Abstract
To elucidate the structure of denatured proteins, we measured the vacuum-ultraviolet circular dichroism (VUVCD) spectra from 260 to 172 nm of three proteins (metmyoglobin, staphylococcal nuclease, and thioredoxin) in the native and the acid-, cold-, and heat-denatured states, using a synchrotron-radiation VUVCD spectrophotometer. The circular dichroism spectra of proteins fully unfolded by guanidine hydrochloride (GdnHCl) were also measured down to 197 nm for comparison. These denatured proteins exhibited characteristic VUVCD spectra that reflected a considerable amount of residual secondary structures. The contents of α-helices, β-strands, turns, poly-L-proline type II (PPII), and unordered structures were estimated for each denatured state of the three proteins using the SELCON3 program with Protein Data Bank data and the VUVCD spectra of 31 reference proteins reported in our previous study. Based on these contents, the characteristics of the four types of denaturation were discussed for each protein. In all types of denaturation, a decrease in α-helices was accompanied by increases in β-strands, PPII, and unordered structures. About 20% β-strands were present even in the proteins fully unfolded by GdnHCl in which β-sheets should be broken. From these results, we propose that denatured proteins constitute an ensemble of residual α-helices and β-sheets, partly unfolded (or distorted) α-helices and β-strands, PPII, and unordered structures.
INTRODUCTION
The structural characterization of denatured proteins is of fundamental importance to understanding the mechanisms of protein folding and stability (1). The free-energy difference between the native state (N-state) and denatured states is both small and sensitively dependent on the denaturing conditions, which lead to various denatured structures. High concentration of urea and guanidine hydrochloride (GdnHCl) generally yields the most complete unfolding. In contrast, an acid-denatured state (A-state) is less thoroughly unfolded and often resembles the molten globule or the initial and early folding intermediate structures (2–4). Cold and heat denaturation induce smaller conformational changes, but there is no definitive view about the resulting structures despite evident thermodynamic differences between these denatured states (5). One conclusion of recent studies is that denatured proteins contain residual secondary structures and that a polypeptide chain is not a true random coil even when urea and GdnHCl are used for the denaturation (6,7). This is also supported by the theoretical studies using alanine-based peptides (8,9). Such residual structures must play an important role in the energetics and kinetics of protein folding, and hence it is crucial to understand how the residual secondary structures differ between the denatured states.
The structures of denatured proteins have been extensively studied using various techniques, including differential scanning calorimetry (DSC) (10), circular dichroism (CD) (11), NMR (12,13), small-angle x-ray scattering (14,15), and Fourier transform infrared spectroscopy (FTIR) (16). NMR, FTIR, and CD can all be used to directly investigate the secondary structures, but CD spectroscopy is the most widely used because of its great advantage that the CD spectra are measurable for any protein at a low concentration under various solvent conditions, and because it is sensitive to the local secondary structures, even though unlike NMR, it does not provide atomic resolution. The secondary-structure analysis of native proteins by CD spectroscopy has been considerably improved by 1) the development of software programs (DSSP and Xtlsstr) that are capable of extracting the secondary structures from atomic coordinates (17,18), 2) advancements in the software (CONTIN, SELCON3, and CDSSTR) for analyzing the CD spectra (19–21), and 3) the extension of CD measurements to the vacuum ultraviolet (VUV) region (22). However, few studies have used CD spectroscopy to investigate the detailed secondary structures of denatured proteins. Venyaminov et al. estimated the secondary-structure contents of denatured proteins using a database of 20 reference proteins that included 4 denatured proteins (23). An expanded database with 37 reference proteins including 5 denatured proteins has been constructed by Sreerama et al. (24). These databases are useful for predicting the secondary structures of denatured proteins, but the technical difficulties limit the shortest wavelength of most CD spectra of denatured proteins to ∼185 nm.
