VirD2–T-strand particles (T-complexes) as suggested by a chromatin immunoprecipitation (ChIP) assay (Cascales and Christie, 2004b), wherein anti-VirD2 antibodies immunoprecipitated abundant levels of the T-strand from WT cell extracts, about threefold lower amounts from mutants lacking one or both virC genes, and no detectable substrate from extracts of a virD2 mutant (Figure 1C).
VirD1 is important for VirD2-mediated border cleavage in vivo and a probable relaxosome component at T-DNA borders (Yanofsky et al, 1986; Stachel et al, 1987). Yet, it is unknown whether VirD1 remains associated with the VirD2–T-strand particle upon its release from the Ti plasmid. As shown with the ChIP assay, the anti-VirD1 antibodies precipitated T-strands from WT cells and virC mutants at levels similar to those recovered with the anti-VirD2 antibodies, suggesting that VirD1 binds at least transiently to the free VirD2–T-strand particle (Figure 1C). The anti-VirD1 and -VirD2 antibodies did not precipitate the Ti plasmid even though both proteins likely bind T-DNA border repeats directly or indirectly as components of the relaxosome. Conceivably, the relaxosome assembles only transiently or at levels below a threshold required for detection by ChIP.
In contrast to results shown in Figure 1C, the anti-VirC1 and -VirC2 antibodies precipitated abundant levels of the Ti plasmid from WT extracts (Figure 1D). These antibodies also precipitated the Ti plasmid from virD2 mutant extracts, indicating that the VirC proteins bind the T plasmid independently of relaxase binding or T-DNA processing. The VirC1 antibodies might precipitate the Ti plasmid via formation of a VirC1—overdrive complex (Toro et al, 1989), but it is interesting to note that these antibodies precipitated about fivefold less Ti plasmid from a virC2 mutant than a WT strain (Figure 1D), suggesting that VirC1 binding at overdrive or another Ti plasmid target sequence(s) is stimulated by VirC2. Because both anti-VirC antibodies precipitated the Ti plasmid, we could not use ChIP to assess whether the VirC proteins also associate with the free T-complex. Finally, by use of a modified ChIP assay termed transfer DNA immunoprecipitation (TrIP), we showed that even though the virC mutant strains generate low levels of the transfer intermediate (Figure 1B), these mutants displayed an overall WT pattern of close contacts between the T-DNA substrate and six VirB/D4 channel subunits (Supplementary Figure S2; Cascales and Christie, 2004b). The VirC proteins thus act by stimulating early T-DNA processing reactions and do not contribute to T-DNA substrate transfer per se through the VirB/D4 T4S channel.
VirC1 and VirC2 form a complex
Next, we assayed for interactions among the VirC and VirD proteins, testing first for a VirC1–VirC2 partner interaction by co-immunoprecipitation. Due to background crossreactivity of the VirC2 antibodies, we tagged VirC2 with a FLAG-epitope and confirmed that the VirC2FL protein was fully functional in virulence assays (data not shown). The anti-VirC1 antibodies co-precipitated native VirC1 and VirC2FL, as did the FLAG-epitope antibodies in reciprocal experiments (Figure 2A). Similarly, both antibodies co-precipitated a presumptive complex of VirC1K15Q and VirC2FL, although reproducibly less efficiently than native VirC1 and VirC2FL (Figure 2A). This result cannot be attributed to the effects of the Walker mutation on VirC1 stability, as VirC1K15Q accumulated at WT levels (Supplementary Figure S1), suggesting instead that the K15Q mutation partially disrupts VirC1–VirC2 multimer formation. Complexes containing VirC1 and VirC2FL also were recovered from various vir mutant strains as well as a Ti-plasmidless strain producing both proteins from an IncP replicon, establishing that Vir proteins and other factors encoded by the Ti plasmid do not contribute to VirC1–VirC2 complex formation (Supplementary Figure S3 and data not shown).
Figure 2.
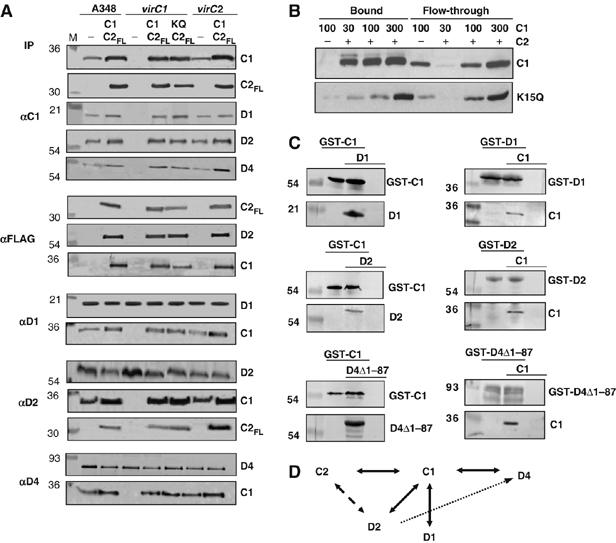
Protein–protein interactions among the VirC and VirD subunits. (A) Co-immunoprecipitation of VirC and VirD proteins. Strains: A348 WT strain; virC1, virC1 mutant Mx365; virC2, virC2 mutant Mx364. Strains were assayed for formation of immunoprecipitable complexes without (−) or with VirC proteins synthesized from an IncP replicon: A348(C1,C2FL), A348(pKAB192); virC1(C1,C2FL), Mx365(pKAB192); virC1(C1KQ,C2FL), Mx365(pKAB193); virC2(C1,C2FL), Mx364(pKAB192). Antibodies used for immunoprecipitation (IP) are listed at left and proteins in the immunoprecipitates detected by immunoblotting at the right. M, Molecular mass markers with sizes (kDa) are listed at left. (B) Co-retention of VirC1 and VirC2-T7 by affinity chromatography. Purified VirC1 or VirC1K15Q in the amounts indicated at top (in ng) were mixed with a T7-tag antibody agarose (−) or the affinity resin pre-bound with His-T7-VirC2 (+). VirC1 and VirC1K15Q in the flow-through and bound fractions were detected by western immunoblotting with anti-VirC1 antibodies. (C) GST-pulldown of VirC1 and VirD proteins. Strain: E. coli BL21(DE3). Plasmids (in parantheses) were introduced into this strain for production of the following protein(s): GST-C1 (pOB1); GST-C1+D1 (pOB1,pKA204); D1 (pKA204); GST-C1+D2 (pOB1,pKA205); D2 (pKA205); GST-C1+D4Δ1-87 (pOB1,pKA207); D4Δ1-87 (pKA207); GST-D1, (pKVD16); GST-D1+C1 (pKVD16,pKA208), C1 (pKA208); GST-D2 (pKA29); GST-D2+C1 (pKA29,pKA208); GST-D4Δ1-87 (pKA28); GST-D4Δ1-87+C1 (pKA28,pKA208). Vir proteins detected by GST-pulldown are shown to the right, Molecular mass markers with sizes (kDa) are listed to the left. (D) A proposed interaction network for the VirC and VirD subunits. Reciprocal interactions detected by co-immunoprecipitation are depicted with solid arrows. Interactions between VirD1 and VirD2 and VirD2 (as a component of the VirD2–T-strand intermediate) and the VirD4 receptor were not detected by co-immunoprecipitation (broken arrow), but likely form transiently or via another factor (see text).
