Abstract
We have investigated the x-ray scattering signal of highly aligned multilayers of the zwitterionic lipid 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine containing pores formed by the antimicrobial peptide alamethicin as a function of the peptide/lipid ratio. We are able to obtain information on the structure factor of the pore fluid, which then yields the interaction potential between pores in the plane of the bilayers. Aside from a hard core with a radius corresponding to the geometric radius of the pore, we find a repulsive lipid-mediated interaction with a range of ∼30 Å and a contact value of 2.4 kBT. This result is in qualitative agreement with recent theoretical models.
INTRODUCTION
The elucidation of lipid-mediated interaction forces between membrane proteins and the corresponding lateral distribution in the plane of the membrane is an important step toward a quantitative understanding of the functional mechanisms of membrane proteins and membrane peptides. Experimentally, the lateral structure and organization of multicomponent membranes is as important as it is difficult to probe. While fluorescence microscopy in biological and model membranes can be used to monitor domains and the distribution of proteins typically at the micron scale (1) and down to a few hundred nanometers at best, atomic force microscopy can resolve lateral structures down to a nanometer (2), but only in relatively stiff systems and rarely in the fluid state of the membrane. To this end, x-ray or neutron scattering from aligned fluid bilayers is an excellent tool to probe correlations between proteins or peptides in the bilayer. In this work, we show how the lateral and vertical intensity profiles of a peptide pore correlation peak can be analyzed as a function of peptide concentration to determine the corresponding interaction forces.
A well-known example of biological function deriving from lipid-peptide interaction and self-assembly is the activity of a family of short and amphiphilic membrane active polypeptides denoted as antimicrobial peptides. These molecules bind to microbial cell membranes, subsequently causing an increase in membrane permeability and cell lysis. One such molecule is alamethicin, a 20-amino-acid peptide from the fungus Trichoderma viride; it is well known that alamethicin acts by creating pores in the cell membrane (3). This conclusion has been reached by a convergence of multiple experimental techniques: oriented circular dichroism (4,5) and nuclear magnetic resonance (NMR) (6) have shown that, above a certain concentration, the binding state of the peptides changes from adsorbed parallel to the membrane to inserted into the bilayer. At the same time, a notable increase in membrane conductivity ((7), and references therein) and permeability (8) was measured. Pore formation is usually a highly cooperative process (9,10); this was confirmed for alamethicin and a membrane-mediated interaction between peptides was invoked to explain the phenomenon (5).
Although determining the interaction between (adsorbed or inserted) monomers is very difficult and, to our knowledge, has never been attempted, the interaction between already formed pores within the membrane can be studied using neutron or x-ray scattering from oriented multilamellar stacks, a method pioneered by Huang and collaborators (11–13).
For the case of alamethicin, they observed a lateral correlation peak, which was attributed to liquidlike ordering of pores in the plane of the membrane and was modeled based on hard disk interaction, with very satisfactory results. However, in these studies, at most, two peptide-to-lipid concentrations P/L were investigated for each system.
Building upon this work, we gathered detailed information on the quasi two-dimensional fluid of pores in the lipid bilayer, using high-resolution synchrotron scattering from aligned multilamellar stacks of alamethicin/DMPC mixtures. We measured the two-dimensional scattering distribution for an entire concentration series P/L and performed a simultaneous lineshape analysis on all recorded curves.
We found that the in-plane interaction potential consists of a hard core, with a radius that agrees very well with the geometrical outer radius of the pore, and an additional repulsive contribution, which can be described as a Gaussian, with a range of 31.5 Å and a contact value of 2.41 kBT. The results are in qualitative agreement with recent theoretical models (14,15).
In principle, this method is readily applied to any peptide/lipid system, provided that well-oriented multilayer samples can be prepared. Thus, the role of different parameters such as bilayer composition, temperature, nature of the aqueous medium, etc. can be systematically studied.
MATERIALS AND METHODS
Sample preparation and environment
The lipid 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine (DMPC) was purchased from Avanti Polar Lipids (Birmingham, AL) with a purity of at least 99% and alamethicin was bought from Sigma Aldrich (St. Louis, MO) with a purity of at least 98.9%. Without further purification, the products were dissolved in a TFE/CHCl3, 1:1 vol/vol mixture at a concentration of 60 mg/ml for the lipid and 15 mg/ml for the peptide. The stock solutions were then mixed (and solvent added as necessary) to give the desired molar lipid/peptide ratio P/L, at a final lipid concentration of 20 mg/ml. The resulting solutions were then kept at 4°C for at least 24 h before preparing the samples. More details on sample preparation and on the choice of solvents are given by the literature (16–18).
Rectangular silicon substrates (15 × 25 mm2) were cut from 0.4-mm-thick commercial wafers (Silchem, Freiberg, Germany) and cleaned by sonicating them (during 15 min) twice in methanol and then twice in ultrapure water (specific resistance ≥18 MΩ cm, Millipore, Bedford, MA). Finally, they were abundantly rinsed in ultrapure water and dried under nitrogen flow.
An amount of 0.2 ml of the solution was pipetted onto the substrates under a laminar flux hood, where they were subsequently left to dry at room temperature for a few hours and then exposed to high vacuum at 40°C overnight to remove any remaining traces of solvent. They were finally stored at 4°C until the measurement. From the amount of lipid deposited, the thickness of the film can be estimated at ∼3000 lipid bilayers.
