SUMMARY
Plasmodium malariae, a protozoan parasite that causes malaria in humans, has a global distribution in tropical and subtropical regions and is commonly found in sympatry with other Plasmodium species of humans. Little is known about the genetics or population structure of P. malariae. In the present study, we describe polymorphic genetic markers for P. malariae and present the first molecular epidemiological data for this parasite. Six microsatellite or minisatellite markers were validated using 76 P. malariae samples from a diverse geographical range. The repeat unit length varied from 2 to17 bp, and up to 10 different alleles per locus were detected. Multiple genotypes of P. malariae were detected in 33 of 70 samples from humans with naturally acquired infection. Heterozygosity was calculated to be between 0·236 and 0·811. Allelic diversity was reduced for samples from South America and, at some loci, in samples from Thailand compared with those from Malawi. The number of unique multilocus genotypes defined using the 6 markers was significantly greater in Malawi than in Thailand, even when data from single genotype infections were used. There was a significant reduction in the multiplicity of infection in symptomatic infections compared with asymptomatic ones, suggesting that clinical episodes are usually caused by the expansion of a single genotype.
Keywords: Plasmodium malariae, malaria, genetic markers, microsatellites, minisatellites, population genetics
INTRODUCTION
Plasmodium malariae is one of the 4 recognized species of Plasmodium causing malaria in humans. This species is distributed in tropical and subtropical regions and is most frequently found in sympatry with other Plasmodium species. P. malariae is often found as a mixed infection with P. falciparum or P. vivax. Historically, this parasite has been considered of minor importance because of its moderate pathogenicity and low prevalence. In endemic regions, the prevalence of P. malariae, based on standard microscopic examination of Giemsa-stained blood smears, can vary from less than 4% to more than 20% in different epidemiological settings (Molineaux and Grammiccia, 1980; Greenwood et al. 1987; Smith et al. 1993, 2001; Trape et al. 1994). These values are 3-15 times less than the prevalence of P. falciparum in the same geographical regions. However, longitudinal microscopy studies reveal cumulative prevalences in children of 50 to 80% for periods of 1-24 months (Molineaux and Grammiccia, 1980; Trape et al. 1994; Bruce et al. 2000a). Studies using PCR-based diagnostic methods have shown that prevalences based on microscopy are grossly underestimated due to its inability to detect low-level P. malariae parasitaemia, particularly when this species is present with one or more other species of Plasmodium (see Kawamoto et al. 1999; Singh et al. 1999). In studies in Africa, high prevalences of P. malariae were revealed by PCR analyses in surveys in which infection was not detectable by microscopy; for example, 36% in Gabon (Blampain-Azzibrouck et al. 1999) and 25% in Kenya (Tobian et al. 2000). In studies in which P. malariae was detected by microscopy and PCR, higher prevalences (8-30 times) were recorded using the latter technique (Zhou et al. 1998; Mehlotra et al. 2000; Scopel et al. 2004). No figures are published for the prevalence of P. malariae infection worldwide, but a minimum estimate of 67 million infections per year can be calculated by comparison with the prevalence of P. falciparum (see Greenwood et al. 1991; Breman et al. 2001).
P. malariae differs from other Plamodium species of humans in numerous biological features that underlie parasite transmission and virulence, including reduced growth rate (Gilles and Warrell, 1993), mature red blood cell preference (Garnham, 1966), late gametocyte formation (Garnham, 1966), extended duration of infection (up to 40 years in the absence of re-infection) (Vinetz et al. 1998; Chadee et al. 2000; Siala et al. 2005) and the absence of hypnozoite stages in the liver. The clinical features associated with P. malariae fever paroxysms are more moderate relative to P. falciparum and P. vivax (see Wernsdorfer and McGregor, 1988). Fevers show quartan periodicity (due to a 72 h erythrocytic cycle) and parasite density is low, usually considerably below 1000 parasites per μl of blood, being microscopically undetectable much of the time. Infection is rarely life threatening, but can result in childhood mortality due to a ‘nephrotic syndrome’, an immunological complication which persists even after successful antimalarial therapy (Kibukamusoke, 1986; Otieno and Mc’Ligeyo, 1988; Abdurrahman et al. 1990). Despite its relatively low morbidity in humans, P. malariae, in most endemic regions, is likely to contribute to the overall morbidity caused by Plasmodium species, manifested as anaemia (May et al. 2000), low birth weight and reduced resistance to other infections, but its individual contribution has rarely been considered. Importantly, a number of studies have described the effect of prior or concurrent P. malariae infection on those with other Plasmodium species. Interactions can modify the within-host dynamics of Plasmodium species (see Bruce et al. 2000b; Bruce and Day, 2003), reduce the density of asexual stages of P. falciparum (see Alifrangis et al. 1999) and result in an increased production of P. falciparum gametocytes (McKenzie et al. 2002). Prior infection with P. malariae has been shown to reduce fever episodes caused by experimental, malaria therapy P. falciparum infections in humans with limited malaria exposure (Collins and Jeffery, 1999) and also in humans living in areas with intense malaria transmission (Black et al. 1994; Smith et al. 2001).
Due to the difficulty of undertaking laboratory studies of P. malariae, few research projects have focused on this parasite, despite its global importance. Long-term in vitro culture is not available for P. malariae (see Millet et al. 1988; Lingnau et al. 1994) and large numbers of parasites can be obtained only from costly non-human primate infections (Collins et al. 1990, 1997). Limited availability of P. malariae DNA has resulted in a paucity of information about the genetics or genome of this parasite (Carlton et al. 1999). Phylogenetic analyses suggest that P. malariae is distantly related to both P. falciparum and P. vivax (see Qari et al. 1996), with estimated divergence times being up to 174 million years ago (Escalante et al. 1995). P. brasilianum, a malaria parasite of New World monkeys, is both morphologically and genetically indistinguishable from P. malariae (see Cochrane et al. 1985; Ayala et al. 1998; Fandeur et al. 2000). P. brasilianum and P. malariae are likely to represent the same species but appear to have undergone a host switch within the last 500 to 15 000 years (Coatney et al. 1971; Ayala et al. 1998; Escalante et al. 1998; Fandeur et al. 2000), most likely as an anthroponosis in the earlier time-frame. Morphological and genetic variants of human P. malariae described in the Far East may represent subspecies (Liu et al. 1998; Kawamoto et al. 2002).
In order to address knowledge gaps relating to P. malariae, we have isolated polymorphic genetic markers that can be used to investigate multiple aspects of the biology, epidemiology and population structure of this parasite. Microsatellite markers for other species of Plasmodium, including P. falciparum (see Anderson et al. 1999) and P. vivax (see Gomez et al. 2003; Leclerc et al. 2004), have been used for understanding their population genetic structures (Anderson et al. 2000), in the study of the emergence and spread of drug resistance (Nair et al. 2003; Roper et al. 2004; Nash et al. 2005), and in distinguishing new and recrudescent infections (Nyachieo et al. 2005; Mwangi et al. 2006). In this study, we isolated non-coding, microsatellite and minisatellite markers, as they are less likely to be under selection pressure from the immune system of humans, have a high degree of polymorphism and also show a high evolutionary rate relative to expressed genes (Goldstein and Schlotterer, 1999), thus providing useful information regarding genetic changes considered to be relevant to recent Plasmodium population dynamics. These markers provide a means of genetically characterizing P. malariae samples for the first time. Such characterization is considered essential for in vivo drug and vaccine efficacy studies, for addressing questions about the duration and rate of relapse of P. malariae in endemic regions, in the study of within-host dynamics of the parasite and for exploring the population biology of P. malariae. We have validated the markers using a panel of 76 P. malariae isolates from individuals with varying clinical presentation and from a diverse geographical range. We present herein the first molecular epidemiological data for P. malariae and use them to explore the genetic diversity within P. malariae in relation to geographical origin, give insights into its population structure and describe the relationship between multiplicity of infection and clinical presentation.