VirC1 and VirC2 were purified using glutathione-S-transferase (GST) and T7 epitope tags, respectively (see Materials and methods). The GST tag was cleaved from VirC1, and the purified protein was shown to bind T7-VirC2 (Figure 2B). Similarly, purified VirC1K15Q bound T7-VirC2, although again less efficiently than the native protein. These experiments were carried out in the presence of added ATP, but similar results were obtained in the presence of other NTP's and in the absence of NTP (data not shown). VirC1 and VirC2 thus interact independently of NTP energy, although NTP binding or hydrolysis still might affect the dynamics of multimer formation.
VirC1 and VirC2 form a network of interactions with VirD1, VirD2 relaxase, and the VirD4 substrate receptor
The anti-VirC1 antibodies also precipitated VirC1 and each of the VirD proteins, VirD1, VirD2, and VirD4 from WT extracts (Figure 2A). Reciprocally, the anti-VirD1, -VirD2, and -VirD4 antibodies co-precipitated the cognate protein and VirC1. Further studies showed: (i) the VirC1K15Q mutant also formed complexes with the VirD proteins, (ii) the VirC proteins interacted directly or indirectly with VirD1 independently of VirD2 relaxase, (iii) the VirC proteins interacted with VirD2 independently of the VirB T4S channel subunits, and (iv) VirC1 interacted with VirD4 independently of the VirB subunits or other Ti plasmid-encoded factors (Figure 2A and Supplementary Figure S3A and B).
The above findings prompted a test for pairwise interactions between VirC1 and each of the VirD proteins. To this end, we co-produced the GST–VirC1 fusion protein and each VirD protein in heterologous Escherichia coli cells and assayed for interactions by glutathione affinity chromatography. As shown in Figure 2C, VirD1 and VirD2 were retained on glutathione-coated sepharose beads only when co-produced with GST–VirC1. Similarly, VirD4Δ1-87, a derivative deleted of the N-terminal TM domain that confers instability of native VirD4 in E. coli, was retained on the glutathione beads only when co-produced with GST–VirC1. In the reciprocal experiments, native VirC1 was retained on the affinity matrix only when co-produced with GST-tagged forms of VirD1, VirD2, and VirD4Δ1-87 (Figure 2C).
Taken together, our findings support a VirC/VirD interaction network as depicted in Figure 2D. Most noteworthy, VirC1 uniquely interacts in pairwise fashion with the other processing factors and the VirD4 receptor. Curiously, we were unable to detect a VirD1–VirD2 interaction despite evidence that VirD1 is important for VirD2 nicking at T-DNA borders in vivo (de Vos and Zambryski, 1989), or a VirD2–VirD4 interaction despite evidence that the T-strand component of the T-complex interacts with VirD4 (Cascales and Christie, 2004b). These observations suggest the interaction network is more complex than presented, possibly because some partner contacts form transiently or another unidentified factors(s) mediates certain interactions.
Spatial positioning of the VirC and VirD proteins: evidence for VirC1 recruitment of VirD2 relaxase to the cytoplasmic membrane and cell poles
We explored the possibility that VirC1 couples the relaxase or the processed VirD2–T-strand to the VirD4 receptor. Because the VirB/D4 T4S system localizes at A. tumefaciens cell poles, initial studies focused on defining spatial patterns for the Dtr factors. As shown by immunofluorescence microscopy (IFM), VirC1 and VirC2FL accumulated primarily at one pole of WT cells although some clustering also was evident at the opposite pole (Figure 3A). The VirC proteins localized at the same cell pole and independently of each other (Figure 3A and Supplementary Figure S4). Moreover, both Walker A mutants, VirC1K15Q and VirC1K15E (Figure 3A and Supplementary Figure S4), displayed WT spatial patterns, suggesting that polar positioning does not require NTP binding or hydrolysis. In complementary studies, VirC1 and VirC2 tagged with the green fluorescent protein (GFP) also accumulated primarily at one cell pole, independently of each other and other Ti-encoded proteins (Supplementary Figure S5 and data not shown). In contrast to reports for GFP-tagged ParA, Soj, and MinD proteins (Quisel et al, 1999; Raskin and de Boer, 1999; Ebersbach and Gerdes, 2004; Lim et al, 2005), we were unable to detect mid-cell localization or oscillatory behavior of the VirC proteins bearing GFP tags at either terminal end.
Figure 3.
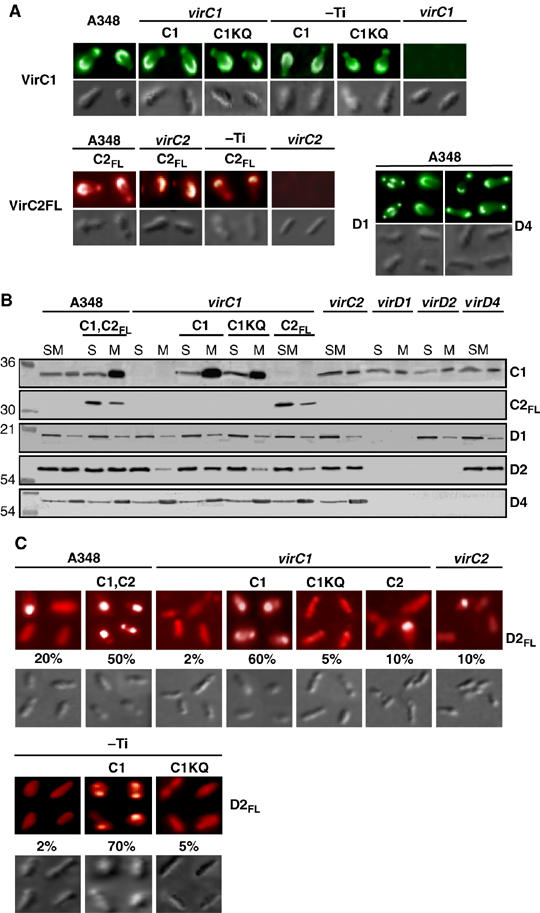
VirC polar localization and VirC1-mediated polar recruitment of VirD2 relaxase. (A) Localization of VirC1 and VirC2FL at A. tumefaciens poles. Cells were induced for 16-18 h with 200 μM AS and assayed for Vir protein localization by IFM. Strains: A348; virC1, Mx365; virC2, Mx364; and pTi-, Ti-plasmidless strain A136 carrying the virA/virG two-component regulatory system in the chromosome. Proteins listed were synthesized from the following plasmids: C1 (pKAB187), C1KQ (pKAB189) C2FL (pKAB194). Upper colored panels: VirC1, VirC1KQ, VirD1, and VirD4 were detected with Alexa fluor® 488 goat-anti-rabbit IgG as the 2o antibody. VirC2FL was detected with Rhodamine Red™-X goat anti-mouse IgG. Lower panels: corresponding DIC images by Nomarski microscopy. (B) Effects of VirC1 on VirD2 membrane binding. Subcellular fractions of AS-induced cells were analyzed for Vir protein content by western immunoblotting. Strains and Vir proteins synthesized from IncP replicons: as described in (A) plus virD1, Mx306; virD2, Mx311; virD4, Mx355; C1, C2FL (pKAB192). Mutations in Mx365, Mx306, Mx311 are polar on downstream genes (Stachel and Nester, 1986). (S) total soluble (cytoplasmic) and (M) membrane fractions. Molecular mass markers at left with sizes listed in kilodaltons. (C) VirC1 polar recruitment of VirD2. Strains: as described in (A, B), but also producing FLAG-tagged VirD2 (D2FL) from plasmid pKA196. Upper: AS-induced cells were examined by IFM for VirD2FL localization with Rhodamine Red™-X goat anti-mouse IgG. Lower: corresponding DIC images by Nomarski microscopy. The percentage of cells (>1000 cells examined) displaying polar fluorescence is listed.