Before the measurement, the samples were placed in the experimental chamber and the hydrating solution was gently injected so as to avoid washing the lipid film off the substrate. Two types of sample chambers were used, the first one machined out of Plexiglas and with an optical path of ∼1.7 cm, and the second one made of Teflon and with an optical path of ∼1.1 cm. Both chambers have 0.3 mm thick kapton windows and were mounted on a metal heating stage temperature-controlled by water flow from a heating bath (Julabo, Seelbach, Germany).
For all P/L values, the hydrating solution was 100 mM NaCl brine containing 31% w/w PEG 20,000 (Fluka Chemie, Buch, Switzerland), yielding an osmotic pressure of ∼1.68 106 Pa. (This value was obtained from the data of Prof. Peter Rand, at the Membrane Biophysics Laboratory of Brock University, Canada: http://aqueous.labs.brocku.ca/osfile.html.) Additionally, for P/L = 1/12.5 we also performed measurements at 12.1 and 5.8% PEG concentration, corresponding to 1.2 105 and 3.5 104 Pa, respectively. The temperature was kept at 30°C for all experiments.
Measurement
The measurements were performed at the undulator beamline ID1 of the European Synchrotron Radiation Facility (ESRF, Grenoble, France). The photon energy was set at 19 keV by a double-bounce Si111 monochromator and the higher-order harmonics were cut by reflection on two Rh-coated mirrors. At this energy, the transmission of 1 cm of water is of 0.45, so the presence of the experimental cell does not pose any attenuation problems.
Three types of measurements were performed:
Reflectivity scans (in the vertical scattering plane) up to a z component of the scattering vector
 (see Fig. 4) give access to the electronic density profile of the bilayers along the z direction (19,20). However, they correspond to averaging over the plane of the bilayer, so all lateral information is lost.
(see Fig. 4) give access to the electronic density profile of the bilayers along the z direction (19,20). However, they correspond to averaging over the plane of the bilayer, so all lateral information is lost.CCD images are taken (using a Peltier-cooled camera, 1242 × 1152 pixels, from Princeton Scientific Instruments, Princeton, NJ) at a fixed incidence angle αi of the x-ray beam onto the sample and correspond to sections through the reciprocal space with the Ewald sphere (see Fig. 3); they provide a global image of the q-space and the position of the pore signal can be quickly determined.
Quantitative measurements were performed using a point-detector (Cyberstar scintillation detector from Oxford Danfysik, Oxford, UK). Transversal (along qy) scans were taken through the pore scattering signal, with wide open slits in the vertical direction, covering a qz range between 0.14 and 0.18 Å−1. For some samples, longitudinal scans (along qz) were also taken. Their trajectories in q-space are shown in Fig. 3 (right) as dotted lines.
FIGURE 4.
Reflectivity spectra of aligned DMPC multilayers containing alamethicin. The data is only shown for four P/L concentrations. Curves vertically shifted for clarity, with the P/L ratio increasing from bottom to top.
FIGURE 3.
Structure of the reciprocal space for a lamellar stack. Reflectivity scans are performed along the vertical z axis, while the CCD images—an example of which is shown to the right—represent slices through the reciprocal space along the Ewald sphere. The characteristic features can easily be identified: the intense diffuse sheets around the very sharp Bragg peaks; the extended and narrow circle arcs going through the Bragg peaks are defect-induced Debye-Scherrer rings. The intensity increase close to the horizon is due to dynamic effects. Finally, the off-axis signal exhibiting a maximum at ∼qy = 0.1−1 is due to the presence of alamethicin pores.
Analysis
The alamethicin pores are dispersed in the lamellar phase matrix. Since the “pure” lamellar phase gives a signal confined to the vicinity of the Bragg peaks, from the Babinet principle it ensues that the off-axis scattering is the same as for a system where the density profile of the lamellar phase is subtracted, and one is left with fictitious “pore-bilayer” objects in a completely transparent medium. Furthermore, as the pores represent a collection of identical and similarly oriented objects (up to an azimuthal averaging), the classical separation of the scattering intensity in a structure factor multiplied by a form factor can be applied (21), yielding: 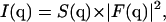 with
with
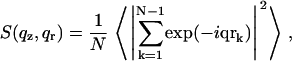 |
(1) |
where N is the number of objects and object “0” is taken as the origin of the coordinates. If there is no in-plane ordering, S only depends on the absolute value of the in-plane scattering vector 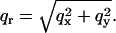 The form factor is given by
The form factor is given by
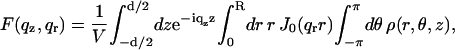 |
(2) |
where ρ is the electron density and V is the integration volume (corresponding to the size of the object).
The first step in computing the structure factor is determining the numerical density of pores in the plane of the bilayer (or, conversely, the area per pore). For the alamethicin, we use the values given in the literature for an eight-monomer pore in DLPC (12): the peptides are modeled as cylinders of 11 Å in diameter; however, the effective cross-section of a peptide is only 66 Å2 (the rest being occupied by lipid chains). For the DMPC (Table 6 in (22),), we consider that the area per molecule at 30°C and in the absence of applied pressure is A0 = 59.6 Å2; in our experiments, the osmotic pressure reduces it to A = 59 Å2, the area compressibility modulus being KA = 0.234 N/m (Table 1 in (23)). We only detect a significant off-axis signal (assignable to the pores) for P/L ≥ 1:25, concentration at which >80% of the peptide is in the inserted state, and this ratio increases to >90% for P/L = 1:20 (5,24). (These results were obtained for DPhPC; the peptide is in the inserted state at all measured concentrations in DLPC (12).) Thus, we consider that all alamethicin molecules are involved in pore formation.
TABLE 1.