The short-wavelength limit of CD spectroscopy can be extended by using a synchrotron as an excellent high-flux source of photons, yielding signal/noise ratios in the VUV region that cannot be attained by a conventional CD spectrophotometer (25–28). We recently constructed a vacuum-ultraviolet circular dichroism (VUVCD) spectrophotometer at the Hiroshima Synchrotron Radiation Center (HSRC) that is able to measure CD spectra down to 140 nm and −30°C by keeping all of the optical devices under a high vacuum (29,30). We successfully measured the VUVCD spectra of 31 globular proteins in the wavelength region from 260 to 160 nm using this spectrophotometer and showed the considerable improvement attainable in predictions of the contents and segment numbers of the secondary structures by extending CD spectra to the VUV region (31,32). In this study, this spectrophotometer was applied to measure the VUVCD spectra of three proteins—metmyoglobin (metMb), staphylococcal nuclease (SNase), and thioredoxin (Trx)—in the N- and A-states and the cold- and heat-denatured states (C- and H-states, respectively) down to 172 nm. The CD spectra of proteins fully unfolded by GdnHCl were also measured down to 197 nm for comparison. These proteins were adopted because their denaturation has been widely investigated using various techniques and under diverse conditions due to favorable structural aspects such as monomeric proteins and the absence of disulfide bonds (16,33,34). The characteristic differences between four types of denatured structures were revealed based on the estimated contents of α-helices, β-strands, turns, poly-L-proline type II (PPII), and unordered structures, which allow us to propose a generalized denatured structure.
MATERIALS AND METHODS
Materials
MetMb from horse heart and Trx (reduced form) from Escherichia coli were purchased from Sigma (St. Louis, MO). These proteins were used without further purification. SNase was overexpressed with E. coli and purified according to a previously reported method (35). Analytical-grade GdnHCl was obtained from Junsei Chemical (Tokyo, Japan), and all other chemicals were analytical-grade products obtained from Sigma. MetMb and SNase were dialyzed against double-distilled water at 4°C and adjusted to pH 3.9 and pH 3.8 with acetate acid, respectively, and to pH 2.0 with hydrochloric acid. Trx was dissolved in double-distilled water and adjusted to pH 1.8 with hydrochloric acid. The protein solutions thus obtained were centrifuged at 14,000 × g rpm for 15 min and filtered by a membrane filter (DISMIC 25AS020AS, ADVANTEC, Tokyo, Japan) to remove the aggregates before performing CD measurements. Protein concentrations were determined by absorption measurements (V-560, Jasco, Tokyo, Japan) using extinction coefficients of 17.9, 9.3, and 11.4 dL (g cm)−1 at 280 nm for metMb, SNase, and Trx, respectively (16,36,37).
CD measurements
The VUVCD spectra of proteins were measured from 260 to 172 nm under a high vacuum (10−4 Pa) using the VUVCD spectrophotometer constructed at the HSRC and an assembled optical cell (29,30). The temperature of the cell was controlled in the range from −15°C to 68°C using a Peltier temperature-control unit. The details of the optical devices of the spectrophotometer and the optical cell are available elsewhere (30), and their performances were assessed by monitoring the CD spectrum of ammonium (+)-camphor-10-sulfonate solution, which exhibits positive and negative peaks at 291 and 192 nm with a 1:2 intensity ratio, respectively. The path length of the cell was adjusted with a Teflon spacer to 50 and 25 μm. All of the VUVCD spectra were recorded at protein concentrations of 0.1%–0.3% with a 0.25-mm slit, a 16-s time constant, a 4-nm min−1 scan speed, and 4–9 accumulations. The spectra were calibrated by normalizing the ellipticity to those measured using a conventional CD spectrophotometer (J-720W, Jasco) in the overlapping wavelength region. The ellipticity was reproducible within an error of 5%, with this error being mainly attributable to noise and to inaccuracy in the length of the light path.