VirD1 also accumulated predominantly at one cell pole in about 50% of the cells examined, but the remaining cells displayed a more complex pattern of bipolar and diffuse mid-cell localization (Figure 3A). The VirD4 receptor accumulated at both poles as shown previously (Kumar and Das, 2002; Atmakuri et al, 2003), although a substantial fraction of cells consistently displayed brighter fluorescence at one pole suggestive of a preference for clustering at one pole (Figure 3A).
The VirC1, VirC2, VirD1, and VirD4 proteins also displayed interesting differences in subcellular fractionation studies (Figure 3B). VirC1 partitioned approximately equally with cytoplasmic and membrane fractions of WT cells, albeit when overproduced the bulk of VirC1 partitioned with the membrane fraction possibly as a result of cooperative binding or aggregation at the membrane. VirC2 and VirD1 were mainly cytoplasmic, although appreciable amounts of both proteins associated with membrane. With the exception of VirC1 (see Discussion), these factors lack obvious hydrophobic domains and thus likely bind the membrane peripherally or indirectly through interaction with a membrane protein. By contrast, the VirD4 substrate receptor possesses an N-terminal transmembrane domain (Kumar and Das, 2002), and as expected, partitioned mainly with the membrane (Figure 3B).
The spatial and subcellular fractionation profiles of the ancillary processing factors were unaffected by production of other Vir proteins (Figure 3A and B, data not shown). In striking contrast, both membrane association and polar positioning of the VirD2 relaxase were significantly affected by VirC1 synthesis (Figure 3B). VirD2 partitioned approximately equally with the membrane and cytoplasmic fractions of VirC1-producing cells, but almost exclusively with the cytoplasmic fraction of virC1 mutants. VirD2 also partitioned with the cytoplasmic fractions of strains producing the VirC1K15Q mutant, further suggesting that VirC1 recruits VirD2 to the membrane by an NTP-dependent mechanism. Finally, VirC1 exerted its effects on VirD2 membrane binding independently of its partner protein VirC2 or the VirD4 substrate receptor.
We constructed a FLAG-tagged VirD2 derivative (VirD2FL) to overcome problems encountered with polyclonal anti-VirD2 antibodies in IFM studies. The FLAG-tagged protein was stable and fully functional in mediating T-DNA transfer to susceptible plants (Supplementary Figure S6 and data not shown). Interestingly, VirD2FL localized predominantly at one pole in approximately 20% of WT cells and at the poles of >50% of cells engineered to overproduce VirC1 (Figure 3C). Conversely, among strains lacking VirC1 or producing the VirC1K15Q mutant, fewer than 5% of cells displayed polar foci. These findings strongly implicate VirC1 in recruitment of the VirD2 relaxase to the polar membrane by an NTP-dependent mechanism.
VirC1 recruits the T-strand to the cell pole
The above findings did not distinguish whether VirC1 recruits VirD2 alone or the T-DNA transfer intermediate (VirD2–T-strand complex) to the cell pole. To assay for the latter activity, we modified the fluorescence in situ hybridization (FISH) assay for detection of single-stranded DNA (ssDNA) in cells (See Materials and methods). As shown in Figure 4A and consistent with previous findings (Kahng and Shapiro, 2003), with the standard FISH assay, the chromosome and the Ti plasmid were found at or near the poles of both WT and virC1 mutant strains. By contrast, with the nondenaturing FISH assay and by use of a T-strand-specific DNA probe, the T-strand accumulated primarily at one pole of WT cells, but not at poles or elsewhere in virC1 mutant cells (Figure 4B and C). With the nondenaturing FISH assay and the complementary ssDNA probe, we were unable to detect the nontransferred strand (T-strand complement) in WT or virC mutant cells. This finding and results of other control experiments confirmed that the T-strand probe hybridized with free T-strand molecules, the T-strand complement did not accumulate to detectable levels in cells, and the ssDNA probes specific for the two strands of T-DNA failed to anneal to the ds T-DNA carried on the Ti plasmid (Figure 4B and C).
Figure 4.
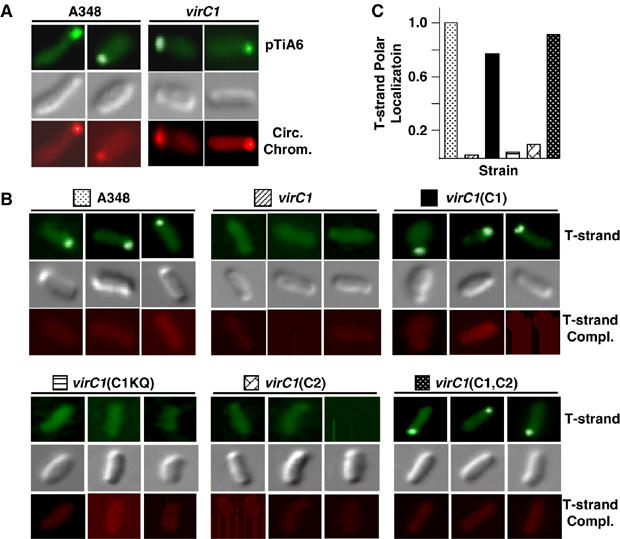
VirC1-mediated polar recruitment of the T-strand intermediate. (A) Polar localization of pTiA6 and the circular chromosome in A348 and the virC1(Mx365) mutant. The Ti plasmid and chromosome were detected by FISH with Alexa Fluor® 488-tagged (green; top) and Alexa Fluor® 555-tagged (red; bottom) DNA probes, respectively. Cells were visualized by Nomarski microscopy (gray; middle). (B) VirC1 polar recruitment of the T-strand. AS-induced cells were examined for localization of the ss T-strand by a modified, nondenaturing FISH assay (Materials and methods). Strains: A348; virC1, Mx365; virC1(C1), Mx365(pKAB187); virC1(C1KQ), Mx365(pKAB189); virC1(C2), Mx365(pKA114); virC1(C1,C2), Mx365(pKAB188). The T-strand was detected with a ssDNA-specific probe fluorescently tagged with Alexa Fluor® 488 (green; top). Non-transferred strand (T-strand complex) was assayed for with the ssDNA-specific complementary probe fluorescently tagged with Alexa Fluor® 555 (red; bottom). Cells were visualized by Nomarski microscopy (gray; middle). (C) Relative numbers of cells displaying T-strand polar positioning. The histogram represents the numbers of cells from strains analyzed in (B) displaying polar-localization of the T-strand. WT cells with polar foci are normalized to 1.0 and the numbers of vir mutant cells with polar foci are presented as a fraction of the WT profile. Values were obtained by examination of least 500 cells for each strain.