Fit results with different models
| P/L | Param. | Same R, same U0 | Same R, U0 = 0 | Free R, same U0 | Free R, U0 = 0 |
|---|---|---|---|---|---|
| 1:25 | R [Å] | 18.3 | 19.2 | 24.3 | 24.8 |
| 1:20 | R | 18.3 | 19.2 | 23.5 | 24.1 |
| 1:15 | R | 18.3 | 19.2 | 21.7 | 22.1 |
| 1:15 | R | 18.3 | 19.2 | 22.8 | 23.1 |
| 1:12.5 | R | 18.3 | 19.2 | 21.4 | 21.7 |
| 1:10 | R | 18.3 | 19.2 | 19.4 | 19.8 |
| 1:7.5 | R | 18.3 | 19.2 | 17.7 | 17.9 |
| U0 [kBT] | 4.74 | 0 | 1.56 | 0 | |
| ξ [Å] | 31.5 | — | 34.8 | — | |
| Nparam | 10 | 8 | 16 | 14 | |
| χ2/Npnts | 15.14 | 22.0 | 8.93 | 9.44 |
Fit conditions refer to the hard core radius R being the same for all scans or allowed to take different values for different individual scans (free) and to the presence or absence (U0 = 0) of the additional interaction. The values U0 and ξ are the amplitude and range of the additional interaction. The value χ2/Npnts is an indication of the fit quality. The value Nparam is the number of fit parameters, including the seven intensity prefactors, one for each scan.
The area per pore is determined assuming that the peptides are straight cylinders placed at the vertices of a regular hexa-, hepta-, or octagon. The water pore is the inscribed cylinder (tangent to the peptides), with a radius Rim = 5.5, 7.2, and 9 Å, for m = 6, 7, and 8, respectively,
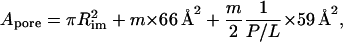 |
(3) |
where the 1/2 factor in the third term (corresponding to the area taken by the lipid molecules) accounts for the two monolayers. For simplicity, the kink in the peptide (see Hydrophobic Mismatch) and the polydispersity in aggregation number (25) were neglected. To determine the form factor, we used the molecular dynamics (MD) results (available on the web site of Dr. Peter Tieleman, Department of Biological Sciences, University of Calgary: http://moose.bio.ucalgary.ca/downloads/; used with permission) of Tieleman et al. (26), who studied alamethicin pores of different sizes in a POPC bilayer.
The form factor was computed according to Eq. 2, for a 30 × 30 Å2 patch containing the pore and for a similar patch containing only lipids, obtained by tiling four times a 15 × 15 Å2 patch from the same simulation. (The patch sizes are 40 × 40 Å2 and 20 × 20 Å2, respectively, in the case of the eight-monomer pore.) The effective form factor used is the difference of the two. We neglected the difference between POPC and DMPC when using the resulting form factor in our fits. Since the MD simulations indicate that the hexamer is the most stable configuration in POPC (26) and neutron scattering results find 8–9 monomers per pore in DLPC (12), we considered the pore configurations with six, seven, and eight monomers. As we shall see later, the seven-monomer configuration gives the best fits, so all results presented in the following correspond to this configuration.
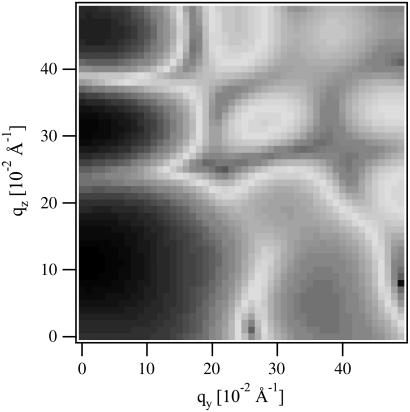
In Fig. 1 we present a (y, z) cross-section through the form factor |F(q)| of a pore, after subtraction of the pure bilayer background and azimuthal averaging. Directions x and y are equivalent.
FIGURE 1.
Absolute value of the form factor |F(qy,qz)| for a seven-monomer alamethicin pore after subtraction of the pure bilayer background and azimuthal averaging.
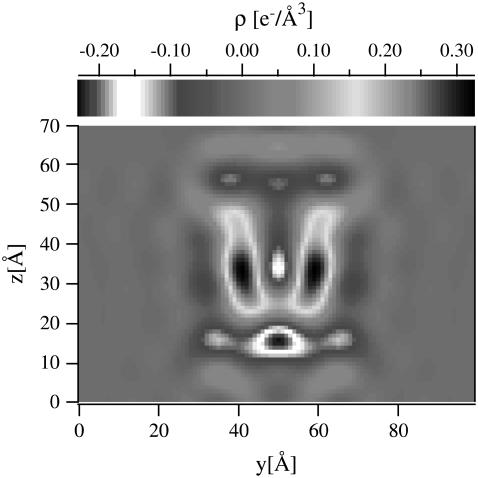
For a visual representation of the scattering object (pore-bilayer) we performed a Fourier transform of F(q) back to real space, yielding the density profile shown in Fig. 2. The peptide monomers are clearly visible as higher density streaks.
FIGURE 2.
Reconstruction of the electron density profile of the seven-monomer alamethicin pore (after subtracting the pure bilayer background) by Fourier transforming back to real space the form factor displayed in Fig. 1.
RESULTS
Structure of the scattered signal
To serve as an illustration for the discussion of the results, we show in Fig. 3 a diagram of the reciprocal space structure for a multilamellar stack on solid support, as well as an actual CCD image (which amounts to a cut by the Ewald sphere). The off-axis signal (exhibiting a maximum at ∼qy = 0.1 Å−1) is due to the alamethicin pores; for clarity, it is not represented in the diagram on the left. To bring up this (very weak) signal, the Bragg sheets are severely overexposed. The image was taken at an incidence angle αi = 0.55°, for a sample with P/L = 1:20.