The CD spectra of denatured proteins in GdnHCl solution were measured from 260 to 197 nm using a conventional CD spectrophotometer at a protein concentration of 0.5%. These spectra were recorded with a 4-s time constant, a 50-nm min−1 scan speed, and 16 accumulations. A cell with a path length of 50 μm was used for the measurements from 260 to 210 nm, and no spacer was used for the measurements from 210 to 197 nm to reduce the light absorption by GdnHCl. The spectra obtained without the spacer were normalized to those measured using a 50-μm spacer in the overlapping wavelength region. The near-ultraviolet (UV) CD spectra of metMb were recorded with a 1-mm path length at a protein concentration of 0.3% from 320 to 250 nm. The other conditions were the same as those for the far-UV CD measurements.
Secondary-structure analysis of denatured proteins
The secondary structures of denatured proteins were analyzed using the improved SELCON3 program (31,38) and the VUVCD spectra of the following 31 reference proteins with known x-ray structures (32) (their Protein Data Bank (PDB) codes are in parentheses): metMb (1WLA), hemoglobin (1G08), human serum albumin (1E78), cytochrome c (1HRC), peroxidase (1ATJ), α-lactalbumin (1F6S), lysozyme (1HEL), ribonuclease A (1FS3), insulin (4INS), lactate dehydrogenase (9LDT), glucose isomerase (1OAD), lipase (3LIP), conalbumin (1OVT), transferrin (1LFG), catalase (7CAT), subtilisin A (1SBC), α-amylase (1BAG), papain (9PAP), ovalbumin (1OVA), β-lactoglobulin (1B8E), pepsin (4PEP), trypsinogen (1TGN), α-chymotrypsinogen (2CGA), soybean trypsin inhibitor (1AVU), concanavalin A (2CTV), SNase (1EY0), Trx (2TRX), carbonic anhydrase (1G6V), elastase (3EST), avidin (1AVE), and xylanase (1ENX). The secondary structures of these proteins in crystal form were assigned using the Xtlsstr program (18) based on two dihedral angles (ζ and τ) and three distances (two hydrogen bonds and one nonhydrogen bond) between adjacent amide groups. The 310-helices were grouped as α-helices and the nonhydrogen-bonded turns were treated as turns. PPII, which is grouped as an unordered structure in the DSSP program (17), was also assigned separately because it has been suggested that this structure is abundant in the denatured state (6,7). Thus the secondary structures of proteins were classified into five components: α-helices, β-strands, turns, PPII, and unordered structures.
RESULTS
The VUVCD spectra of metMb, Trx, and SNase were measured down to 172−175 nm for the N-state and the three denatured states (A-, C-, and H-states) and down to 197 nm for the GdnHCl-unfolded state (U-state). The spectra for the partially acid-denatured state (PA-state) were also measured for comparison. The solvent conditions inducing denaturation differed between the individual proteins. The temperature dependence of CD spectra showed that metMb (pH 3.9), SNase (pH 3.8), and Trx (pH 1.8) were in the H-state at 68°C, 45°C, and 68°C, respectively. It is known that metMb and SNase are in the A-state at pH 2.0 and in the U-state when 4 M GdnHCl is used (2,16,39) and that Trx is in the U-state when 6 M GdnHCl is used (40). All the VUVCD spectra obtained were superimposed on those in the far-UV region as measured by a conventional spectrophotometer. The VUVCD spectra were constant during the data-acquisition period (∼2 h) at all solvent conditions and temperatures, indicating that the synchrotron radiation (0.7 GeV) did not damage the proteins. This indicated that the obtained VUVCD spectra were suitable for studying the protein structures under various denaturing conditions.