Further studies showed that VirC1 mediates polar accumulation of the T-strand independently of VirC2 synthesis, and that the K15Q mutation abolished substrate recruitment to cell poles (Figure 4B). In principle, our inability to detect polar positioning of the T-strand in virC mutants could be due to the low amounts of T-strand generated by these mutants (see Figure 1B). However, this was not the case because mutants producing VirC1, but not VirC2, produced low levels of T-strand (Figure 1B), yet clearly displayed polar localization of T-strands (Figure 4B). We conclude that VirC1 alone mediates accumulation of the T-DNA transfer intermediate at cell poles and that recruitment is energized by NTP binding or hydrolysis.
DivIVA–VirC1 recruits the T-strand intermediate to both poles and the mid-cell
To confirm that VirC1 functions as a spatial determinant for the T-strand, we adapted a cytology-based two-hybrid assay (Ding et al, 2002). VirC1 was fused to the cell division protein, DivIVA, and as expected, the fusion protein displayed a spatial pattern distinct from that of native VirC1 by accumulating at both poles and the mid-cell (compare Figures 3A and 5A). With nondenaturing FISH and the T-strand-specific probe, we next assayed for repositioning of the T-strand in the DivIVA–VirC1-producing cells. Remarkably, we detected the T-strand at both poles or the poles and the mid-cell in >80% of cells examined (Figure 5B). The remaining cells displayed unipolar localization, the pattern observed in cells producing native VirC1 (Figure 5B).
Figure 5.
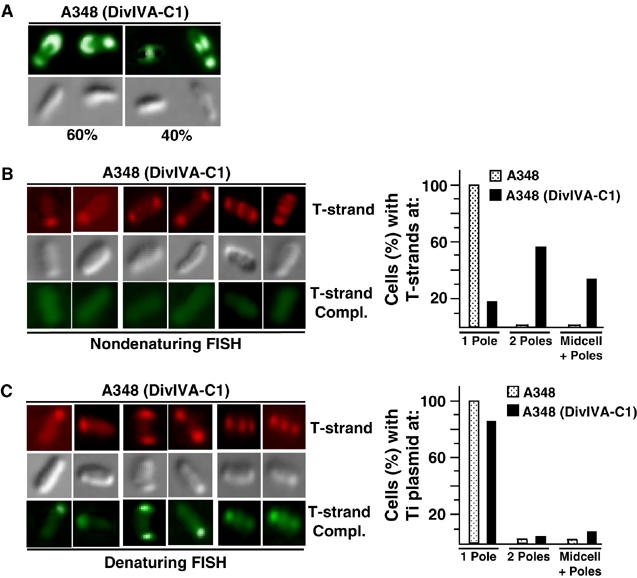
DivIVA–VirC1 recruits the VirD2–T-strand complex to mid-cell from cell poles. (A) DivIVA–VirC1 localizes to mid-cell. A348(pKAB202) cells producing DivIVA–VirC1 were induced for 16–18 h in ABIM pH 5.5 with 200 μM AS and subjected to IFM. VirC1 was detected using secondary antibodies tagged with Alexa fluor® 488 goat-anti-rabbit IgG (green; top panels). Corresponding Nomarski microscopy images (lower panels). Numbers below bottom panels represent the percentage of cells (>1000 examined) with polar or mid-cell fluorescence. (B, C) DivIVA–VirC1 relocalizes the VirD2–T-strand to mid-cell. Cells from A348 expressing DivIVA–VirC1 were induced 24 h in ABIM pH 5.5, treated with 1.5% formaldehyde, fixed on 0.1% poly-L-lysine-coated cover glass and subjected to FISH analyses. Under nondenaturing conditions (B), only the T-strand was detected by hybridizing with Alexa Fluor® 555-tagged (red; top), whereas under denaturing conditions (C) T-strand and Ti plasmid (T-strand complement) were detected with Alexa Fluor® 555-tagged (red; top) and Alexa Fluor® 488-tagged (green; bottom) ssDNA-specific probes, respectively (see Materials and methods). Corresponding cells were visualized by Nomarski microscopy (gray; middle). The histograms represents the percentages of cells (>500 examined) of A348(DivIVA–VirC1) exhibiting unipolar, bipolar, or mid-cell+polar positioning of the VirD2–T-strand complex (T-strand-specific probe, nondenaturing FISH; (B)) or Ti plasmid (T-strand complement probe, denaturing FISH; (C)).
In view of our finding that VirC1 binds the Ti plasmid (Figure 1D), we tested whether DivIVA–VirC1 also relocalized the Ti plasmid. With the standard FISH assay, we detected the Ti plasmid primarily at or near the poles of DivIVA–VirC1-producing cells, although a small fraction (5–10%) reproducibly displayed a mid-cell localization (Figure 5C and data not shown). The DivIVA–VirC1 fusion protein thus appears to spatially reposition the Ti plasmid at a low but detectable level.
Discussion
Spatial positioning of the VirB/D4 T4S system
Presently, there are many examples of protein complexes or machines that localize at discrete sites (cell poles, division sites, distributed foci) or display dynamic oscillatory behavior in bacteria (Hahn et al, 2005; Kidane and Graumann, 2005; Shih and Rothfield, 2006). Included among the spatially organized surface organelles are several DNA-translocation machines—the A. tumefaciens VirB/D4 (Lai et al, 2000; Atmakuri et al, 2003; Judd et al, 2005) and plasmid R27 (Gunton et al, 2005) conjugation systems, the conjugation-like Legionella pneumophila Dot/Icm T4S system (J Vogel, personal communication), and a Bacillus subtilis competence system (Hahn et al, 2005). Here, we showed that VirC1, a member of the ParA/MinD ATPase superfamily, stimulates T-DNA transfer through the A. tumefaciens VirB/D4 T4S system by functioning as a central organizer for relaxosome assembly and as a spatial determinant for the processed T-complex. These functions, mediated through contacts with the Dtr processing factors (VirC2, VirD1, VirD2) and the substrate receptor (VirD4), respectively, and driven by NTP energy, generate multiple copies of the T-complex per cell and promote substrate docking with the VirB/D4 translocation channel. A contribution of a ParA/MinD-like protein to the DNA substrate—T4S receptor docking reaction has not been described previously, but might be widespread given that other mobile elements, most notably some integrated conjugative elements and bacteriophages, encode homologs of this ATPase superfamily (see below).
The A. tumefaciens T-DNA transfer system provides an interesting example of a surface organelle whose polar assembly and function represent the outcome of several independent positioning events. Adding to previous findings that the VirB proteins, the T-pilus, and the VirD4 receptor accumulate and function at cell poles (Lai et al, 2000; Kumar and Das, 2002; Atmakuri et al, 2003; Judd et al, 2005), we showed here that the T-DNA processing factors VirC1, VirC2, and VirD1, also position at the A. tumefaciens poles independently of each other and the VirB and VirD4 components, and that VirC1 actively recruits the VirD2 relaxase to cell poles. This is the first evidence for spatial positioning of conjugative Dtr factors and, by extrapolation, the relaxosomal complex at specific sites at the cell envelope. The underlying mechanism(s) responsible for spatial positioning of the various VirB/D4 T4S system components is not known. An active recruitment mechanism(s) might direct polar localization, although the finding that the Ti plasmid itself is polarly localized (Figure 4A; Kahng and Shapiro, 2003) raises the possibility that localized synthesis of the Vir proteins might simply achieve a concentration threshold necessary for VirB/D4 machine biogenesis at these sites.