Perfectly aligned samples
We checked the quality of the samples and their alignment by performing reflectivity measurements (19,20). The mosaicity can be estimated at ∼0.01° from the rocking scans. The reflectivity curves are shown in Fig. 4 for four different P/L values. Seven Bragg peaks are generally visible, and the smectic period d changes very little with P/L (18). The reflectivity yields the electron density profile along the director of the lamellar phase ρ(z) (averaged in-plane), but the analysis is rather involved (18,27–30) and we shall not go into further detail here.
No interaction from one bilayer to the next
A question of paramount importance is whether the pores interact from one bilayer to the next (along the z direction); we need to answer it to choose the theoretical model employed for describing the data (two-dimensional versus three-dimensional interaction) and, furthermore, to determine the biological relevance of our measurements.
An effective way of determining the interbilayer interactions (13) is by measuring the scattering pattern at different swelling values and comparing the qz variation between different curves and against the expected form factor of the scattering object. We performed this investigation by exposing concentrated samples (P/L = 1:12.5) to different osmotic pressures (see Sample Preparation and Environment). Fig. 5 shows detector scans through the peaks along qz (various symbols), as well as a cut through the simulated form factor in Fig. 1 (solid line).
FIGURE 5.
Sections along qz through the pore signal of samples with P/L = 1:12.5 at different osmotic pressures (symbols) for qy = 0.1 Å−1 and cut through the square of the simulated form factor |F(qz)|2 (solid line). The sharp peaks appearing in the top and bottom curves are due to the Debye-Scherrer rings (see Fig. 3).
The first observation is that the measured curves are very similar; furthermore, their shape is qualitatively similar to that of the simulated form factor, if we neglect the presence of a slowly varying background, presumably due to thermal fluctuations of the lamellar phase (see next section). We can therefore conclude that there is no interaction between pores from one bilayer to the next.
Thus, for the purpose of studying pore interaction, the bilayers in the solid-supported stacks we investigate can be considered independent, as one would require for modeling the cell membrane. Although the Ala monomers can be charged at neutral pH (31,32), the 100 mM NaCl concentration (similar to that of biological media) reduces the Debye length to ∼10 Å, effectively screening the electrostatic potential; the only remaining interaction is that mediated by the bilayer.
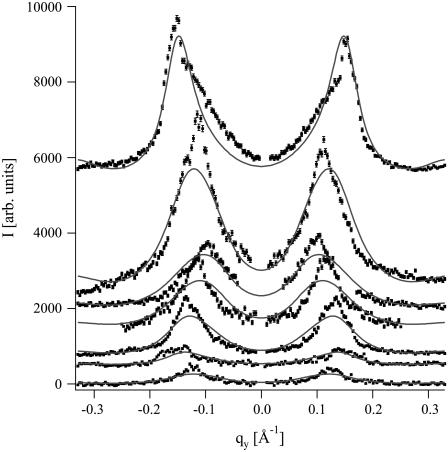
Pore signal
Fig. 6 shows the detector scans along qy (out of the plane of incidence) for four different P/L values (indicated alongside the curves). A very intense and sharp component in qy = 0 (due to the specular beam) was removed for clarity. Scattering from the thermal fluctuations gives rise to a wide “bump” centered at the origin; to remove it, we fit the scans with a three-Lorentzian model (illustrated for the lower curve) and subtract the central component from the measured data.
FIGURE 6.
Detector scans along qy through the pore signal for four P/L concentrations (integrated in the range 0.14 < qz < 0.18 Å−1). For the bottom scan we also show the three-Lorentzian fit to the data; the central “bump” is subtracted before further treatment.
As a measure of fit quality we use the χ2 function divided by the number of points Npnts. The standard deviation σn for each experimental point is determined considering a Poisson distribution for the measured signal (before background subtraction) 
Hard disk model
The simplest model for the interaction is that of hard disks confined in the plane. Using the “fundamental measure” approach, Rosenfeld ((33), Eq. 6.8) provided a simple analytical expression for Shd(qr), which is accurate over the entire concentration range we explore; the complete formula is given in the Appendix.
First, the fits were performed for each scan individually, the hard disk radius R and the number density of pores n = 1/Apore being free parameters. For each scan, we tried the form factor for the six-, seven-, and eight-monomer pores. One fit example is displayed in Fig. 7 (for P/L = 1:12.5), and the values of the fit parameters are shown in Fig. 8 for all scans.
FIGURE 7.
Fit of the data for P/L = 1/12.5 with a hard-disk model. (Diamonds) Point detector scan. The fit, shown as a gray line, is the product of the form factor |F(qz)|2 for the seven-monomer pore (solid line) the structure factor for a hard disk system (dashed line), with radius R = 20.95 Å and number density n = 1/Apore = 3.59 10−4 Å−2. For clarity, only the qy > 0 range is displayed.
FIGURE 8.
Values of the fit parameters (density and hard disk radius) obtained from individual fits of the scans, with the form factor of 6- (open diamonds), 7- (solid dots), and 8-monomer pores (open triangles). For comparison, the density obtained as n = 1/Apore according to formula (1) is shown as dotted, solid, and dashed lines, for the 6-, 7-, and 8-monomer pores, respectively.
The first conclusion is that the best agreement between the value of n obtained by fitting and that calculated using Eq. 1 is obtained assuming a seven-monomer pore. The agreement is slightly worse for the hexamer and clearly off for the octamer; this can also be seen from the values of the χ2 function for the different individual fits (data not shown). We can therefore assume that we are dealing with seven-monomer pores.