MATERIALS AND METHODS
Parasite and host samples and DNA extraction
Two strains of Plasmodium malariae were isolated from patients who had acquired P. malariae infections in Uganda (Collins et al. 1984) or Brazil (John Barnwell, unpublished data). The UgandaI/CDC strain, after isolation from an infant infected by blood transfusion, was maintained by passage in splenectomized chimpanzees (Pan troglodytes) and New World monkeys. The infected red blood cells from the Brazil isolate (Pm-Brazil I) were isolated directly from a patient in North-East Brazil. P. brasilianum isolates from Peru (Pb-Peru I/NYU) and Colombia (Pb-Colombia I/NYU) were isolated from infections arising in newly splenectomized New World monkeys (Saimiri peruviensis and S. sciureus, respectively) and maintained by passage in splenectomized Saimiri boliviensis boliviensis (a related species of primate). Blood leukocytes and platelets were removed from the samples using acid-washed glass beads and CF11 cellulose columns or Plasmodipur filters (Euro-Diagnostica, Arnhem, The Netherlands), and genomic DNA was extracted using standard phenol/chloroform extraction protocols (Barnwell et al. 1983, 1989). Laboratory strains of P. falciparum 3D7 and HB3 were used as controls. These isolates were grown in vitro (Trager and Jenson, 1976) and parasites were prepared using saponin lysis and a standard DNA phenol/chloroform extraction (Bruce et al. 1999). DNA from the monkey-adapted P. vivax isolate Salvador I and P. vivax from symptomatic travellers returning to the UK was isolated as reported previously (Bruce et al. 1999).
Blood samples from human patients with naturally acquired, asymptomatic infections of P. falciparum, P. malariae or P. ovale were collected by ‘fingerprick’ in Malawi from residents of villages within a 25 km radius of the Dedza and Mangochi District Hospitals. Samples from symptomatic human infections known to be positive by microscopy for only P. malariae (320-385 parasites per μl of blood) or P. malariae and P. falciparum (P. malariae range 20-5000 per μl, P. falciparum range 6000-8500 per μl) from Gambian children (1-9 years of age) were collected as part of a clinical trial (Sutherland et al. 2003). Samples from Thai patients in the Maela Refugee Camp, Tak Province, presenting with symptomatic infections positive by microscopy for only for P. malariae (1440-8096 parasites per μl of blood) were collected as part of ongoing drug resistance studies (Alan Brockman, personal communication); DNA from humans with naturally acquired infection was extracted, using a Chelex method (Kyes et al. 1993), from blood stored either on filter paper (20-50 μl of blood resulting in an extract volume of 200 μl) or as whole blood or red blood cell pellets stored frozen. Host DNA samples from humans, chimpanzees (Pan troglodytes) and squirrel monkeys (Saimiri boliviensis) were isolated from leukocytes using standard phenol/chloroform extraction protocols (Sambrook and Russell, 2001) and used as negative controls in the molecular analyses.
Ethics approval for experimental infections in animals was granted by the Institutional Animal Care and Use Committees of Emory University and the Centres for Disease Control and Prevention, Atlanta, USA, and by the Animal Ethics Committee of Glasgow University. Collection of parasite samples from humans in Malawi was approved by the National Health Sciences Research Committee, Ministry of Population and Health, Malawi, and the Ethics Committee for Non-clinical Research Involving Human Subjects of Glasgow University. Ethics approval for the use of the samples from Gambia was obtained from the Joint Gambia Government/MRC Laboratories Ethics Committee. Ethics approval for the collection of the samples from Thailand was granted by the Ethics Committees of the Faculty of Tropical Medicine, Mahidol University, Bangkok and the Karen Refugee committee, Mae Sot, Thailand.
Diagnosis of Plasmodium species in naturally acquired human infections
Each of the 4 human species of Plasmodium in naturally acquired infections was detected by microscopic examination of Giemsa-stained blood smears. The species present were determined by examination of thin smears, and their density was measured by counting the number of parasites corresponding to 200 or 500 leukocytes in thick blood smears. The number of parasites per μl of blood was calculated, assuming a leukocyte density of 8000 per μl of blood. To confirm the microscopy results and detect parasites present below the detection limit of microscopy (∼40 parasites per μl of blood), a nested PCR based on the amplification of small subunit ribosomal RNA (ssrRNA) was also carried out essentially as described previously by other authors (Snounou et al. 1993; Singh et al. 1999). In brief, the genus-specific primers Plu1 and Plu5 were used in a primary reaction, followed by a nested reaction using specific primers for each of the 4 species of Plasmodium from humans. Primary reactions were carried out in 50 μl containing 1·25 U of Thermoprime Plus Taq polymerase (ABgene), and the nested reactions were carried out in 25 μl containing 0·6 U of Thermoprime Plus Taq polymerase (transferring 2 μl of primary product). Both reactions contained the same reagents in the same concentrations: 75 mM Tris-HCl (pH 8·8), 20 mM (NH4)2SO4, 4·0 mM MgCl2, 0·01% Tween 20 (Sigma), 0·2mM each of dATP, dCTP, dGTP and dTTP (ABgene) and 250 nM of each primer pair. The approximate amounts of template added to the primary PCR were 0·1 ng of purified DNA or 5 μl of DNA extract from filter-paper blood samples. Amplicons were separated on 1·6% agarose gels in TBE buffer (0·045 M Tris-borate, 0·001 M EDTA, pH 8·0), stained with ethidium bromide and detected by ultraviolet transillumination. A 100 bp ladder (Promega) was used as a size marker on these gels.
Isolation of microsatellite loci
Sequences containing microsatellite loci were isolated from a genomic library of Plasmodium brasilianum of Colombian origin (Pb-Colombia I/NYU). Genomic DNA was digested with RsaI (New England Biolabs, Beverly, MA, USA). Fragments of 300-1500 bp were selected, gel-purified using β-agarase I, according to the manufacturer’s instructions (New England Biolabs) and cloned into the vector pZERO (Invitrogen). Approximately 100 000 colonies were transferred to N+ nitrocellulose filters (Amersham Biosciences) and screened using radio-isotope labelled probes containing polyT, TA, CA and TAA repeats of 100-300 bp in length (Armour et al. 1994). Equimolar concentrations of each probe were mixed, and 200 ng of the mixture was labelled with α32P dCTP (Perkin Elmer) using random priming (Prime-It II Kit, Stratagene). Unincorporated nucleotides were removed using a Microspin G-25 column (Amersham Biosciences). Filters were washed to a stringency of 50°C, 2 × SSC, 0·1% SDS. Inserts of positive colonies were sequenced in both directions using Big Dye chemistry on a 377 sequencer (Applied Biosystems).
Sequencing of P. malariae microsatellite alleles
Polymorphism of P. malariae microsatellite alleles was initially assessed by aligning sequence data from the P. brasilianum library strain and from PCR products from orthologous loci in P. malariae Uganda I/CDC and P. malariae Brazil I, for which large amounts of genomic DNA were available. PCR primers were designed from non-repetitive flanking regions of microsatellite loci using Staden software (Staden et al. 2000). PCR products for sequencing were amplified using the primary reaction primers (see Table 2), with the following alterations: Extensor Hi-Fidelity PCR Enzyme mix (containing Thermoprime Taq and a proof-reading enzyme, ABgene) with buffer 1 and 40 ng of genomic DNA were used. Amplicons were cloned using TOPO TA Cloning® Kit (Invitrogen) according to the manufacturer’s instructions. DNA from transformed colonies was prepared using the Minprep Kit (QIAgen), and inserts were sequenced in both directions using M13 primers. Sequences were assembled using the Staden software, and alignments were made using ClustalW (Chenna et al. 2003). Loci showing polymorphism in repetitive domains were selected for further analysis using a panel of P. malariae DNA samples and controls from various geographical origins. Sequence homology to other Plasmodium species in the non-repetitive regions flanking each polymorphic locus was assessed by comparison with sequences available in GenBank™ using BLASTn.
Table 2.
Sequences of primers used in a semi-nested PCR for the amplification of microsatellite loci from Plasmodium malariae (The first 2 sequences for each locus were used for the primary reaction. The second and third were used in the semi-nested PCR. The third primer was 5′-labelled with a fluorescent tag for the detection of PCR products.)