Mechanistic similarities between VirC1 and other deviant Walker-type ATPases
VirC1 contributes in novel ways to conjugative DNA transfer, yet shares features of the ParA and MinD ATPases. The broad biological function of these homologs is to spatially organize cognate DNA elements or the division machinery within the cell by mechanisms energized by NTP binding and hydrolysis. VirC1 resembles ParA proteins in forming a partner interaction with the product of a co-transcribed gene and binding as a complex to a cis-acting DNA target sequence (Toro et al, 1988; Bignell and Thomas, 2001). VirC1 binds directly to overdrive in vitro, yet VirC2 apparently stimulates VirC1 DNA binding to this or another Ti plasmid sequence(s) in vivo (Figure 1B). The ParA proteins also are targeted to cognate parS sequences through interactions with ParB DNA-binding proteins (Ebersbach and Gerdes, 2005). The VirC1/VirC2/overdrive nucleoprotein complex thus might correspond to a ParA/ParB/par partitioning complex adapted for conjugative DNA processing. VirC1 is unusual among the ParA homologs in its capacity to bind DNA directly (Figure 1D), although it is interesting that the Thermus thermophilus Soj ATPase binds both ds and ssDNA, the latter occurring at open transcription complexes to repress transcription (Leonard et al, 2005). ParB proteins also have been shown to silence gene expression by polymerizing along DNA away from centromeric sequences (Shih and Rothfield, 2006). Binding and possible spreading of one or both VirC proteins at the T-DNA borders or along the T-strand might be physiologically relevant to the VirC-mediated T-DNA processing or substrate recruitment reactions.
Although ATP binding or hydrolysis modulate the activities of VirC1 (this study) and the ParA, MinD, and Soj proteins in different ways (see Shih and Rothfield, 2006), all of these homologs require NTP energy to function as spatial determinants. Nucleotide-dependent, pole-to-pole oscillation is a remarkable feature common among the ParA, Soj, and MinD proteins (Ebersbach and Gerdes, 2005), but could not be detected for VirC1 under our experimental conditions. Even so, a dynamic mode of action or the formation of filaments extending through the cytoplasm remain appealing mechanisms for how VirC1 might recruit T-complexes from a cytosolic pool to the polar membrane.
One noteworthy sequence motif missing in the ParA and Soj proteins that is present in the related MinD proteins is a C-terminal amphipathic helix shown to be important for MinD binding to the polar membrane (Supplementary Figure S7; Hu and Lutkenhaus, 2003). Intriguingly, VirC1 also binds the polar membrane (Figure 3) and our recent in silico analysis identified a potential amphipathic helix at the extreme C terminus of VirC1 (Supplementary Figure S7). Like MinD—and in contrast to the ParA proteins, which do not bind membranes—VirC1 might bind the polar membrane through its C-terminal amphipathic helix, and we are currently investigating this possibility.
VirC1 and VirC2 stimulate T-strand production
Conjugative DNA processing reactions have been noted to mechanistically resemble rolling circle (RC) replication systems of bacterial plasmids and bacteriophages (Waters and Guiney, 1993; Pansegrau and Lanka, 1996; Llosa et al, 2002). Typically, the plasmid systems, identified in both Gram-positive and -negative bacteria, strictly regulate RC initiation events to maintain copy number control (Solar et al, 1998; Khan, 2005). By contrast, certain bacteriophages, for example, phiX174, have adapted RC replication to maximize phage burst size for propagation to new bacterial host cells (Novick, 1998). A striking finding of the present study is that A. tumefaciens cells induced with a plant signal for expression of the vir genes accumulate as many as 50 copies of the T-DNA transfer intermediate in the cell cytosol (Figure 1B). A. tumefaciens thus appears to have evolved a phage-like infection strategy by initiating multiple rounds of RC replication at the T-DNA border sequences to generate a pool of available substrate for plant cell transformation. Substrate amplification is strictly dependent on synthesis of the VirC proteins (Figure 1), and likely evolved to augment the kinetics and magnitude of tumor formation, and also enhance the A. tumefaciens host range (Close et al, 1987). In support of this proposal, it was recently shown that VirC-producing strains transform fungal cells more efficiently than virC mutants (Michielse et al, 2004). We note, however, that even though correlations exist between VirC-dependent T-DNA substrate amplification, virulence, and host range, our quantitative analyses were with A. tumefaciens cells induced in the absence of plant cells. Bacterial contact with the plant host might limit T-complex production through a feedback mechanism, and we are currently exploring this possibility.
VirC2 was not the focus of our present study, yet our findings support an early prediction that both VirC proteins participate as a complex in the T-DNA processing reaction (Toro et al, 1988). We supplied evidence that VirC1 and VirC2 function together to stimulate T-DNA processing and form multimers in vitro and in vivo independently of other transfer components (Figure 2). Elsewhere, it was reported that virC2 mutants transfer T-DNA molecules with aberrant border junctions to the fungus Aspergillus awamor (Michielse et al, 2004). The virC2 mutants tended to translocate T-DNA molecules with intact T-DNA right border sequences, but imprecisely cleaved T-DNA left border sequences. VirC1 and VirC2 were postulated to stimulate relaxosome assembly at both T-DNA borders, which is of interest because overdrive sequences were found adjacent only to T-DNA right borders (Toro et al, 1989). It is thus likely that VirC1–VirC2 multimers binds DNA target sequences other than or in addition to overdrive.
VirC1 as a spatial determinant for the VirD2–T-strand complex
The accumulation of a cytosolic pool of the free T-complex and VirC1-mediated substrate recruitment to the polar membrane likely are physiologically relevant events in the context of A. tumefaciens plant transformation, in view of striking correlations between cytosolic levels of free T-complexes and plant tumor formation (Figure 1) (Close et al, 1987; Toro et al, 1988; Veluthambi et al, 1988). Also, we recently discovered that WT cells engineered to overproduce VirC1 accumulate WT levels of cytosolic T-complexes, but display a supervirulent phenotype in plant infection assays (K Atmakuri, data not shown), which is consistent with the notion that VirC1 functions stoichiometrically to recruit available free T-complexes to the T4S channel. Finally, our finding that DivIVA–VirC1 repositions free T-complexes from a cytosolic pool to division sites (Figure 5) strongly implies that the reactions mediating T-complex generation and recruitment to the membrane can be spatially uncoupled.