A very important result of the individual fits is that the value of R decreases with the P/L concentration from 24.8 to 17.9 Å. One might understand an increase in radius at higher concentration due to the appearance of pores with more than seven monomers, but a decrease is clearly an unphysical result, which might indicate the presence of a “soft” repulsive interaction: as the concentration increases, the pores are forced closer together, overcoming this energy barrier. We therefore redid the fits including such a contribution.
The samples with P/L = 1:12.5 at lower osmotic pressure yield sensibly higher values of R than those corresponding to c = 31% (see Fig. 8, right). For reliability, we decided to ignore these points in further fits. This discrepancy does not correspond to a change in interaction between pores (see No Interaction From One Bilayer to the Next); it originates most probably in the difficulty of obtaining a clear separation between the pore signal and the thermal scattering, which increases substantially with decreasing osmotic pressure (data not shown).
Additional interaction
We now consider a more complex interaction, consisting of hard core repulsion (when the pores are in contact) and an additional, longer-range term, corresponding to a bilayer-mediated interaction. For simplicity, we describe this component as a Gaussian and we account for its effect on the structure factor perturbatively, using the random phase approximation (RPA); see the Appendix for more details.
The experimental data are fitted simultaneously using the same parameters; R is the hard core radius, U0 corresponds to the amplitude of the additional component, and ξ to its range (see Eq. 5 for the definition).
Since the best individual fits were obtained with a seven-monomer pore, we impose the pore density calculated for this model (corresponding to m = 7 in Eq. 3) as well as the form factor (solid line in Fig. 7). We also checked that the fit quality (as given by the χ2 function) is better than for the six- and eight-monomer pores.
The fit results are shown in Fig. 9, and the interaction potential is plotted in Fig. 10. Comparison between the different fit configurations is detailed in Table 1 and Fig. 11. Briefly, the presence of the additional interaction strongly increases the quality of the fit with respect to a fit with a fixed R but U0 = 0 (χ2/Npnts = 15.14, as opposed to 22.0). Although this value seems very large, the fit quality is (visually) adequate and the error bars on the fit parameters quite small: the decimal places in Table 1 are significant. It is very likely that the Poisson distribution severely underestimates the standard deviation on each point. The fit quality is still much worse than that obtained by letting R vary with the P/L ratio, but in this latter situation more fit parameters are used, aside from the unphysical assumption of shrinking pore size. Even for varying R, the additional interaction yields a modest decrease in χ2. In this case we obtain a similar range ξ, but a much lower amplitude U0, most of the effect being “simulated” by the apparent R variation (see Table 1 for the value of the fitting parameters and Fig. 12 for the plots).
FIGURE 9.
Experimental data (dots) and fits (solid lines) with a hard disk model and an additional repulsive contribution. The curves correspond to different peptide concentrations; from top to bottom, Ala/DMPC = 1:7.5, 1:15, 1:25, 1:20, 1:12.5, 1:10, 1:15. All scans are fitted simultaneously, yielding R = 18.3 Å, U0 = 4.74 kBT, and ξ = 31.5 Å.
FIGURE 10.
The interaction potential used for the fits in Fig. 9, consisting of a hard core and an additional Gaussian repulsion, given by Eq. 5, with parameters U0 = 4.74 kBT and ξ = 31.5 Å. The contact value U(2R) = 2.41 kBT.
FIGURE 11.
Schematic representation of interacting pores in a lipid bilayer. The seven monomers border the water pore. The halo shows the range of the lipid-mediated repulsion.
FIGURE 12.
Experimental data (dots) and fits (solid lines) in the various configurations described in Table 1. The top left set, corresponding to the same R and U0, is the same as in Fig. 9.
To summarize, we find that the interaction between seven-monomer pores of alamethicin in DMPC bilayers can be described by a hard core with radius 18.3 Å, in excellent agreement with the geometrically estimated outer radius of the pore (18.2 Å) and an additional repulsive interaction described by Eq. A2, with a range ξ = 31.5 ± 0.27Å and an amplitude U0 = 4.74 ± 0.09 kBT, corresponding to a contact value U(2R) = 2.4 kBT. These very small error bars on the fit parameters should, however, be considered very carefully, since the most important source of error is probably the simplified model for S(q).
DISCUSSION AND CONCLUSION
Very few experimental results point to the existence of lipid-mediated interaction between membrane inclusions; to our knowledge, they were all obtained by freeze-fracture electron microscopy (34–37) and yielded directly the radial distribution function of the inclusions. The data were compared to liquid state theories (38–40) and could be described by a hard-core model with, in some cases, an additional repulsive or attractive interaction.
In contrast, very sustained theoretical efforts aimed at understanding these systems started 30 years ago (41–44); they are either continuum-elasticity theories (45–48) or more detailed models taking into account the molecular structure of the lipid bilayer (14,15,41,49). Two main origins for inclusion interaction have emerged, as discussed below.
Hydrophobic mismatch
A wide consensus has been reached as to the importance of “hydrophobic mismatch,” the difference in length between the hydrophobic part of the protein or peptide and that of the host membrane (50,51). However, the specific way in which this mismatch is accommodated for one particular system is not at all clear, especially when the peptide is longer than the lipid (the case of alamethicin in DMPC), since both bilayer compression/expansion and peptide tilt can be involved (51,52).
The alamethicin/DMPC system was studied by NMR, finding that the peptide is either parallel to the bilayer normal (53) or tilted by 10–20° (54), conclusion supported by simulations (55). Moreover, the peptide exhibits a kink at the Pro14 residue, (54,56,57), making the evaluation even more complicated. If one considers the entire pore as one (rigid) object, the tilt is probably very small, due to its size (58). Thus, the mismatch is likely compensated by bilayer expansion, which propagates over a few tens of Å from the edge of the inclusion (58,59), values comparable to our experimental findings.