| Locus | Primer sequences 5′ to 3′ | Tm(°C) | Fluorescent tag | |
|---|---|---|---|---|
| Pm02 | for | GGG GCA TAA AGG AAA AAC | 58.4 | |
| rev | GAA TTT TTG AAT AAC AAG AAA CC | 57.3 | ||
| for* | GCA TAA TAC AGT GAT GCT TAG | 49.6 | 6-Fam | |
| Pm11 | for | GGG ATA TGA ATT ACA TAC AC | 50.5 | |
| rev | CTT TAT TTG TGG TCG AGG | 55.5 | ||
| for* | GAG GAA AGG TTG AAG TGG | 57.0 | Hex | |
| Pm25 | for | CCA AAT AAG TGA CAT ACA AC | 51.8 | |
| rev | GAG GTA ACT TAA AAA ATT CAC | 51.4 | ||
| for* | CTC CCC ATT TTT CGT TC | 57.9 | Ned | |
| Pm34 | rev | TTG GAC AAT GAA AAA ACT AAG | 55.9 | |
| for2 | GAA TGG AAA AAT TCC TTC AG | 57.6 | ||
| rev* | CAA GAT GAA CAA CTG ATA GGG | 58.6 | Hex | |
| Pm09 | rev | GTT CAT AAC TTT GAT CTT AAC | 51.6 | |
| for | ACG ATA ATA ATA TAA ATG GGG | 43.6 | ||
| rev* | CAT TTG ACC AAT TTA ACA CAT TC | 61.3 | Ned | |
| Pm47 | rev | TGT ATG TTT TAT GTT CAC TTC | 49.5 | |
| for | TAA AGG TGT AGT GTA GAG | 53.8 | ||
| rev* | AAA CCG CTG GCT TTA CG | 59.6 | Hex | |
Assessing the extent of polymorphism and validation of the microsatellite markers
To assess the extent of polymorphism for the selected loci microsatellite alleles were amplified, using a semi-nested PCR reaction, from a panel of 93 test samples. The panel contained the following samples (see Table 1). (1) Positive controls: laboratory strains of P. malariae and P. brasilianum, (2) P. malariae from humans with asymptomatic, naturally acquired infections, (3) P. malariae from humans with symptomatic, naturally acquired infections, (4) heterologous controls, including P. falciparum, P. vivax and P. ovale from laboratory strains or humans with naturally acquired infections, in order to test the species-specificity of the primers and PCR conditions, (5) negative controls, including host DNA samples from human, chimpanzee and Saimiri monkey.
Table 1.
Number and geographical origin of Plasmodium species and strains used in the present study
| Sample type | Origin | Plasmodium species and strain | n | |
|---|---|---|---|---|
| Laboratory isolates | Uganda | P. malariae Uganda I/CDC | 1 | |
| Brazil | P. malariae Brazil I | 1 | ||
| Colombia | P. brasilianum Colombia I/NYU | 1 | ||
| Peru | P. brasilianum Peru I/NYU | 1 | ||
| Asymptomatic human isolates | Malawi | P. malariae only | 15 | |
| P. malariae and P. falciparum | 24 | |||
| P. malariae, P. falciparum and P. ovale | 2 | |||
| Symptomatic human isolates | The Gambia | P. malariae only | 5 | |
| P. malariae and P. falciparum | 1 | |||
| Thailand | P. malariae only | 16 | ||
| P. malariae and P. falciparum | 4 | |||
| P. malariae and P. vivax | 3 | |||
| P. malariae, P. falciparum and P. vivax | 4 | |||
| Plasmodium negative controls | Lab strains | |||
| The Netherlands | P. falciparum 3D7 | 1 | ||
| Honduras | P. falciparum HB3 | 1 | ||
| El Salvador | P. vivax Salvador I | 1 | ||
| Malawi human isolates | P. falciparum only | 3 | ||
| P. falciparum and P. ovale | 2 | |||
| P. ovale only | 1 | |||
| Thai human isolates | P. vivax only | 2 | ||
| UK human isolate - origin India | P. vivax 14/97 | 1 | ||
| Host negative controls | Human | n/a | 1 | |
| Chimpanzee | n/a | 1 | ||
| Saimiri monkey | n/a | 1 | ||
| Total | 93 |
The detection of Plasmodium species in humans with naturally acquired infections was based initially on microscopy and then confirmed by PCR. Due to the high parasitaemia in samples from Gambia and Thailand, DNA extracts were diluted 1: 50 or 1: 20 before use. The sequences of primary and semi-nested PCR primers are listed in Table 2, and the amplification conditions are as follows. Both primary and semi-nested reactions were carried out in 25 μl volumes containing 0·6 units of Thermoprime Plus Taq polymerase, 75 mM Tris-HCl, pH 8·8, 20 mM (NH4)2SO4, 3·0 mM MgCl2, 0.01% Tween®20, 0·2 mM each of dATP, dCTP, dGTP, dTTP (ABgene, UK). A reduction in the efficiency of amplification of these and other microsatellite loci occurs if other polymerase-buffer combinations are used (Marian Bruce and Annette MacCleod, Glasgow University, unpublished observations). Primers were employed at 40 nM each for primary reaction and 80 nM for the semi-nested PCR. Target DNA used in the primary reaction was 0·1 ng of purified DNA or 5 μl of DNA extract from filter paper blood samples. A volume of 2 μl of the primary reaction was used as template in the semi-nested reaction. Temperature cycling conditions were as follows: primary reaction (94 °C for 4 min, 1 cycle; 94 °C for 30 sec, 48 °C for 30 sec, 68 °C for 60 sec, 35 cycles; 68 °C for 2 min, 1 cycle); semi-nested reaction (94 °C for 4 min, 1 cycle; 94 °C for 30 sec, 52 °C for 30 sec, 68 °C for 60 sec, 19 cycles; 68 °C for 2 min, 1 cycle). PCR products were separated on a 3730 capillary sequencer (Applied Biosystems) following dilution (1/50-1/100). The analysis of electropherograms was carried out using Gene Mapper v3.7 software (Applied Biosystems). Alleles were scored by eye and sizes measured by comparison with a size standard HD400 (Applied Biosystems). To prevent miss-scoring of ‘stutter peaks’ (Goldstein and Schlotterer, 1999), multiple alleles were scored only if the peak height was greater than one third of the dominant peak. Alleles were ‘binned’ according to size and bin sizes were assigned to within 2 bp. The dominant allele in samples containing multiple alleles was defined as that with the highest peak height. To test the reproducibility of amplification and sizing of alleles, seminested PCR amplification and analysis from all loci and all samples was carried out in duplicate. For laboratory isolates, this was done up to 8 times. Multilocus genotypes from the 6 polymorphic loci were constructed using the dominant allele from each locus. The number of distinct P. malariae genotypes detectable per sample, defined as the ‘multiplicity of infection’ (MOI; see Smith et al. 1999), was considered to be the greatest number of alleles detected at any single locus.
RESULTS
Isolation, sequencing and PCR amplification of microsatellite loci
In order to isolate genetic markers for Plasmodium malariae, a library containing P. brasilianum genomic DNA was constructed. This genetically closely-related parasite species was used in place of P. malariae as the only available P. malariae genomic DNA was insufficiently pure for library construction (contaminated with ∼1 host leukocyte per 100 parasites). Phylogenetic analysis has shown that P. brasilianum (found in New World monkeys) and P. malariae are genetically indistinguishable (Escalante and Ayala, 1994) and are likely to be a single species exhibiting host polymorphism (Ayala et al. 1998; Fandeur et al. 2000).
The genomic library was screened with probes containing repetitive sequences, in order to isolate non-coding, microsatellite markers. Thirty-six positive colonies were isolated from the screen and their inserts sequenced. Six were found to have homology to host DNA, 2 contained no repeats and 1 had a repeat at the end of the sequence. PCR primers were designed to the sequences of the other 27 clones and orthologous sequences were amplified and sequenced from the DNA from each of 2 laboratory isolates of P. malariae (Pm-Uganda I/CDC; Collins et al. 1984 and Pm-Brazil I). On the basis of observed and potential for polymorphism, 11 loci were selected for further use and analyses using a semi-nested PCR (see Table 2) employing a panel of 76 test samples of P. malariae from a wide geographical range, 2 P. brasilianum isolates and 15 controls including other Plasmodium species from humans and host DNA samples (Table 1).
Identification and validation of polymorphic markers
The amplification of alleles from the panel of test samples revealed that 6 of 11 loci tested displayed size polymorphism. The characteristics of polymorphic loci are shown in Table 3. Sequence data for these loci from P. brasilianum Colombia I/NYU are available at GenBank™ (Accession numbers DQ787845-DQ787850). The chromosomal position or the linkage of these markers is not known due to the absence of P. malariae genome data. None of the sequences adjacent to the markers show significant homology with any other genomic sequences from Plasmodium species for which extensive sequence data are available; therefore, they cannot be positioned by inferred synteny (Carlton et al. 1999).
Table 3.
Allelic characteristics for 6 polymorphic microsatellite loci detected in 76 Plasmodium malariae and 2 P. brasilianum samples (cf. Table 1)
| Locus | Repeat unit1 | Allele size range (bp) | Total no. of alleles detected | No. of distinct alleles | Heterozygosity2 | Adjusted heterozygosity3 |
|---|---|---|---|---|---|---|
| Pm02 | (CATA)23 | 167-205 | 103 | 10 | 0.839 | 0.811 |
| Pm11 | (TAAAACAAAAA)11 | 256-309 | 87 | 6 | 0.542 | 0.504 |
| Pm25 | (GT)17 | 100-135 | 97 | 10 | 0.662 | 0.524 |
| Pm34 | (CA)25 | 204-231 | 90 | 9 | 0.607 | 0.546 |
| Pm09 | (GCAAAATAACAAAAAGA)11 | 299-351 | 68 | 5 | 0.331 | 0.236 |
| Pm47 | (CAATT)7 | 326-341 | 72 | 4 | 0.542 | 0.490 |
The number of repeat units is given for the lab isolate P. malariae Brazil I.