This last finding also is of particular interest in view of current models depicting transfer of conjugative plasmids. The relaxosome typically is presented as being physically coupled to the VirD4-like receptor through relaxase- or other Dtr factor-mediated interactions. For example, F plasmid TraM, which partially associates with the membrane, is important for F plasmid processing and interacts with the TraD receptor (Di Laurenzio et al, 1992; Lu and Frost, 2005). Coupling might permit simultaneous substrate processing with translocation through the T4S channel. However, although this coupling activity might be important for conjugative plasmids carrying a single oriT sequence, the T-DNA is in fact an integrated mobile element flanked by two oriT-like sequences. Other integrated mobile elements, including conjugative transposons and bacteriophages, also encode ParA-like proteins whose functions are not known (Bignell and Thomas, 2001). Except for those bacteriophages whose lifestyles include excision and stable maintenance of a plasmid prophage, a Par system is unlikely to function in partitioning or stabilization of integrated mobile elements. It is enticing to suggest that, reminiscent of VirC1 action, at least some of these integrated elements appropriated an ancestral Par/Min ATPase to spatially couple the excised transfer intermediate to its cognate translocation apparatus. Indeed, this idea gains support from recent investigations of an F-like T4S system encoded on a genetic island (GGI) in the Neisseria gonorrhoeae genome that is responsible for secreting chromosomal DNA to the extracellular milieu. The GGI also encodes a parA-like gene which, when mutated, abolishes T4S-mediated chromosome secretion (Hamilton et al, 2005).
Summary
In summary, we propose a working model in which polar localization of the VirC proteins is important for assembly and activity of the relaxosome and also for T-complex docking with the VirD4 receptor (Supplementary Figure S8). Upon synthesis, VirC1, VirC2, and VirD1 localize and form a multimeric complex at the cell pole, perhaps simply due to a concentration effect resulting from cellular colocalization. This Dtr complex binds the T-DNA border repeats and other cis-acting sequences, including but not limited to overdrive, by a mechanism that might be regulated by VirC1 NTP binding or hydrolysis. VirC1 recruits the VirD2 relaxase to complete relaxosome assembly and activated VirD2 cleaves the T-DNA borders. A T-strand displacement reaction releases the VirD2–T-strand particle from the Ti plasmid to the cytosol or directly to the T4S channel via a VirC1–VirD4 receptor interaction. In our favored model, the ancillary proteins remain stably associated at the T-DNA borders to initiate successive rounds of VirD2 recruitment and T-complex generation. VirC1, which is not associated with the relaxosome, interacts with the free T-complex by an NTP-dependent mechanism and delivers the T-complex to the VirD4 receptor, possibly through a dynamic oscillatory behavior or formation of connecting filaments.
Materials and methods
Bacterial strains, induction conditions, and plasmids
E. coli strain DH5α served as the host for all plasmid constructions. Plasmids are listed in Table I and Supplementary Table SI (see Supplementary data for construction details). E. coli strain BL21(DE3) (Novagen) was used for protein expression. A. tumefaciens strains were derived from WT strain C58. A136 is strain C58 cured of its pTiC58 plasmid, and A348 is strain A136 harboring pTiA6 plasmid, and A348 derivatives include strains with Tn3HoHo1 insertion mutations exerting polar effects on downstream genes: Mx306 (virD1), Mx311 (virD2), Mx355 (virD4), Mx364 (virC2), Mx365 (virC1) (polar on virC2; Stachel and Nester, 1986). Strain PC1000 (ΔvirB) is deleted of the virB operon (see Atmakuri et al, 2004). Strain KA2002 is A136 harboring virA of pTiA6 and virG of pTiBo542 on its circular chromosome.
Table 1.
Plasmids used in this studya
| Plasmidb | Relevant characteristics |
|---|---|
| virC1 and derivatives | |
| pKAB57 | pBSIIKS+ with PvirB-virC1 (XbaI site before stop codon) |
| pKAB58 | pBSIIKS+ with PvirB-virC1-gfp |
| pKAB110 | pBSIIKS+ with PvirB-virC1K15Q-gfp |
| pKAB187 | pBSIIKS+ with PvirB-virC1 |
| pKAB189 | pBSIIKS+ with PvirB-virC1K15Q |
| pKAB202 | pBSIIKS+ with PvirB-divIVA-virC1 |
| pKAB220 | pBSIIKS+ with PvirB-virC1K15E |
| pKA208 | pACYCDuet-1 with PT7-virC1 |
| pOB1 | pGEX-6P-1 with Ptac-GST-virC1 |
| pOB2 | pGEX-6P-1 with Ptac-GST-virC1K15Q |
| virC2 and derivatives | |
| pKA106 | pBSIIKS+ with PvirB-virC2 |
| pKAB113 | pBSIIKS+ with PvirB-virC2-gfp |
| pKA114 | pBBR1MCS2 with PvirB-virC2 |
| pKA115 | pBBR1MCS2 with PvirB-virC2-gfp |
| pKAB194 | pBSIIKS+ with PvirB-FLAG-virC2 |
| pSC1 | pET28b(+) with PT7-His,T7-virC2 |
| virC1,C2 and derivatives | |
| pKAB188 | pBSIIKS+ with PvirB-virC1,virC2 |
| pKAB190 | pBSIIKS+ with PvirB-virC1K15Q,virC2 |
| pKAB192 | pBSIIKS+ with PvirB-virC1,FLAG-virC2 |
| pKAB193 | pBSIIKS+ with PvirB-virC1K15Q,FLAG-virC2 |
| virD1 and derivatives | |
| pKA204 | pACYCDuet-1 with PT7-virD1 |
| pKVD16 | pKVD10 with Ptac-GST-virD1 |
| virD2 and derivatives | |
| pKA29 | pKVD10 with Ptac-GST-virD2 |
| pKAB195 | pBSIIKS+ with PvirB-FLAG-virD2 |
| pKA196 | pBBR1MCS2 with PvirB-FLAG-virD2 |
| pKA205 | pACYCDuet-1 with PT7-His-virD2 |
| virD4 derivatives | |
| pKA28 | pKVD10 with Ptac-GST-virD4Δ1-87 |
| pKA207 | pACYCDuet-1 with PT7-virD4Δ1-87 |
| see Supplementary Data and Supplementary Table S1 for construction details. | |
| Plasmids with ‘KAB' nomenclature were ligated to pBBR1MCS2 or pXZ153 BHR plasmids for introduction into A. tumefaciens. | |
Quantitation of Ti plasmid copy number and T-strands
A. tumefaciens cells (100 ml) were induced by shaking at 22°C in induction medium (ABIM) with 200 μM AS. Total DNA was extracted from equal numbers of cells from each strain at times indicated as described previously (Cascales and Christie, 2004b). Ti plasmid copy numbers and T-strand levels were determined by subjecting equal amounts of total DNA to 20 cycles of PCR amplification using primers specific for T-DNA (gene 7 of TL-DNA), Ti plasmid (ophDC locus) (Cascales and Christie, 2004b) and circular chromosome (chvE gene, this study). On the 21st cycle, 1.0 μCi of [α-32P]dGTP (Amersham Biosciences) was added for a single round of PCR amplification, as described previously (Cascales and Christie, 2004b). PCR products were column-purified with Qiaquick PCR purification kit (Qiagen) to remove unincorporated nucleotides and incorporated radioactivity was measured with a Beckman coulter counter. The values of cells obtained were normalized to OD 1.0 (A600). Ti plasmid copy numbers were estimated as the ratio of the amount (counts per min (c.p.m.)) of radioactivity incorporated into the Ti-plasmid-specific amplicon (ophDC locus) to that obtained for the chromosomal specific amplicon (chvE), multiplied by the factor 0.53 (513/973; obtained as a ratio of number of G+C bases in PCR products of chvE (513) to ophDC locus (973)). Similarly, the number of T-strands generated per molecule of Ti-plasmid was obtained as a ratio of cpm of radioactivity incorporated into T-DNA-specific amplicon (gene 7) to that obtained for the Ti-plasmid-specific amplicon (ophDC locus), multiplied by the factor 3.76 (973/259; obtained as a ratio of number of G+C bases in PCR products of ophDC locus (973) to gene 7 (259)).