Changes in lipid ordering
Another, more subtle effect is that an inclusion modifies the structure of the bilayer by perturbing the configuration of the lipid chains (14,41,60,61). In particular, the results of Lagüe et al. (14,15) are in semiquantitative agreement with our observations: they extracted the lateral density-density response function of the hydrocarbon chains from the MD simulations of a DPPC bilayer (62) and used it to determine the interaction between “smooth” (no hydrophobic mismatch) hard cylinders embedded in the bilayer. For the largest cylinder radius they considered (9 Å, approximately half that of alamethicin pores), they obtain a repulsive lipid-mediated interaction with a maximum value of 10 kBT and extending 20 Å from contact (14). This study was followed by a comparison between different lipids, including DMPC (the lipid used in our experiments) (15); intriguingly, in this case they find a nonmonotonic interaction, attractive close to contact and repulsive for larger distances. Furthermore, this interaction extends further than in the case of DPPC. We did not perform a more detailed comparison between their predictions and our experimental results, since the interaction potential varies considerably with the inclusion radius, but the agreement is certainly encouraging.
Perspectives
For a complete description of the perturbation and the interaction it induces, both hydrophobic mismatch and changes in chain ordering must be taken into account (63,64). It has been pointed out repeatedly (48,64) that the spontaneous curvature of the monolayer radically changes the lipid-mediated interaction. To date, no consistent picture has emerged, due to theoretical difficulties but also to the lack of experimental data.
The experimental work presented here consisted in determining the lipid-mediated interaction between alamethicin pores in DMPC bilayers; we found it to be repulsive and the overall shape of the potential is in qualitative agreement with recent theoretical predictions (14,15). However, the quality of the fits to the experimental data is not very good; this can stem from technical difficulties and systematic errors, but also from the rough model employed (RPA approximation). Both these aspects will be improved in the future but the results are already significant.
Acknowledgments
The European Synchrotron Radiation Facility is gratefully acknowledged for the provision of synchrotron radiation facilities (experiments SC-1136 and SC-1375) and we thank the staff of ID1 for competent and enthusiastic support.
D.C. has been supported by a Marie Curie Fellowship of the European Community program Improving the Human Research Potential under contract No. HPMF-CT-2002-01903.
APPENDIX
Hard disk model
We used the analytical expression for the structure factor of hard disks given by Rosenfeld ((33), Eq. 6.8)
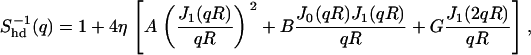 |
(4) |
where q is the in-plane scattering vector, R the hard disk radius, η = nπR2 the packing fraction (with n the number density of the disks), and Jk the Bessel functions of the first kind and order k. The prefactors are given by
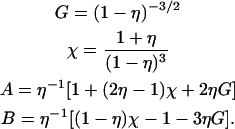 |
Additional repulsive interaction
We added a repulsive component described by a Gaussian, with amplitude U0 and range ξ,
 |
(5) |
considered as a perturbation with respect to the hard disk model, taken into account via the random phase approximation (RPA) (65,66). In this approach, one obtains the direct correlation function of the perturbed system c(r) from that of the reference system cref(r) as
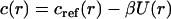 |
(6) |
(66) or, equivalently,
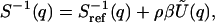 |
(7) |
with 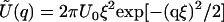 the Fourier transform of U(r).
the Fourier transform of U(r).
Fit parameters
For all practical purposes, we give in Table 1 the value of the fit parameters for the different configurations discussed in the text; throughout, the form factor and the density are those of a seven-monomer pore. The corresponding fits are displayed in Fig. 12. The top left set (same R, same U0) is the same as in Fig. 9.
D. Constantin's present address is Laboratoire de Physique des Solides, Université Paris-Sud, Bât. 510, 91405 Orsay Cedex, France.
G. Brotons' present address is Laboratoire de Physique de l'État Condensé, Université du Maine, Faculté des Sciences et Techniques, Av. O. Messiaen, 72085 Le Mans Cedex 9, France.