VUVCD spectra of metMb
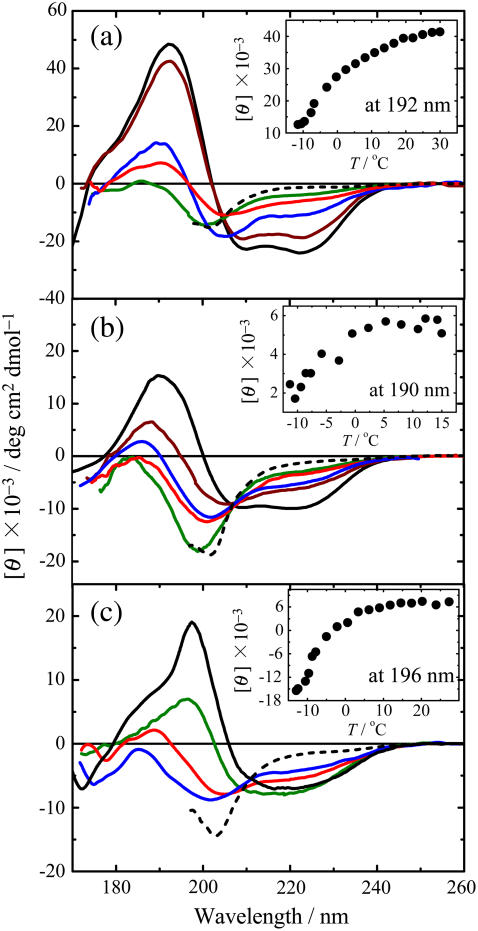
Fig. 1 a shows the VUVCD spectra of metMb at six conformational states: N (pH 5.4, 25°C), PA (pH 3.9, 30°C), A (pH 2.0, 30°C), H (pH 3.9, 68°C), C (pH 3.9, −12°C), and U (4 M GdnHCl, pH 2.0, 25°C). It is evident that these spectra differ significantly from each other, reflecting significant differences in the secondary structures of the denatured proteins. The N-state exhibits a typical spectrum for α-helix proteins, which is characterized by two negative peaks at around 222 and 208 nm, a positive peak at around 192 nm, and a shoulder at around 175 nm (31). The A-state shows two negative peaks at around 225, 205, and 180 nm and a small positive peak at around 185 nm. The H-state is characterized by a positive peak at around 191 nm and two negative peaks at around 205 and 176 nm. The plot of molar ellipticity at 192 nm against temperature in the inset of Fig. 1 a indicates that the ellipticity decreases with decreasing temperature, saturating at around −12°C. This indicates that metMb is susceptible to cold denaturation, with the spectrum at −12°C (Fig. 1 a) ascribable to the C-state. This spectrum has a positive peak at around 190 nm and two negative peaks at around 222 and 205 nm, suggesting the presence of a considerable amount of residual secondary structures in the C-state. The U-state exhibits no characteristic peak at around 220 nm, as expected for a largely unfolded conformation.
FIGURE 1.
VUVCD spectra of metMb (a), SNase (b), and Trx (c) in various conformational states: N (solid black line), H (red line), C (blue line), A (green line), PA (brown line), and U (dotted black line). Inset shows the temperature dependence of the molar ellipticity at 192, 190, and 196 nm for metMb, SNase, and Trx, respectively. The experimental conditions are detailed in the Materials and Methods.
The VUVCD spectra for the N-, PA-, H-, and A-states show an isoellipticity point at 204 nm. On the other hand, the C-state appears to exhibit a different isoellipticity point with the N-state, at 206 nm. This was also the case for the partially cold-denatured state at 0°C (data not shown). These results suggest that the mechanism of cold denaturation differs from those of the other denaturation. There also appears to be other isoellipticity points at 170−175 nm, although they are not clearly discernible due to the short-wavelength limit of CD measurements.