Heterozygosity cannot be measured directly from Plasmodium sp. Blood-stage parasites as they are haploid but is estimated as 1—(sum of the squares of the allele frequencies).
Adjusted heterozygosity was calculated from a restricted data set containing only the dominant allele in each sample.
The species-specificity of the PCR primers and conditions of amplification were tested using DNA samples from other species of Plasmodium and host DNA (Table 1). There was no amplification from DNA samples from humans, chimpanzees or Saimiri monkeys at any of the 6 loci. Similarly, no PCR products were detected from samples containing the other 3 human Plasmodium species, originating either from laboratory isolates (P. falciparum and P. vivax) or naturally acquired infections with varying levels of parasitaemia (P. falciparum 1840-760 000 parasites per μl of blood; P. vivax, 1400 and 2400 parasites per μl of blood; P. ovale, <40 parasites per μl of blood; mixed species samples <40-130 000 parasites per μl of blood).
The efficiency and reproducibility of the PCRs were investigated using results from replicate runs for each sample: All 4 laboratory isolates gave exactly the same single allele (sized to the nearest 1 bp) at each locus on up to 8 replicate runs. Of the 74 P. malariae samples from humans with naturally acquired infection (Table 1), 1 Gambian and 3 Malawian samples gave no results for any loci despite being positive for the P. malariae diagnostic PCR (Table 4). The efficiency of amplification for replicate runs was the lowest for loci Pm09 and Pm47 (79·7 and 78·4%, respectively) and the highest for Pm11 and Pm25 (both 91·9%). In samples for which results were obtained for both replicates, the concordance was based on a comparison of allele sizes. Concordance ranged from 71·1 to 94·3% for the 6 loci (Table 4), but this increased to >93% for all loci when samples with ‘consistent’ results were included. Consistent samples were considered to be those where alleles of the same size were detected for both replicates, but with the addition of 1 or more other alleles in one of the replicates. Blood stages of Plasmodium are haploid; therefore, the detection of multiple alleles in a sample from 1 individual human indicates the presence of multiple genotypes.
Table 4.
Efficiency of amplification and reproducibility of allele scoring in duplicate analysis at 6 polymorphic Plasmodium malariae loci in 74 naturally acquired human infections
| Locus | Pm02 | Pm11 | Pm25B | Pm34 | Pm09 | Pm47 |
|---|---|---|---|---|---|---|
| N | 74 | 74 | 74 | 74 | 74 | 74 |
| Both replicates negative1 | 7 | 6 | 6 | 12 | 15 | 16 |
| Only 1 positive replicate | 14 | 21 | 18 | 17 | 24 | 13 |
| % Efficiency of amplification over both replicates | 90.5 | 91.9 | 91.9 | 83.8 | 79.7 | 78.4 |
| Both replicates positive | 53 | 47 | 50 | 45 | 35 | 45 |
| % Samples with identical results2 | 71.7 | 89.4 | 84.0 | 73.3 | 94.3 | 91.1 |
| % Samples with consistent results3 | 98.1 | 97.9 | 100.0 | 93.3 | 100.0 | 95.6 |
The number of samples negative for both replicates includes 4 samples that gave negative results for all 6 loci.
Calculated as a percentage of samples in which both replicates gave positive results.
Samples where allele(s) detected in one replicate was concordant with the other but with additional allele(s). Calculated as a percentage of samples in which both replicates gave positive results.
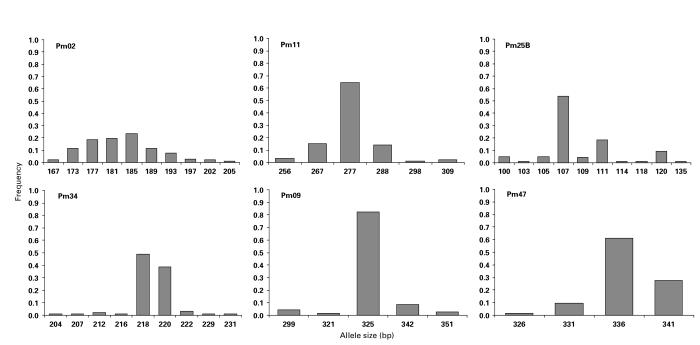
Allelic diversity, geographical distribution of alleles
The number of distinct alleles detected at each of the 6 polymorphic loci was between 4 and 10 (Table 3). The repeat unit size for the 6 loci falls within the range of 2-17 bp. Sequencing of alleles revealed the presence of imperfect repeats at some loci; therefore, bin sizes do not follow exactly the repeat unit size. For some loci, all of the possible alleles within the allele size range were not detected (Fig. 1). Allele frequencies (Fig. 1) were calculated as a proportion of the total number of alleles detected for each locus using P. malariae and P. brasilianum samples (Table 3). The number of alleles detected was greater than the number of samples, due to the presence of multiple alleles per sample in some cases, indicative of the presence of multiple genotypes within a host individual. Heterozygosity was calculated using either all of the allelic data or a restricted data set in which the only dominant allele representing each sample was scored. Values ranged from 0·331 to 0·839 using the whole data set or 0·236 to 0·811 using frequencies only for the dominant allele (Table 3).
Fig. 1.
Allele frequencies for the 6 polymorphic Plasmodium malariae microsatellite loci.
Allelic diversity was restricted in the 3 samples from South America (Table 1). Two samples, P. brasilianum Colombia I/NYU and P. malariae Brazil I, were identical for all loci (i.e. Pm02-185, Pm11-288, Pm25-107, Pm34-220, Pm09-299, Pm47-331). The third sample, P. brasilianum Peru I, differed from the others at 1 locus (Pm02-181). All 3 samples had the allele Pm09-299 that was detected exclusively in samples from South America. The allele Pm47-331, recorded in all 3 South American samples, was rare amongst the other samples, being present in 2 other samples from Malawi. Allelic diversity was also low at some loci for samples from Thailand. All 18 samples from Thailand, for which results were available, were ‘mono-allelic’ for Pm09-325. Allele Pm25-107 was found in 23 of 27 Thai samples that amplified at this locus. In accordance with this ‘allelic restriction’, heterozygosity was lower in Thailand than in Malawi at all loci.
Multilocus genotyping
Genotypes based on all 6 loci were available for 56 of 78 samples. From the 56 samples, 48 different multilocus genotypes were recorded. Most genotypes (n=41) were detected only once, 6 different genotypes were detected in 2 samples and 1 genotype was recorded in 3 samples. In all cases, genotypes detected more than once were only found within the same geographical region. Unique genotypes were recorded in 29 of 33 samples from Malawi, 7 of 14 from Thailand, 3 of 5 from Gambia, and 1 of 3 from South America. The proportion of unique genotypes was significantly greater in Malawi compared with Thailand (χ2=7·88, P<0·005; only these two countries had sufficient sample sizes for statistical analysis). A restricted data set containing samples with a single genotype was also used in this analysis, and 22 different multilocus genotypes were detected in 27 samples: 18 of 22 were detected only once, 3 different genotypes were recorded in 2 samples and 1 genotype was detected in 3 samples. The proportion of samples with unique genotypes in this restricted data set was significantly greater in Malawi (7 of 7) than in Thailand (7 of 12) (χ2=3·96, P<0·01).
Multiple genotypes
Multiple genotypes of P. malariae were detected in 33 of 70 samples from humans with naturally acquired infection (Table 5).
Table 5.
Multiple genotypes detected by analysis of up to 6 microsatellite loci in 70 samples of naturally acquired human Plasmodium malariae infections
| Clinical status | Origin | n (n with data for all 6 loci) | >1 genotype (%) | Mean multiplicity of infection±S.E. |
|---|---|---|---|---|
| Asymptomatic | Malawi | 38 (33) | 27 (71.0) | 2.13±0.15 |
| Symptomatic | The Gambia | 5 (5) | 1 (20.0) | 1.20±0.20 |
| Symptomatic | Thailand | 27 (14) | 5 (18.5) | 1.22±0.10 |
| Total | 70 (52) | 33 (47.1) | 1.71±0.11 |
The greatest number of genotypes in a single sample was 4. The frequency distribution of the MOI is shown in Fig. 2. The MOI was compared between samples from asymptomatic and symptomatic individuals, for which data was available at all 6 loci (Fig. 3). The mean MOI was significantly greater for asymptomatic infections (Malawi, 2·24; S.E.±0·16) compared with symptomatic ones (Gambia and Thailand, 1·21; S.E.±0·12) (t = −5·17, P<0·00001).