References
- 1.Korlach, J., P. Schwille, W. Webb, and G. Feigenson. 1999. Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA. 96:8461–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oesterhelt, F., D. Oesterhelt, M. Pfeiffer, A. Engel, H. Gaub, and D. Muller. 2000. Unfolding pathways of individual bacteriorhodopsins. Science. 288:143–146. [DOI] [PubMed] [Google Scholar]
- 3.Duclohier, H., and H. Wróblewski. 2001. Voltage-dependent pore formation and antimicrobial activity by alamethicin and analogues. J. Membr. Biol. 184:1–12. [DOI] [PubMed] [Google Scholar]
- 4.Olah, G. A., and H. W. Huang. 1988. Circular dichroism of oriented α helices. I. Proof of the exciton theory. J. Chem. Phys. 89:2531–2538. [Google Scholar]
- 5.Chen, F.-Y., M.-T. Lee, and H. W. Huang. 2002. Sigmoidal concentration dependence of antimicrobial peptide activities: a case study on alamethicin. Biophys. J. 82:908–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bechinger, B. 1999. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta. 1462:157–183. [DOI] [PubMed] [Google Scholar]
- 7.Bechinger, B. 1997. Structure and functions of channel-forming peptides: magainins, cecropins, melittin and alamethicin. J. Membr. Biol. 156:197–211. [DOI] [PubMed] [Google Scholar]
- 8.Lau, A. L. Y., and S. I. Chan. 1976. Voltage-induced formation of alamethicin pores in lecithin bilayer vesicles. Biochemistry. 15:2551–2555. [DOI] [PubMed] [Google Scholar]
- 9.Shai, Y. 1999. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta. 1462:55–70. [DOI] [PubMed] [Google Scholar]
- 10.Huang, H. 2000. Action of antimicrobial peptides: two-state model. Biochemistry. 39:8347–8352. [DOI] [PubMed] [Google Scholar]
- 11.He, K., S. J. Ludtke, H. W. Huang, and D. L. Worcester. 1995. Antimicrobial peptide pores in membranes detected by neutron in-plane scattering. Biochemistry. 34:15614–15618. [DOI] [PubMed] [Google Scholar]
- 12.He, K., S. J. Ludtke, D. L. Worcester, and H. W. Huang. 1996. Neutron scattering in the plane of the membranes: structure of alamethicin pores. Biophys. J. 70:2659–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang, L., T. Weiss, T. Harroun, W. Heller, and H. Huang. 1999. Supramolecular structures of peptide assemblies in membranes by neutron off-plane scattering: method of analysis. Biophys. J. 77:2648–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagüe, P., M. J. Zuckermann, and B. Roux. 2000. Lipid-mediated interactions between intrinsic membrane proteins: a theoretical study based on integral equations. Biophys. J. 79:2867–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagüe, P., M. J. Zuckermann, and B. Roux. 2001. Lipid-mediated interactions between intrinsic membrane proteins: dependence on protein size and lipid composition. Biophys. J. 81:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludtke, S. J., K. He, and H. W. Huang. 1995. Membrane thinning caused by magainin 2. Biochemistry. 34:16764–16769. [DOI] [PubMed] [Google Scholar]
- 17.Yang, L., T. M. Weiss, R. I. Lehrer, and H. W. Huang. 2000. Crystallization of antimicrobial pores in membranes: magainin and protegrin. Biophys. J. 79:2002–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, C., D. Constantin, and T. Salditt. 2004. Biomimetic membranes of lipid-peptide model systems prepared on solid support. J. Phys. Cond. Matt. 16:S2439–S2453. [Google Scholar]
- 19.Tolan, M. 1999. X-Ray Scattering from Soft-Matter Thin Films. Springer Verlag, Berlin.
- 20.Als-Nielsen, J., and D. McMorrow. 2001. Elements of Modern X-Ray Physics. John Wiley & Sons.
- 21.Chaikin, P. M., and T. C. Lubensky. 1995. Principles of Condensed Matter Physics. Cambridge University Press, Cambridge.
- 22.Nagle, J., and S. Tristram-Nagle. 2000. Structure of lipid bilayers. Biochim. Biophys. Acta. 1469:159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rawicz, W., K. C. Olbrich, T. McIntosh, D. Needham, and E. Evans. 2000. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 79:328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, Y., K. He, S. J. Ludtke, and H. W. Huang. 1995. X-ray diffraction study of lipid bilayer membranes interacting with amphiphilic helical peptides: diphytanoyl phosphatidylcholine with alamethicin at low concentration. Biophys. J. 68:2361–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantor, R. S. 2002. Size distribution of barrel-stave aggregates of membrane peptides: influence of the bilayer lateral pressure profile. Biophys. J. 82:2520–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tieleman, D. P., B. Hess, and M. S. P. Sansom. 2002. Analysis and evaluation of channel models: simulations of alamethicin. Biophys. J. 83:2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lösche, M. 2002. Surface-sensitive x-ray and neutron scattering characterization of planar lipid model membranes and lipid/peptide interactions. Curr. Topics Membr. 52:117–161. [Google Scholar]
- 28.Salditt, T., C. Li, A. Spaar, and U. Mennicke. 2002. X-ray reflectivity of solid-supported multilamellar membranes. Eur. Phys. J. E. 7:105–116. [Google Scholar]
- 29.Salditt, T., and G. Brotons. 2004. Biomolecular and amphiphilic films probed by surface sensitive x-ray and neutron scattering. Anal. Bioanal. Chem. 379:960–973. [DOI] [PubMed] [Google Scholar]
- 30.Salditt, T. 2005. Thermal fluctuations and stability of solid-supported lipid membranes. J. Phys. Cond. Matt. 17:R287–R314. [Google Scholar]
- 31.Epand, R., and H. Vogel. 1999. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta. 1462:11–28. [DOI] [PubMed] [Google Scholar]
- 32.Tieleman, D. P., H. J. C. Berendsen, and M. S. P. Sansom. 1999. An alamethicin channel in a lipid bilayer: molecular dynamics simulations. Biophys. J. 76:1757–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenfeld, Y. 1990. Free-energy model for the inhomogeneous hard-sphere fluid in D dimensions: structure factors for the hard-disk (D = 2) mixtures in simple explicit form. Phys. Rev. A. 42:5978–5988. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, B. A., and D. M. Engelman. 1983. Bacteriorhodopsin remains dispersed in fluid phospholipid bilayers over a wide range of bilayer thicknesses. J. Mol. Biol. 166:203–210. [DOI] [PubMed] [Google Scholar]