VUVCD spectra of SNase
Fig. 1 b shows the VUVCD spectra of SNase in the N- (pH 6.5, 25°C), PA- (pH 3.8, 15°C), H- (pH 3.8, 45°C), C- (pH 3.8, −12°C), A- (pH 2.0, 15°C), and U- (4 M GdnHCl, pH 2.0, 25°C) states. The N-state exhibits a positive peak at around 190 nm and two negative peaks at around 222 and 208 nm, reflecting a considerable number of α-helices. The H-state is characterized by a small positive peak at around 185 nm and two negative peaks at 225 and 200 nm. The temperature dependence of the ellipticity at 190 nm (inset of Fig. 1 b) appears to indicate that SNase is not completely cold denatured at −12°C. However, we can expect that the CD spectrum at −12°C would correspond to the C-state of this protein, judging from the results of CD and DSC measurements (5) and from the temperature dependence of the excess heat capacity calculated for globular proteins with different stabilities (10). The similarity of the spectra observed for the H- and C-states suggests that the conformations would not be largely different at the two denatured states. The A- and U-states also exhibit similar spectra, as found for metMb. All these denatured states including the PA-state exhibit an isoellipticity point at around 208 nm, suggesting that different types of denaturation follow the same two-state transition mechanism.
VUVCD spectra of Trx
Fig. 1 c shows the VUVCD spectra of Trx in the N- (pH 5.8, 25°C), A- (pH 1.8, 25°C), H- (pH 1.8, 68°C), C- (pH 1.8, −9°C), and U- (6 M GdnHCl, pH 2.0, 25°C) states. This spectrum for the N-state of this protein differs greatly from those of other globular proteins containing comparable amounts of secondary structures (32): the crossover point is significantly red shifted to 207 nm, and a broad negative peak has a shoulder at around 230 nm. This abnormality may be partly due to exciton coupling between the aromatic residues (41,42), since Tyr-70 is located at the closest ring-carbon distances from three surrounding phenylalanines (Phe-12, 4.2 Å; Phe-27, 5.0 Å; and Phe-81, 3.9 Å), which are mutually within 5.4 Å. The spectrum for the A-state (pH 1.8) is also very different from those for metMb and SNase: it has a positive peak at around 198 nm, whereas both metMb and SNase have a negative peak. The same spectrum was observed at pH 0.5 (data not shown), confirming the complete acid denaturation of Trx at pH 1.8. The crossover at around 230 nm between the spectra of the A- and N-states may be attributable to exciton coupling between aromatic residues broken by acid denaturation. The H-state is characterized by a positive peak at around 189 nm and two negative peaks at around 225 and 205 nm. A similar spectrum was observed at −9°C, where the cold denaturation of Trx may not be complete as suggested by the temperature dependence of the ellipticity at 196 nm (inset of Fig. 1 c). This represents the first evidence for cold denaturation of E. coli Trx, although Trx h from Chlamydomonas reinhardtii has previously been shown to be susceptible to cold denaturation (43). The smaller CD intensity for the C-state suggests that the secondary structures of Trx are more extensively broken at the C-state than at the H-state, in contrast to the situation for metMb and SNase. No isoellipticity point was observed among the spectra of the A-, C-, H-, and U-states, indicating that the unfolding mechanism differs with the type of denaturation.
Secondary-structure analysis of denatured proteins
Fig. 1 indicates that the VUVCD spectra of the three proteins were successfully measured down to 172 nm under various denaturing conditions. This extension of the short-wavelength limit in CD measurements yields much more information on the secondary structures and allows us to estimate quantitatively the contents of the various secondary-structure components (such as α-helices and β-strands) in the denatured proteins. In this study, we took into consideration the contribution of PPII, a left-handed threefold helix (44) because the short stretches of PPII helical conformation exist in unordered polypeptides such as poly-L-lysine and poly-L-glutamic acid at pH 7.0 (45). Thus the spectra of five secondary-structure components—α-helices, β-strands, turns, PPII, and unordered structures—were determined by deconvolution analysis of the VUVCD spectra of 31 reference proteins down to 160 nm (31,32) using the SELCON3 program (38) and their PDB data. The VUVCD spectra of the five components averaged across the 31 proteins are shown in Fig. 2. These five component spectra were revealed for the first time down to 172 nm in this study, although the four component spectra except for PPII have been obtained down to 160 nm using the DSSP program (32). The PPII spectrum appears similar to the spectrum for the unordered structure in the four component spectra (32) because PPII is assigned as the unordered structure in the DSSP program.