Transfer DNA immunoprecipitation assay
The TrIP assay and the quantification of the T-strand immunoprecipitated (QTrIP) as a protein–DNA complex were performed as described previously (Cascales and Christie, 2004b).
Protein crosslinking, immunological methods, and cell fractionation
A. tumefaciens cultures (10 ml) were induced at 22°C for 16 h in ABIM with 200 μM AS. Equal numbers of cells (approx. 8.0 OD600) were pelleted by centrifugation at 8000 r.p.m. for 5 min at room temperature and washed once in cold 50 mM Hepes (pH 8.0). Cells resuspended in 500 μl of cold 50 mM Hepes (pH 8.0) were crosslinked with dithiobis(succinimidyl propionate) (DSP) (20 mg ml−1 stock, final concentration 0.5 μg μl−1) for 1 h on ice. Reactions were quenched with 0.5 M L-lysine (pH 8.5) (62.5 mM final concentration) for 15 min on ice, and cells were washed and processed for membrane solubilization and co-immunoprecipitation as described (Cascales and Christie, 2004a) with minor modifications (see Supplementary data). Vir proteins were visualized by SDS–PAGE, transfer to nitrocellulose membranes, and immunostaining with goat anti-rabbit (for anti-VirC1, -VirD1, -VirD2, -VirD4, -VirB9, -ChvE antibodies) or goat anti-mouse (for anti-FLAG antibody) secondary antibodies conjugated to alkaline phosphatase or horseradish peroxidase (Bio-Rad). Cells were lysed with French Pressure cell and fractionated into soluble (cytoplasmic) and total membrane material as described previously (Atmakuri et al, 2004).
GST-pulldown assay
E. coli BL21(DE3) strains harboring appropriate plasmids were grown overnight (O/N) at 37°C. Cells from 1 ml of culture were resuspended in 200 ml LB and incubated with shaking at room temperature to an OD600=0.4 in the presence of 0.5 mM isopropyl-β-D-thiogalactoside. Cells were washed, resuspended in 3 ml of 1 × PBS, and exposed to the crosslinker dithiobis(succinimidyl propionate) (DSP) (20 mg ml−1 stock, final concentration 0.5 μg μl−1) for 1 h on ice. Reactions were quenched with 0.5 M L-lysine (pH 8.5) (62.5 mM final concentration) for 15 min on ice, washed with STE buffer and cells were lysed by French-press treatment. The total cellular extract was solubilized by treatment with 3% lauryldimethylamine oxide O/N at 4°C, and unsolubilized material was removed by centrifugation. GST-Sepharose beads were added to the clarified extract and the resulting mixture was incubated with shaking O/N at 4°C. The beads were washed three times with 1 ml of cold 1 × PBS, resuspended in 80 μl of 25 mM Tris–HCl (pH 7.4) and 20 μl of 5 × Laemmli's sample buffer, and samples were analyzed by western blotting and immunostaining.
Purification of recombinant VirC1 and VirC2 proteins
GST–VirC1 and GST–VirC1K15Q were purified from BL21(DE3) harboring pOB1 or pOB2 plasmids, respectively, using a Glutathione Affinity Purification Kit (Amersham Biosciences). His-T7-VirC2 was purified from E. coli strain BL21(DE3) carrying pSC1 using a T7-Tag™ Affinity Purification Kit (Novagen) (Supplementary data).
In vitro interaction studies
Purified T7-His-VirC2 was bound to T7-Tag™ antibody agarose by incubating for 30 min at 21°C on a Nutator. The agarose beads were washed three times (500 g, 5 min, 4°C) with T7 wash buffer and blocked by incubating (30 min, 21°C, rocking) with 5% nonfat dry milk (w/v) in T7 wash buffer. The affinity matrix was washed once and then incubated (rocking, 30 min, 21°C) with purified VirC1 or VirC1K15Q in 1 × PBS. The matrix was again washed extensively and bound VirC1 or VirC1K15Q was eluted by heating to 95°C for 5 min in SDS–PAGE sample buffer.
Microscopy and image analyses
Freshly grown A. tumefaciens cells were used for microscopy analyses. Cells were induced in ABIM plus AS at 22°C for 3 h for detection of GFP fusion proteins by fluorescence microscopy or 12–16 h for detection of native proteins by IFM. For GFP analyses, 1.5 μl of cells were mixed with 1 μl of 0.1% poly-L-lysine on a microscopic slide and observed with an Olympus BX60 microscope equipped with a × 100 oil immersion phase-contrast objective (Ding et al, 2002). For IFM analyses, cells were fixed and analyzed with Alexa fluor® 488 goat-anti-rabbit IgG or Rhodamine Red™-X goat anti-mouse IgG (Molecular Probes) essentially as described previously (Supplementary data). Each experiment was replicated at least three times.
Fluorescence in situ hybridization
FISH was performed essentially as described (Kahng and Shapiro, 2003) with the FISH Tag™ DNA multicolor kit from Molecular Probes, Invitrogen. ssDNA probes specific for circular chromosome, Ti-plasmid, and TL-DNA (transferred strand) were prepared (Supplementary data). All probes were ∼500 bp in length. DpnI-digested products were column-purified, ethanol-precipitated and labeled with either of the multicolor fluorescent tags (Alexa fluor® 488, Alexa fluor® 555) according to the manufacturer's instructions. Routinely, ∼3–4 μg ml−1 of probe DNA was used for hybridization. For detection of circular chromosomal and Ti-plasmid DNA, cells were exposed to 50% formamide and heat-treated at 95°C for 5 min to denature total cellular DNA before fixation. For detection of the single-stranded T-DNA transfer intermediate, these denaturation steps were omitted during processing (Supplementary data). After hybridization and stringency washes, cells were mounted on microscopic slide with a drop of SlowFade® Gold antifade reagent, sealed with base coat (Revlon) and visualized using an Olympus BX60 microscope equipped with a 100 × oil immersion phase-contrast objective.
Supplementary Material
Supplementary Figures
Supplementary Table
Supplementary data
Acknowledgments
We thank members of our laboratories for helpful comments and critiques of this manuscript. We thank Dr W Margolin for use of his fluorescence microscope facility and Dr Margolin and members of his laboratory for helpful comments. The contributions of Gape Machao, Molly Sharlach, Virginia Newman, Shannon Chiu, and M Esa Seegulam are gratefully acknowledged. This work was supported by NIH grant GM48746 (PJC) and NSF grant MCB-0416471 (LB).