- 35.Chen, Y. S., and W. L. Hubbell. 1973. Temperature- and light-dependent structural changes in rhodopsin-lipid membranes. Exp. Eye Res. 17:517–532. [DOI] [PubMed] [Google Scholar]
- 36.James, R., and D. Branton. 1973. Lipid- and temperature-dependent structural changes in Acholeplasma laidlawii cell membranes. Biochim. Biophys. Acta. 323:378–390. [DOI] [PubMed] [Google Scholar]
- 37.Abney, J. R., J. Braun, and J. C. Owicki. 1987. Lateral interactions among membrane proteins: implications for the organization of gap junctions. Biophys. J. 52:441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson, L. T., B. A. Lewis, D. M. Engelman, and S. I. Chan. 1983. Pair distribution functions of bacteriorhodopsin and rhodopsin in model bilayers. Biophys. J. 43:167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson, L. T., J. Edelman, and S. I. Chan. 1984. Statistical mechanics of lipid membranes, protein correlation functions and lipid ordering. Biophys. J. 45:863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun, J., J. R. Abney, and J. C. Owicki. 1987. Lateral interactions among membrane proteins: valid estimates based on freeze-fracture electron microscopy. Biophys. J. 52:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marčelja, S. 1976. Lipid-mediated protein interaction in membranes. Biochim. Biophys. Acta. 455:1–7. [DOI] [PubMed] [Google Scholar]
- 42.Owicki, J. C., M. W. Springgate, and H. M. McConnell. 1978. Theoretical study of protein-lipid interactions in bilayer membranes. Proc. Natl. Acad. Sci. USA. 75:1616–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owicki, J. C., and H. M. McConnell. 1979. Theory of protein-lipid and protein-protein interactions in bilayer membranes. Proc. Natl. Acad. Sci. USA. 76:4750–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kralchevsky, P. A. 1997. Lateral forces acting between particles in liquid films or lipid membranes. Adv. Biophys. 34:25–39. [DOI] [PubMed] [Google Scholar]
- 45.Huang, H. W. 1986. Deformation free energy of bilayer membrane and its effect on gramicidin channel lifetime. Biophys. J. 50:1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helfrich, P., and E. Jakobsson. 1990. Calculation of deformation energies and conformations in lipid membranes containing gramicidin channels. Biophys. J. 57:1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goulian, M., R. Bruinsma, and P. Pincus. 1993. Long-range forces in heterogeneous fluid membranes. Europhys. Lett. 22:145–150. [Google Scholar]
- 48.Aranda-Espinoza, H., A. Berman, N. Dan, P. Pincus, and S. Safran. 1996. Interaction between inclusions embedded in membranes. Biophys. J. 71:648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.May, S., and A. Ben-Shaul. 1999. Molecular theory of lipid-protein interaction and the Lα–HII transition. Biophys. J. 76:751–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mouritsen, O. G., and M. Bloom. 1984. Mattress model of lipid-protein interactions in membranes. Biophys. J. 46:141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Killian, J. 1998. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta. 1376:401–416. [DOI] [PubMed] [Google Scholar]
- 52.Lee, A. 2003. Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta. 1612:1–40. [DOI] [PubMed] [Google Scholar]
- 53.North, C. L., M. Barranger-Mathys, and D. S. Cafiso. 1995. Membrane orientation of the N-terminal segment of alamethicin determined by solid-state 15N NMR. Biophys. J. 69:2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bak, M., R. P. Bywater, M. Hohwy, J. K. Thomsen, K. Adelhorst, H. J. Jakobsen, O. W. Sørensen, and N. C. Nielsen. 2001. Conformation of alamethicin in oriented phospholipid bilayers determined by 15N solid-state nuclear magnetic resonance. Biophys. J. 81:1684–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kessel, A., D. S. Cafiso, and N. Ben-Tal. 2000. Continuum solvent model calculations of alamethicin-membrane interactions: thermodynamic aspects. Biophys. J. 78:571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fox, R. O., Jr., and F. M. Richards. 1982. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-Å resolution. Nature. 300:325–330. [DOI] [PubMed] [Google Scholar]
- 57.Breed, J., P. C. Biggin, I. D. Kerr, O. S. Smart, and M. S. P. Sansom. 1997. Alamethicin channels—modeling via restrained molecular dynamics simulations. Biochim. Biophys. Acta. 1325:235–249. [DOI] [PubMed] [Google Scholar]
- 58.Venturoli, M., B. Smit, and M. M. Sperotto. 2005. Simulation studies of protein-induced bilayer deformations, and lipid-induced protein tilting, on a mesoscopic model for lipid bilayers with embedded proteins. Biophys. J. 88:1778–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nielsen, C., M. Goulian, and O. S. Andersen. 1998. Energetics of inclusion-induced bilayer deformations. Biophys. J. 74:1966–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sintes, T., and A. Baumgärtner. 1997. Protein attraction in membranes induced by lipid fluctuations. Biophys. J. 73:2251–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.May, S., and A. Ben-Shaul. 2000. A molecular model for lipid-mediated interaction between proteins in membranes. Phys. Chem. Chem. Phys. 2:4494–4502. [Google Scholar]
- 62.Feller, S. E., R. M. Venable, and R. W. Pastor. 1997. Computer simulation of a DPPC phospholipid bilayer: structural changes as a function of molecular surface area. Langmuir. 13:6555–6561. [Google Scholar]
- 63.Marčelja, S. 1999. Toward a realistic theory of the interaction of membrane inclusions. Biophys. J. 76:593–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bohinc, K., V. Kralj-Igliè, and S. May. 2003. Interaction between two cylindrical inclusions in a symmetric lipid bilayer. J. Chem. Phys. 119:7435–7444. [Google Scholar]
- 65.Andersen, H. C., and D. Chandler. 1970. Mode expansion in equilibrium statistical mechanics. I. General theory and application to the classical electron gas. J. Chem. Phys. 53:547–554. [Google Scholar]
- 66.Hansen, J.-P., and I. R. McDonald. 1986. Theory of Simple Liquids, 2nd Ed. Academic Press, New York.