FIGURE 2.
Component VUVCD spectra of five secondary structures deconvoluted from 31 reference protein spectra (31,32): α-helices (thick), β-strands (dot-dashed line), turns (thin line), PPII (dashed line), and unordered structures (dotted line).
Based on the spectra of these components, the secondary-structure contents in the denatured proteins were estimated using the SELCON3 program coupled with the VUVCD spectra down to 172 nm for the N-, A-, C-, and H-states, and the far-UV CD spectra down to 197 nm for the U-state (Fig. 1). In this analysis, the temperature dependence of the CD spectra for the C- and H-states was not considered because the intrinsic temperature dependence of chromophores is generally difficult to evaluate and it does not appear to cause significant errors in secondary-structure estimation. Nölting et al. suggested that the temperature dependence of CD would mainly arise from changes in the relative populations of the states with similar free energy but different conformations (46). Then the secondary-structure contents estimated for the H- and C-states might be regarded as the average ensemble of conformations with small energy differences. Since the cold denaturation of SNase and Trx is not complete at −12°C and −9°C, respectively, the contents of α-helices, β-strands, and turns might be slightly overestimated for the C-states of both of these proteins.
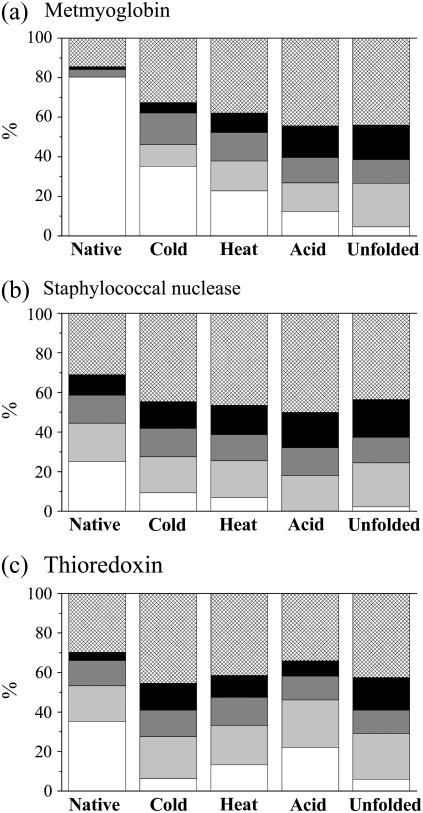
The secondary-structure contents estimated for the three proteins in the N-, A-, C-, H-, and U-states are listed in Table 1. The numbers within parentheses in this table are the secondary-structure contents of native proteins assigned by the Xtlsstr method from the x-ray structures. The root mean-square deviation (δ) and the Pearson correlation coefficient (r) between the x-ray and VUVCD estimates of the secondary-structure contents were 0.058 and 0.92, respectively, indicating the accuracy of the VUVCD estimation. For ease of comparison, the results in Table 1 are shown as proportional histograms of the secondary-structure contents in Fig. 3, in which each secondary-structure content is normalized to a total content of 100% and the histogram for the N-state is depicted with the results from the Xtlsstr method (i.e., the values within parentheses in Table 1).
TABLE 1.