References
- Atmakuri K, Cascales E, Christie PJ (2004) Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol 54: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Ding Z, Christie PJ (2003) VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol Microbiol 49: 1699–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell C, Thomas CM (2001) The bacterial ParA-ParB partitioning proteins. J Biotechnol 91: 1–34 [DOI] [PubMed] [Google Scholar]
- Cascales E, Christie PJ (2004a) Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci USA 101: 17228–17233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ (2004b) Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304: 1170–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Winans SC (1991) Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J Bacteriol 173: 1139–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Winans SC (2005) VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound-released chemical signals. Proc Natl Acad Sci USA 102: 14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E (2005) Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59: 451–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ, Tait RC, Rempel HC, Hirooka T, Kim L, Kado CI (1987) Molecular characterization of the virC genes of the Ti plasmid. J Bacteriol 169: 2336–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos G, Zambryski P (1989) Expression of Agrobacterium nopaline-specific VirD1, VirD2, and VirC1 proteins and their requirement for T-strand production in E. coli. Mol Plant Microbe Interactions 2: 43–52 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Frost LS, Paranchych W (1992) The TraM protein of the conjugative plasmid F binds to the origin of transfer of the F and ColE1 plasmids. Mol Microbiol 6: 2951–2959 [DOI] [PubMed] [Google Scholar]
- Ding Z, Zhao Z, Jakubowski SJ, Krishnamohan A, Margolin W, Christie PJ (2002) A novel cytology-based, two-hybrid screen for bacteria applied to protein–protein interaction studies of a type IV secretion system. J Bacteriol 184: 5572–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Gerdes K (2004) Bacterial mitosis: partitioning protein ParA oscillates in spiral-shaped structures and positions plasmids at mid-cell. Mol Microbiol 52: 385–398 [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Gerdes K (2005) Plasmid segregation mechanisms. Annu Rev Genet 39: 453–479 [DOI] [PubMed] [Google Scholar]
- Gunton JE, Gilmour MW, Alonso G, Taylor DE (2005) Subcellular localization and functional domains of the coupling protein, TraG, from IncHI1 plasmid R27. Microbiology 151: 3549–3561 [DOI] [PubMed] [Google Scholar]
- Hahn J, Maier B, Haijema BJ, Sheetz M, Dubnau D (2005) Transformation proteins and DNA uptake localize to the cell poles of Bacillus subtilis. Cell 122: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP (2005) Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol 55: 1704–1721 [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J (2003) A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol Microbiol 47: 345–355 [DOI] [PubMed] [Google Scholar]
- Judd PK, Kumar RB, Das A (2005) Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc Natl Acad Sci USA 102: 11498–11503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahng LS, Shapiro L (2003) Polar localization of replicon origins in the multipartite genomes of Agrobacterium tumefaciens and Sinorhizobium meliloti. J Bacteriol 185: 3384–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SA (2005) Plasmid rolling-circle replication: highlights of two decades of research. Plasmid 53: 126–136 [DOI] [PubMed] [Google Scholar]
- Kidane D, Graumann PL (2005) Intracellular protein and DNA dynamics in competent Bacillus subtilis cells. Cell 122: 73–84 [DOI] [PubMed] [Google Scholar]
- Koonin EV (1993) A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol 229: 1165–1174 [DOI] [PubMed] [Google Scholar]
- Kumar RB, Das A (2002) Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol Microbiol 43: 1523–1532 [DOI] [PubMed] [Google Scholar]
- Lai EM, Chesnokova O, Banta LM, Kado CI (2000) Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J Bacteriol 182: 3705–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Grossman AD (2006) The chromosome partitioning proteins Soj (ParA) and SpoOJ (ParB) contribute to accurate chromosome partitioning, separation of sister origins, and regulation of replication initiation in Bacillus subtilis. Mol Microbiol 60: 853–869 [DOI] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Lowe J (2005) Bacterial chromosome segregation: structure and DNA binding of the Soj dimer—a conserved biological switch. EMBO J 24: 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GE, Derman AI, Pogliano J (2005) Bacterial DNA segregation by dynamic SopA polymers. Proc Natl Acad Sci USA 102: 17658–17663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M, Gomis-Ruth FX, Coll M, de la Cruz F (2002) Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 45: 1–8 [DOI] [PubMed] [Google Scholar]
- Lu J, Frost LS (2005) Mutations in the C-terminal region of TraM provide evidence for in vivo TraM-TraD interactions during F-plasmid conjugation. J Bacteriol 187: 4767–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullen CA, Binns AN (2006) Agrobacterium tumefaciens plant cell interactions and activities required for interkingdom macromolecular transfer. Annu Rev Cell Dev Biol 22: 101–127 [DOI] [PubMed] [Google Scholar]
- Michielse CB, Ram AF, Hooykaas PJ, van den Hondel CA (2004) Agrobacterium-mediated transformation of Aspergillus awamori in the absence of full-length VirD2, VirC2, or VirE2 leads to insertion of aberrant T-DNA structures. J Bacteriol 186: 2038–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP (1998) Contrasting lifestyles of rolling-circle phages and plasmids. Trends Biochem Sci 23: 434–438 [DOI] [PubMed] [Google Scholar]
- Pansegrau W, Lanka E (1996) Enzymology of DNA transfer by conjugative mechanisms. Prog Nucleic Acid Res Mol Biol 54: 197–251 [DOI] [PubMed] [Google Scholar]
- Peralta EG, Hellmiss R, Ream W (1986) Overdrive, a T-DNA transmission enhancer on the A. tumefaciens tumour-inducing plasmid. EMBO J 5: 1137–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quisel JD, Lin DC, Grossman AD (1999) Control of development by altered localization of a transcription factor in B. subtilis. Mol Cell 4: 665–672 [DOI] [PubMed] [Google Scholar]
- Raskin EM, de Boer PA (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA 96: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Pansegrau W, Lanka E (1995) Initiation of Agrobacterium tumefaciens T-DNA processing. Purified proteins VirD1 and VirD2 catalyze site- and strand-specific cleavage of superhelical T-border DNA in vitro. J Biol Chem 270: 1269–1276 [DOI] [PubMed] [Google Scholar]
- Schroder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E (2002) TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol 184: 2767–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih YL, Rothfield L (2006) The bacterial cytoskeleton. Microbiol Mol Biol Rev 70: 729–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solar GD, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R (1998) Replication and control of bacterial plasmids. Microbiol Mol Biol Rev 62: 434–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel SE, Nester EW (1986) The genetic and transcriptional organization of the vir region of the A6 Ti. EMBO J 5: 1445–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel SE, Timmerman B, Zambryski P (1987) Activation of Agrobacterium tumefaciens vir gene expression generates multiple single-stranded T-strand molecules from the pTiA6 T-region: requirement for 5′ virD gene products. EMBO J 6: 857–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N, Datta A, Carmi OA, Young C, Prusti RK, Nester EW (1989) The Agrobacterium tumefaciens virC1 gene product binds to overdrive, a T-DNA transfer enhancer. J Bacteriol 171: 6845–6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N, Datta A, Yanofsky M, Nester EW (1988) Role of the overdrive sequence in T-DNA border cleavage in Agrobacterium. Proc Natl Acad Sci USA 85: 8558–8562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluthambi K, Ream W, Gelvin SB (1988) Virulence genes, borders, and overdrive generate single-stranded T-DNA molecules from the A6 Ti plasmid of Agrobacterium tumefaciens. J Bacteriol 170: 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters VL, Guiney DG (1993) Processes at the nick region link conjugation, T-DNA transfer and rolling circle replication. Mol Microbiol 9: 1123–1130 [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Porter SG, Young C, Albright LM, Gordon MP, Nester EW (1986) The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell 47: 471–477 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table
Supplementary data