Fig. 2.
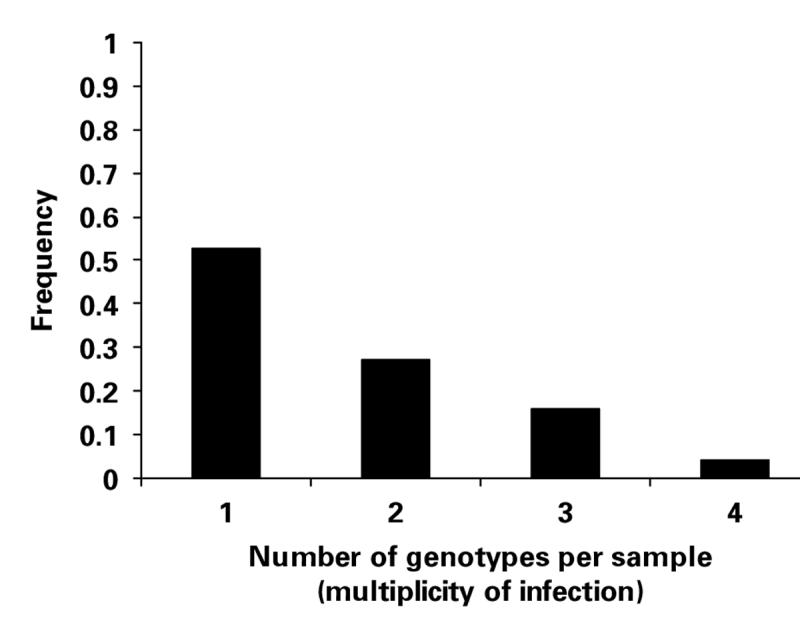
Number of Plasmodium malariae genotypes per sample recorded for 70 samples from humans with naturally acquired infections for which data for any of the 6 loci were available.
Fig. 3.
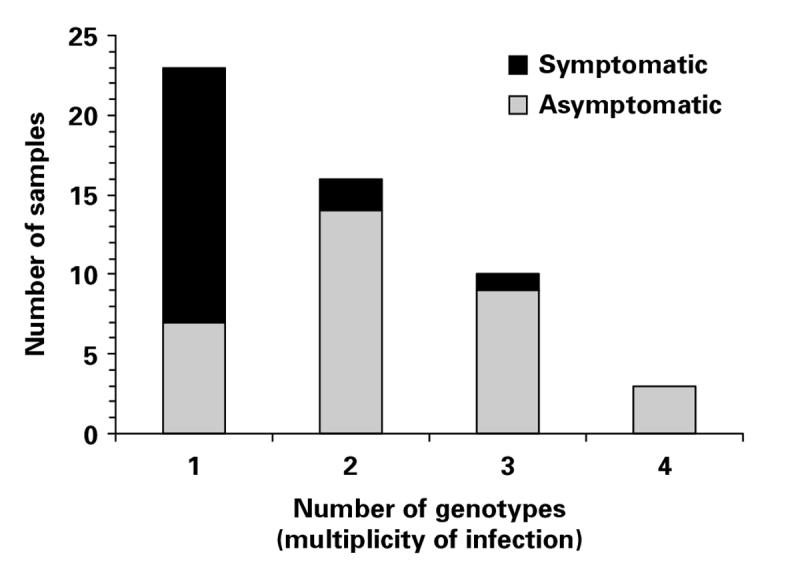
Comparison of the number of Plasmodium malariae genotypes per sample in symptomatic (n=19) and asymptomatic (n=33) samples of naturally acquired human infections for which data were available at all 6 microsatellite loci.
DISCUSSION
In the present study, we have isolated 6 polymorphic genetic markers from Plasmodium malariae. These markers were validated using a series of P. malariae samples from laboratory isolates or from humans with naturally acquired infections. Previous genetic differences between P. malariae strains have been limited to 3 genes, namely the ssrRNA gene (Liu et al. 1998; Kawamoto et al. 2002), the merozoite surface protein 1 gene (msp1) (Fandeur et al. 2000) and the circumsporozoite gene (csp) (Escalante et al. 1995; Tahar et al. 1998; Ayala et al. 1999). These genes have limited usefulness as genetic markers for studying populations. The ssrRNA gene is subjected to gene conversion and has few variants, whereas the gene products of msp1 and csp are major targets of immunity and are thus considered to be under ‘selection pressure’ from the immune system. Multiple, polymorphic, selectively neutral loci have been characterized to enable an assessment of the population genetic structure of P. malariae to be made. Microsatellite markers are useful for multilocus genotyping, and, in the present study, we have used them to distinguish multiple P. malariae genotypes in humans with naturally acquired infections, and have compared the number of genotypes (or multiplicity of infection, MOI) from asymptomatic and symptomatic individuals. This is the first molecular epidemiological study of P. malariae.
The markers are likely to be non-coding as they all lie within repetitive, highly AT-rich regions of the genome, in which there are numerous stop codons (Goldstein and Schlotterer, 1999). In such regions, primer sites are restricted and, therefore, amplicons were often significantly larger than the repetitive regions. Repeats units of 1-6 bp are considered microsatellites and those of a greater size are referred to as minisatellites. The size of the repeat unit is thought to relate to the mechanism/s by which such loci evolve (Armour et al. 1999). For simplicity, we have used ‘microsatellites’ to describe these markers, although two markers Pm11 and Pm09 have repetitive units greater than 6 bp and, thus, formally represent minisatellites.
Differences between loci in the efficiency of amplification (Table 4) are considered to reflect variation in the efficiency of PCR reactions for each locus, as a result of the use of different primers, the quantity and quality of target DNA and potential errors at numerous micro-pipetting steps in the analysis protocol. Four P. malariae samples that gave negative results for all 6 loci were likely to have contained small amounts of target DNA that could be detected by nested PCR of the ssrRNA gene (which is repetitive), but was not sufficient for the detection at the (single copy) microsatellite loci. Differences in replicate results are thought to arise from variation in the dynamics of the PCR-amplification of multiple, low copy DNA targets and has been observed by other authors investigating P. falciparum (see Färnert et al. 2001). Almost half (47·1%) of natural human P. malariae samples were shown to contain multiple genotypes. Allele frequencies were calculated, allowing for multiple genotypes per sample. However, data from multiple genotypes can overestimate heterozygosity, because the frequency of minority alleles is over-estimated. Therefore, heterozygosity was calculated from both the whole data set and from a restricted data set in which only the dominant allele recorded in each sample was scored. For some loci, the 2 values did not differ significantly (Pm02 and Pm11) but in others a substantial difference was observed. We did not assess the presence of null alleles in our study. Null alleles are difficult to distinguish from ‘pipetting errors’ without additional multiple amplification replicates or internal controls and cannot be adequately assessed in samples containing multiple genotypes.
Allelic diversity was restricted in the 3 samples from South America, and all contained the unique allele Pm09-299. Two of these samples showed identical multilocus genotypes, and the other varied from these only at 1 locus. This restricted allelic diversity may be a result of a ‘bottleneck’ from recent colonization events. Malaria is thought to have been brought to South America with European settlers and later with African slaves, and this has been suggested as an explanation of restricted diversity for P. falciparum in South America (Anderson et al. 2000). However, 2 of 3 samples from South America were P. brasilianum from New World monkeys, a species morphologically and genetically indistinguishable from P. malariae (see Cochrane et al. 1985; Ayala et al. 1998; Fandeur et al. 2000). The restricted diversity could also be the result of a single host-switch event of P. malariae from humans to New World Monkeys (Fandeur et al. 2000) or a selective ‘sweep’ associated with a host-switch. In some settings in South America, it is thought that some P. malariae infections in humans may result from monkey to human transmission of P. brasilianum. Analysis of more South American samples from both monkey and human hosts in defined geographical settings is required to test these hypotheses.
The restriction of allelic diversity in Thai samples compared with Malawian samples was also seen at 2 loci. One explanation for the reduced diversity at some loci may be a ‘selective sweep’ due to linkage of the microsatellite loci to drug resistance loci. This phenomenon has been observed for P. falciparum microsatellite loci closely linked to the pyrimethamine resistance locus (Nair et al. 2003; Pearce et al. 2005). P. malariae is often clinically asymptomatic but will have increased exposure to drug pressure, as a result of treatment of humans with infections with multiple species of Plasmodium. The prevalence of mixed-species infections is high in South East Asia (Zhou et al. 1998), and this is the only region in which drug resistance has been reported in P. malariae (see Maguire et al. 2002). Low genetic diversity could also be a result of subspeciation within P. malariae populations. Morphologically and genetically different forms of P. malariae have been detected in South East Asia (Kawamoto et al. 2002), but it is not known whether any of the Thai samples in this study represent one of these forms.