Secondary-structure contents of metMb, SNase, and Trx in the native and four denatured states as determined by CD spectroscopy
| Contents (%) of
|
||||||
|---|---|---|---|---|---|---|
| State | α-Helix | β-Strand | Turn | PPII | Unordered | |
| Metmyoglobin | ||||||
| Native (pH 5.4, 25°C) | 79.8 ± 0.9 (80.3)† | −2.2 ± 0.6 (0.0) | 9.3 ± 1.2 (3.9) | 1.2 ± 0.1 (1.3) | 15.9 ± 0.5 (14.5) | |
| Denatured | ||||||
| Cold | (pH 3.9, −12°C) | 36.7 ± 0.3 | 11.6 ± 0.4 | 16.6 ± 0.2 | 5.5 ± 0.1 | 34.0 ± 0.4 |
| Heat | (pH 3.9, 68°C) | 23.2 ± 1.2 | 15.4 ± 0.7 | 14.8 ± 0.6 | 10.0 ± 1.0 | 38.6 ± 1.1 |
| Acid | (pH 2.0, 30°C) | 12.0 ± 1.1 | 14.3 ± 1.5 | 12.6 ± 1.1 | 15.7 ± 0.7 | 43.6 ± 1.0 |
| Unfolded | (4 M GdnHCl)* | 4.4 ± 1.8 | 21.5 ± 0.9 | 11.7 ± 0.9 | 17.0 ± 0.6 | 42.8 ± 1.6 |
| Staphylococcal nuclease | ||||||
| Native (pH 6.5, 25°C) | 30.6 ± 0.9 (25.2)† | 15.3 ± 1.7 (19.3) | 13.2 ± 0.5 (14.1) | 7.5 ± 0.4 (10.4) | 34.7 ± 1.3 (31.0) | |
| Denatured | ||||||
| Cold | (pH 3.8, −12°C) | 9.6 ± 1.0 | 18.6 ± 1.1 | 14.6 ±0.5 | 13.8 ± 0.6 | 45.7 ± 1.3 |
| Heat | (pH 3.8, 45°C) | 7.0 ± 1.1 | 18.2 ±2.0 | 13.1 ± 0.5 | 14.6 ± 0.9 | 45.9 ± 1.6 |
| Acid | (pH 2.0, 15°C) | −0.1 ± 1.4 | 18.1 ± 1.9 | 14.3 ± 0.7 | 17.8 ± 0.8 | 50.5 ± 1.3 |
| Unfolded | (4 M GdnHCl)* | 2.2 ± 0.4 | 22.0 ± 0.6 | 12.8 ± 0.3 | 18.8 ± 0.5 | 43.1 ± 1.1 |
| Thioredoxin | ||||||
| Native (pH 5.8, 25°C) | 31.1 ± 0.9 (35.3)† | 12.8 ± 1.9 (18.0) | 15.8 ± 0.7 (12.8) | 4.3 ± 1.1 (4.2) | 36.0 ± 1.5 (29.7) | |
| Denatured | ||||||
| Cold | (pH 1.8, −9°C) | 9.0 ± 1.7 | 18.8 ± 3.0 | 11.6 ± 0.9 | 14.6 ± 0.6 | 46.2 ± 1.3 |
| Heat | (pH 1.8, 68°C) | 14.9 ± 1.3 | 18.2 ± 2.2 | 12.6 ± 0.8 | 12.2 ± 0.6 | 41.6 ± 1.4 |
| Acid | (pH 1.8, 25°C) | 25.1 ± 2.0 | 17.3 ± 0.9 | 11.4 ± 1.3 | 9.3 ± 0.7 | 32.3 ± 0.4 |
| Unfolded | (6 M GdnHCl)* | 6.0 ± 0.8 | 22.9 ± 1.5 | 11.7 ± 0.7 | 16.3 ± 0.5 | 42.1 ± 1.7 |
pH 2.0, 25°C.
Values in parentheses are secondary-structure contents from x-ray analysis.
FIGURE 3.
Histogram of the secondary-structure contents of native and denatured metMb (a), SNase (b), and Trx (c): α-helices (white rectangle), β-strands (light gray rectangle), turns (dark gray rectangle), PPII (black rectangle), and unordered structures (hatched rectangle).
DISCUSSION
These VUVCD spectroscopy experiments have clearly revealed that the investigated denatured proteins contain many residual structures. The results of secondary-structure analyses (Table 1 and Fig. 3) quantitatively demonstrate characteristic differences in the secondary structures of the three proteins at four denatured states. The characteristic structures of the denatured proteins are useful for deriving the general features of protein folding and stability.