The extent of multispecies Plasmodium infections in asymptomatic patients could be indicative of an accumulation of chronic, long-lived, low-density infections over time rather than a high rate of acquisition of multiple genotypes in a short time-period. An analysis of how MOI is related to the age of individuals is required to further investigate this question. The lower number of genotypes in symptomatic compared with asymptomatic samples suggests that the clinical symptoms arising from increased P. malariae parasitaemia results from the growth of a single P. malariae genotype rather than the accumulation of multiple lower density infections. The samples from asymptomatic patients used herein contained ‘pure’ P. malariae infection or P. malariae in combination with other Plasmodium species. Most ‘asymptomatic samples’ from Malawi contain a high proportion of multiple species (>90%) and, when tested, gave similar results to those described here (multiple genotypes 74·6%, mean MOI 2·16; Marian Bruce, unpublished observations). Most ‘symptomatic samples’ originated from a low endemicity region in Thailand (Luxemburger et al. 1996) where most malaria infections are symptomatic. In contrast, the ‘asymptomatic samples’ originated from Malawi, which has much higher rates of malaria transmission, where individuals are commonly infected with multiple species and genotypes from infancy and where there is a high level of naturally acquired immunity in humans. However, the 4 samples from symptomatic patients from Gambia were also due to a single genotype. This strengthens the argument that symptoms arise from single genotypes, even between different endemic regions. A more rigorous comparison would include samples from symptomatic and asymptomatic individuals living in the same endemic region, although obtaining sufficient numbers for such a comparison would be challenging.
Direct comparison of the MOI data for P. malariae with those for other Plasmodium species is not informative, due to multiple confounding factors, including the age structure of the population sampled, level of malaria endemicity and the type, number and heterozygosity of the genetic markers used. However, MOI values for P. malariae described here fall within documented ranges for P. falciparum (see Babiker and Walliker, 1997) and P. vivax (see Cole-Tobian et al. 2005), despite the significant differences in the epidemiology of P. malariae. The MOI is a useful indicator of the degree of malaria transmission and is important for understanding the potential for recombination in Plasmodium species. Only when different genotypes are ingested by the mosquito vector in a single bloodmeal can recombination occur. The rate of recombination between different genotypes influences the generation of diversity and the rate of spread of drug resistance through a population (Hastings and Watkins, 2005). A major aim of molecular and population genetic studies of Plasmodium is to determine how the rate of recombination, genetic diversity and the rate of transmission are linked to the acquisition of clinical immunity in human populations living in endemic regions. This description of genetic markers for P. malariae enables, for the first time, the investigation of the molecular epidemiology and population genetics of this little studied, yet widely distributed pathogen of humans. The first molecular epidemiology data for P. malariae presented herein, shows a high level of multi-genotype carriage in humans, suggesting that regular genetic exchange within P. malariae populations is probable, despite the lower prevalence and density of this parasite relative to other species of Plasmodium. Further analysis of a larger number of samples from a greater geographical range, different epidemiological settings, a variety of age groups and from potential primate reservoirs is necessary to further characterize the epidemiology and population structure of P. malariae.
We are grateful to Colin Sutherland, Ali Alloueche, Rosalynn Ord, Alan Brockman and Geoff Pasvol for the provision of P. malariae samples. Thanks to Malcolm Molyneux for assistance in the collection of Malawian samples. We are indebted to Keith Scott, Liz Peat and Julie Galbraith for excellent technical support. Our thanks go to Andy Tait and Annette MacLeod, Glasgow University, for useful discussions. Thanks also to Nick Helps and Joanne Quinney at the University of Dundee Sequencing Service for processing of microsatellite samples. The technical post of Keith Scott was funded by the University of Glasgow. This research was funded by a Wellcome Trust Junior Biodiversity Fellowship (Reference 060446) to Marian Bruce.
REFERENCES
- Abdurrahman MB, Aikhionbare HA, Babaoye FA, Sathiakumar N, Narayana PT. Clinicopathological features of childhood nephrotic syndrome in northern Nigeria. Quarterly Journal of Medicine. 1990;75:563–576. [PubMed] [Google Scholar]
- Alifrangis M, Lemnge MM, Moon R, Theisen M, Bygbjerg I, Ridley RG, Jakobsen PH. IgG reactivities against recombinant Rhoptry-Associated Protein-1 (rRAP-1) are associated with mixed Plasmodium infections and protection against disease in Tanzanian children. Parasitology. 1999;119:337–342. doi: 10.1017/s0031182099004825. [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Molecular Biology and Evolution. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- Anderson TJC, Su X-Z, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterisation of Plasmodium falciparum from fingerprick blood samples. Parasitology. 1999;119:113–125. doi: 10.1017/s0031182099004552. [DOI] [PubMed] [Google Scholar]
- Armour JA, Neumann R, Gobert S, Jeffreys AJ. Isolation of human simple repeat loci by hybridization selection. Human Molecular Genetics. 1994;3:599–565. doi: 10.1093/hmg/3.4.599. [DOI] [PubMed] [Google Scholar]
- Armour JAL, Alegre SA, Miles S, Williams LJ, Badge RM. Minisatellites and mutation processes in tandemly repetitive DNA. In: Goldstein DB, Schlotterer C, editors. Microsatellites: Evolution and Applications. Oxford: Oxford University Press; 1999. pp. 24–33. [Google Scholar]
- Ayala FJ, Escalante AA, Rich SM. Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum. Parassitologia. 1999;41:55–68. [PubMed] [Google Scholar]
- Ayala FJ, Lal AA, Escalante AA, Rich SM. Evolutionary relationships of human malaria parasites. In: Sherman IW, editor. Malaria: Parasite Biology, Pathogenesis and Protection. Washington D.C.: ASM Press; 1998. pp. 285–300. [Google Scholar]
- Babiker HA, Walliker D. Current views on the population structure of Plasmodium falciparum: implications for control. Parasitology Today. 1997;13:262–267. doi: 10.1016/s0169-4758(97)01075-2. [DOI] [PubMed] [Google Scholar]
- Barnwell JW, Howard RJ, Coon HG, Miller LH. Splenic requirement for antigenic variation and expression of the variant antigen on the erythrocyte membrane in cloned Plasmodium knowlesi malaria. Infection and Immunity. 1983;40:985–994. doi: 10.1128/iai.40.3.985-994.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwell JW, Nichols ME, Rubinstein P. In vitro evaluation of the role of the Duffy blood group in erythrocyte invasion by Plasmodium vivax. Journal of Experimental Medicine. 1989;169:1795–1802. doi: 10.1084/jem.169.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J, Hommel M, Snounou G, Pinder M. Mixed infections with Plasmodium falciparum and P. malariae and fever in malaria. Lancet. 1994;343:1095. doi: 10.1016/s0140-6736(94)90203-8. [DOI] [PubMed] [Google Scholar]
- Blampain-Azzibrouck G, Lekoulou F, Snounou G, Ravollet JC, Ntoumi F. Short communication: Plasmodium falciparum and P. malariae infections in isolates from sickle cell gene carriers living in a hyperendemic area of Gabon. Tropical Medicine and International Health. 1999;4:872–874. doi: 10.1046/j.1365-3156.1999.00500.x. [DOI] [PubMed] [Google Scholar]
- Breman JG, Egan A, Keusch GT. The intolerable burden of malaria: a new look at the numbers. American Journal of Tropical Medicine and Hygiene. 2001;64:iv–vii. doi: 10.4269/ajtmh.2001.64.iv. [DOI] [PubMed] [Google Scholar]
- Bruce MC, Day KP. Cross-species regulation of Plasmodium parasitaemia in semi-immune children from Papua New Guinea. Trends in Parasitology. 2003;19:271–277. doi: 10.1016/s1471-4922(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Bruce MC, Galinski MR, Barnwell JW, Snouou G, Day KP. Polymorphism at the Msp3α locus of P. vivax: global and local diversity. American Journal of Tropical Medicine and Hygiene. 1999;61:518–525. doi: 10.4269/ajtmh.1999.61.518. [DOI] [PubMed] [Google Scholar]
- Bruce MC, Donnelly CA, Alpers MP, Galinski MR, Barwell JW, Walliker D, Day KP. Cross-species interactions between malaria parasites in humans. Science. 2000b;287:845–848. doi: 10.1126/science.287.5454.845. [DOI] [PubMed] [Google Scholar]
- Bruce MC, Donnelly CA, Packer M, Lagog M, Gibson N, Narara A, Walliker D, Alpers MP, Day KP. Age- and species-specific duration of infection in asymptomatic malaria infections in an endemic population in Papua New Guinea. Parasitology. 2000a;121:247–256. doi: 10.1017/s0031182099006344. [DOI] [PubMed] [Google Scholar]
- Carlton JMR, Galinski MR, Barnwell JW, Dame JB. Karyotype and synteny among the chromosomes of all four species of human malaria parasite. Molecular and Biochemical Parasitology. 1999;101:23–32. doi: 10.1016/s0166-6851(99)00045-6. [DOI] [PubMed] [Google Scholar]
- Chadee DD, Tilluckdharry CC, Maharaj P, Sinanan C. Reactivation of Plasmodium malariae infection in a Trinidadian man after neurosurgery. New England Journal of Medicine. 2000;342:1924. doi: 10.1056/NEJM200006223422520. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coatney GR, Collins WE, Warren M, Contacos PG. The Primate Malarias. Washington D.C.: U.S. Government Printing Office; 1971. [Google Scholar]
- Cochrane AH, Barnwell JW, Collins WE, Nussenzweig RS. Monoclonal antibodies produced against sporozoites of the human parasite Plasmodium malariae abolish infectivity of sporozoites of the simian parasite Plasmodium brasilianum. Infection and Immunity. 1985;50:58–61. doi: 10.1128/iai.50.1.58-61.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole-Tobian JL, Biasor M, King CL. High complexity of Plasmodium vivax infections in Papua New Guinean children. American Journal of Tropical Medicine and Hygiene. 2005;73:626–633. [PubMed] [Google Scholar]
- Collins W, Jeffery G. A retrospective examination of sporozoite- and trophozoite-induced infections with Plasmodium falciparum in patients previously infected with heterologous species of Plasmodium: effect on development of parasitologic and clinical immunity. American Journal of Tropical Medicine and Hygiene. 1999;61:36–43. doi: 10.4269/tropmed.1999.61-036. [DOI] [PubMed] [Google Scholar]
- Collins WE, Schwartz IK, Skinner JC, Broderson JR. Studies on the UgandaI/CDC strain of Plasmodium mlarariae in Bolivian Aotus monkeys and various anophelines. Journal of Parasitology. 1984;70:677–681. [PubMed] [Google Scholar]
- Collins WE, Richardson BB, Sullivan JS, Morris CL, Galland GG. Infection of Anopheles freeborni mosquitoes on New World monkeys infected with the Uganda I/CDC strain of Plasmodium malariae. Journal of Parasitology. 1997;83:1099–1103. [PubMed] [Google Scholar]
- Collins WE, McClure HM, Strobert E, Filipski V, Skinner JC, Stanfill PS, Richardson BB, Morris C. Infection of chimpanzees with the Uganda I/CDC strain of Plasmodium malariae. American Journal of Tropical Medicine and Hygiene. 1990;42:99–103. doi: 10.4269/ajtmh.1990.42.99. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium derived from rRNA gene sequences. Proceedings of the National Academy of Sciences, USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Barrio E, Ayala FJ. Evolutionary origin of human and primate malarias: evidence from the circumsporozoite protein gene. Molecular Biology and Evolution. 1995;12:616–626. doi: 10.1093/oxfordjournals.molbev.a040241. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proceedings of the National Academy of Sciences, USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fandeur T, Volney B, Peneau C, de Thoisy B. Monkeys of the rainforest in French Guiana are natural reservoirs for P. brasilianum/P. malariae malaria. Parasitology. 2000;120:11–21. doi: 10.1017/s0031182099005168. [DOI] [PubMed] [Google Scholar]
- Färnert A, Arez AP, Babiker HA, Beck HP, Benito A, Bjorkman A, Bruce MC, Conway DJ, Day KP, Henning L, Mercereau-Puijalon O, Ranford-Cartwright LC, Rubio JM, Snounou G, Walliker D, Zwetyenga J, do Rosario VE. Genotyping of Plasmodium falciparum infections by PCR: a comparative multicentre study. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2001;95:225–232. doi: 10.1016/s0035-9203(01)90175-0. [DOI] [PubMed] [Google Scholar]
- Garnham PCC. Malaria Parasites and Other Haemosporidia. Oxford: Blackwell; 1966. [Google Scholar]
- Gilles HM, Warrell DA. Bruce-Chwatt’s Essential Malariology. London: Edward Arnold; 1993. [Google Scholar]
- Goldstein DB, Schlotterer C. Microsatellites: Evolution and Applications. Oxford, UK: Oxford University Press; 1999. [Google Scholar]
- Gomez JC, McNamara DT, Bockarie MJ, Baird JK, Carlton JM, Zimmerman PA. Identification of a polymorphic Plasmodium vivax microsatellite marker. American Journal of Tropical Medicine and Hygiene. 2003;69:377–379. [PMC free article] [PubMed] [Google Scholar]
- Greenwood BM, Marsh K, Snow R. Why do some African children develop severe malaria? Parasitology Today. 1991;8:239–242. doi: 10.1016/0169-4758(91)90096-7. [DOI] [PubMed] [Google Scholar]
- Greenwood BM, Bradley AK, Greenwood AM, Byass P, Jammeh L, Marsh K, Tulloch S, Oldfield FST, Hayes R. Mortality and morbidity from malaria among children in a rural area of The Gambia, West Africa. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1987;81:478–486. doi: 10.1016/0035-9203(87)90170-2. [DOI] [PubMed] [Google Scholar]
- Hastings IM, Watkins WM. Intensity of malaria transmission and the evolution of drug resistance. Acta Tropica. 2005;94:218–229. doi: 10.1016/j.actatropica.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Kawamoto F, Liu Q, Ferreira M, Tantular I. How prevalant are Plasmodium ovale and P. malariae in East Asia? Parasitology Today. 1999;15:422–426. doi: 10.1016/s0169-4758(99)01511-2. [DOI] [PubMed] [Google Scholar]
- Kawamoto F, Win TT, Mizuno S, Lin K, Kyaw O, Tantulart IS, Mason DP, Kimura M, Wongsrichanalai C. Unusual Plasmodium malariae-like parasites in southeast Asia. Journal of Parasitology. 2002;88:350–357. doi: 10.1645/0022-3395(2002)088[0350:UPMLPI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Kibukamusoke JW. The hazard of malarial nephropathy. Parasitology Today. 1986;2:119–121. doi: 10.1016/0169-4758(86)90043-8. [DOI] [PubMed] [Google Scholar]
- Kyes S, Craig AG, Marsh K, Newbold CI. Plasmodium falciparum: a method for the amplification of S antigens and its application to laboratory and field samples. Experimental Parasitology. 1993;77:473–483. doi: 10.1006/expr.1993.1108. [DOI] [PubMed] [Google Scholar]
- Leclerc MC, Durand P, Gauthier C, Patot S, Billotte N, Menegon M, Severini C, Ayala FJ, Renaud F. Meager genetic variability of the human malaria agent Plasmodium vivax. Proceedings of the National Academy of Sciences, USA. 2004;101:14455–14460. doi: 10.1073/pnas.0405186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingnau A, Doehring-Schwerdtfeger E, Maier WA. Evidence for 6-day cultivation of human Plasmodium malariae. Parasitology Research. 1994;80:265–266. doi: 10.1007/BF00932687. [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhu S, Mizuno S, Kimura M, Liu P, Isomura S, Wang X, Kawamoto F. Sequence variation in the small-subunit rRNA gene of Plasmodium malariae and prevalence of isolates with the variant sequence in Sichuan, China. Journal of Clinical Microbiology. 1998;36:3378–3381. doi: 10.1128/jcm.36.11.3378-3381.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxemburger C, Thwai KL, White NJ, Webster HK, Kyle DE, Maelankirri L, Chongsuphajaisiddhi T, Nosten F. The epidemiology of malaria in a Karen population on the western border of Thailand. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1996;90:105–111. doi: 10.1016/s0035-9203(96)90102-9. [DOI] [PubMed] [Google Scholar]
- Maguire JD, Sumawinata IW, Masbar S, Laksana B, Prodjodipuro P, Susanti I, Sismadi P, Mahmud N, Bangs MJ, Baird JK. Chloroquine-resistant Plasmodium malariae in south Sumatra, Indonesia. Lancet. 2002;360:58–60. doi: 10.1016/S0140-6736(02)09336-4. [DOI] [PubMed] [Google Scholar]
- May J, Falusi AG, Mockenhaupt FP, Ademowo OG, Olumese PE, Bienzle U, Meyer CG. Impact of subpatent multi-species and multi-clonal plasmodial infections on anaemia in children from Nigeria. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2000;94:399–403. doi: 10.1016/s0035-9203(00)90119-6. [DOI] [PubMed] [Google Scholar]
- McKenzie FE, Jeffery GM, Collins WE. Plasmodium malariae infection boosts Plasmodium falciparum gametocyte production. American Journal of Tropical Medicine and Hygiene. 2002;67:411–414. doi: 10.4269/ajtmh.2002.67.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Lorry K, Kastens W, Miller SM, Alpers MP, Bockarie M, Kazura JW, Zimmerman PA. Random distribution of mixed species malaria infections in Papua New Guinea. American Journal of Tropical Medicine and Hygiene. 2000;62:225–231. doi: 10.4269/ajtmh.2000.62.225. [DOI] [PubMed] [Google Scholar]
- Millet P, Collins WE, Fisk TL, Nguyen-Dinh P. In vitro cultivation of exoerythrocytic stages of the human malaria parasite Plasmodium malariae. American Journal of Tropical Medicine and Hygiene. 1988;38:470–473. doi: 10.4269/ajtmh.1988.38.470. [DOI] [PubMed] [Google Scholar]
- Molineaux L, Grammiccia G. The Garki Project: Research on the Epidemiology and Control of Malaria in the Sudan Savanna of West Africa. Geneva: The World Health Organization; 1980. [Google Scholar]
- Mwangi JM, Omar SA, Ranford-Cartwright LC. Comparison of microsatellite and antigen-coding loci for differentiating recrudescing Plasmodium falciparum infections from reinfections in Kenya. International Journal for Parasitology. 2006;36:329–336. doi: 10.1016/j.ijpara.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Nair S, Williams JT, Brockman A, Paiphun L, Mayxay M, Newton PN, Guthmann JP, Smithuis FM, Hien TT, White NJ, Nosten F, Anderson TJ. A selective sweep driven by pyrimethamine treatment in southeast asian malaria parasites. Molecular Biology and Evolution. 2003;20:1526–1536. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- Nash D, Nair S, Mayxay M, Newton PN, Guthmann JP, Nosten F, Anderson TJ. Selection strength and hitchhiking around two anti-malarial resistance genes. Proceedings of the Royal Society of London, B. 2005;272:1153–1161. doi: 10.1098/rspb.2004.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyachieo A, Van Overmeir C, Laurent T, Dujardin JC, D’Alessandro U. Plasmodium falciparum genotyping by microsatellites as a method to distinguish between recrudescent and new infections. American Journal of Tropical Medicine and Hygiene. 2005;73:210–213. [PubMed] [Google Scholar]
- Otieno LS, Mc’Ligeyo SO. Review article: immune nephritides due to malaria. East African Medical Journal. 1988;65:402–406. [PubMed] [Google Scholar]
- Pearce R, Malisa A, Kachur SP, Barnes K, Sharp B, Roper C. Reduced variation around drug-resistant dhfr alleles in African Plasmodium falciparum. Molecular Biology and Evolution. 2005;22:1834–1844. doi: 10.1093/molbev/msi177. [DOI] [PubMed] [Google Scholar]
- Qari SH, Shi YP, Pieniazek NJ, Collins WE, Lal AA. Phylogenetic relationship among the malaria parasites based on small subunit rRNA gene sequences: monophyletic nature of the human malaria parasite, Plasmodium falciparum. Molecular Phylogenetics and Evolution. 1996;6:157–165. doi: 10.1006/mpev.1996.0068. [DOI] [PubMed] [Google Scholar]
- Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning - a Laboratory Manual. 3rd Edn. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Scopel KK, Fontes CJ, Nunes AC, Horta MF, Braga EM. High prevalence of Plamodium malariae infections in a Brazilian Amazon endemic area (Apiacas-Mato Grosso State) as detected by polymerase chain reaction. Acta Tropica. 2004;90:61–64. doi: 10.1016/j.actatropica.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Siala E, Khalfaoui M, Bouratbine A, Hamdi S, Hili K, Aoun K. Relapse of Plasmodium malariae malaria 20 years after living in an endemic area. Presse Medicale. 2005;34:371–372. doi: 10.1016/s0755-4982(05)83926-0. [DOI] [PubMed] [Google Scholar]
- Singh B, Bobogare A, Cox-Singh J, Snounou G, Shukri Abdullah M, Abdul Rahman H. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. American Journal of Tropical Medicine and Hygiene. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- Smith T, Charlwood JD, Kihonda J, Mwankusye S, Billingsey P, Meuwissen J, Lyimo E, Takken W, Teuscer T, Tanner M. Absence of seasonal variation in malaria parasitaemia in an area of intense seasonal transmission. Acta Tropica. 1993;54:55–72. doi: 10.1016/0001-706x(93)90068-m. [DOI] [PubMed] [Google Scholar]
- Smith T, Beck HP, Kitua A, Mwankusye S, Felger I, Fraser-Hurt N, Irion A, Alonso P, Teuscher T, Tanner M. Age dependence of the multiplicity of Plasmodium falciparum infections and of other malariological indices in an area of high endemicity. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1999;93(Suppl 1):15–20. doi: 10.1016/s0035-9203(99)90322-x. [DOI] [PubMed] [Google Scholar]
- Smith T, Genton B, Baea K, Gibson N, Narara A, Alpers MP. Prospective risk of morbidity in relation to malaria infection in an area of high endemicity of multiple species of Plasmodium. American Journal of Tropical Medicine and Hygiene. 2001;64:262–267. doi: 10.4269/ajtmh.2001.64.262. [DOI] [PubMed] [Google Scholar]
- Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, Rosário VE, Thaithong S, Brown KN. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Molecular and Biochemical Parasitology. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods in Molecular Biology. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- Sutherland CJ, Drakeley CJ, Obisike U, Coleman R, Jawara M, Targett GA, Milligan P, Pinder M, Walraven G. The addition of artesunate to chloroquine for treatment of Plasmodium falciparum malaria in Gambian children delays, but does not prevent treatment failure. American Journal of Tropical Medicine and Hygiene. 2003;69:19–25. [PubMed] [Google Scholar]
- Tahar R, Ringwald P, Basco LK. Heterogeneity in the circumsporozoite protein gene of Plasmodium malariae isolates from sub-Saharan Africa. Molecular and Biochemical Parasitology. 1998;92:71–78. doi: 10.1016/s0166-6851(97)00226-0. [DOI] [PubMed] [Google Scholar]
- Tobian AA, Mehlotra RK, Malhotra I, Wamachi A, Mungai P, Koech D, Ouma J, Zimmerman P, King CL. Frequent umbilical cord-blood and maternal-blood infections with Plasmodium falciparum, P. malariae, and P. ovale in Kenya. Journal of Infectious Diseases. 2000;182:558–563. doi: 10.1086/315729. [DOI] [PubMed] [Google Scholar]
- Trager W, Jenson JB. Human malaria parasites in continuous culture. Science. 1976;193:674–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Trape JF, Rogier C, Konate L, Diagne N, Bouganali H, Canque B, Legros F, Badji A, Ndiaye G, Ndiaye P, Brahimi K, Faye O, Druilhe P, Pereira da Silva L. The Dielmo project: a longitudinal study of natural malaria infection and the mechanisms of protective immunity in a community living in a holoendemic area of Senegal. American Journal of Tropical Medicine and Hygiene. 1994;51:123–137. doi: 10.4269/ajtmh.1994.51.123. [DOI] [PubMed] [Google Scholar]
- Vinetz JM, Li J, McCutchan TF, Kaslow DC. Plasmodium malariae infection in an asymptomatic 74-year-old Greek woman with splenomegaly. New England Journal of Medicine. 1998;338:367–371. doi: 10.1056/NEJM199802053380605. [DOI] [PubMed] [Google Scholar]
- Wernsdorfer WH, McGregor I. Malaria: Principles and Practice of Malariology. London: Churchill Livingstone; 1988. [Google Scholar]
- Zhou M, Liu Q, Wongsrichanalai C, Suwonkerd W, Panart K, Prajakwong S, Pensiri A, Kimura M, Matsuoka H, Ferreira MU, Isomura S, Kawamoto F. High prevalence of Plasmodium malariae and Plasmodium ovale in malaria patients along the Thai-Myanmar border, as revealed by acridine orange staining and PCR-based diagnoses. Tropical Medicine and International Health. 1998;3:304–312. doi: 10.1046/j.1365-3156.1998.00223.x. [DOI] [PubMed] [Google Scholar]