Conformation of denatured myoglobin
Myoglobin is a typical helix protein (80% helix content) consisting of eight α-helix segments A−H (33). Its stability and folding mechanism have been widely investigated using various techniques, including NMR, FTIR, DSC, and CD (10,11,33,47), but the denatured structures remain undefined. As indicated in Table 1 and Fig. 3 a, the secondary structures of metMb vary considerably with the type of denaturation. The α-helix content decreases in the order N-state > C-state > H-state > A-state > U-state, whereas the contents of PPII and β-strands increase in the same order.
As indicated in Table 1, the C-state of metMb contains a large number of α-helices (36.7%) and β-strands (11.6%). From an FTIR study on hydrogen exchange, Meersman et al. also suggested that metMb is only partially unfolded by cold denaturation under high pressure, with a rigid core consisting of G and H helices remaining folded (47). However, the helix contents from these two helices amount to only 28.7%, suggesting the presence of additional helix segments at atmospheric pressure.
The H-state has a helix content of 23.2%, which is considerably lower than the percentage of 36.7% for the C-state (Table 1), indicating that the conformational disruption is greater for heat denaturation than for cold denaturation. The heat denaturation was irreversible as revealed by incomplete recovery of the VUVCD spectra when the samples were cooled, then part of the β-strands produced (15.4% at 68°C) may be transformed into intermolecular β-sheets. This β-strand content is much smaller than the FTIR estimate (27% at 90°C) by Meersman et al. (47) in which aggregation would dominate due to the high protein concentration used. On the other hand, the cold denaturation was completely reversible, and no aggregation was observed despite the production of β-strands. Therefore, the formation of β-strands on denaturation is essentially an intramolecular event, and their transformation into β-sheets would depend on the experimental conditions. In this respect, heat- and cold-denaturation processes do not follow the same pathway, as expected from the different isoellipticity points in Fig. 1 a.
Acid denaturation of metMb greatly decreases the helix content to 12.0% and induces a β-strand content of 14.3% (Table 1). Similar results have been observed for A- or molten globule states of apoMb using far-UV CD spectroscopy (2,4,48,50). However, NMR experiments have not provided evidence for β-strands in apoMb at pH 2.3 (51), probably because the fluctuations of the denatured protein are much faster than the detection time of signals associated with the nuclear Overhauser effect. On the other hand, the radius of gyration of acid-denatured apoMb is close to that observed for the urea- or GdnHCl-unfolded form (4,52), indicating that the backbone chain obeys the random-coil statistics (53). These results suggest that the β-strands in the acid-denatured protein do not exist as β-sheets but rather as an ensemble of many short peptide segments with the dihedral angles of β-strands.
The U-state of metMb contains 17.0% PPII, 4.4% helices, and 21.5% β-strands. It is noteworthy that the contents of PPII and β-strands increase with the extent of unfolding. Whittington et al. also observed that the PPII content in urea-unfolded apoMb increases with the urea concentration (7). These findings suggest that the increased backbone flexibility could cause the formation of PPII and β-strands, with the resulting β-strands not forming β-sheets but instead consisting of local structures with the dihedral angles of β-strands, as in the A-state. Thus the U- and A-states of metMb could be regarded as an ensemble of many short peptide segments with the dihedral angles of α-helices, β-strands, and PPII and including some turn structures.
Conformation of denatured SNase
SNase is a monomeric protein that is widely used as a model protein in folding studies and consists of 149 residues without disulfide bonds, with three α-helices (25.2%) and five β-strands (19.3%). As indicated in Table 1 and Fig. 3 b, the helix content decreases in the order N-state > C-state > H-state > A-state, whereas the contents of PPII and unordered structures increase in the same order. However, no significant difference is observed in the overall features of the secondary structures between the C- and H-states, as expected from the CD spectra in Fig. 1 b. Griko et al. also suggested that heat and cold denaturation of SNase lead to an almost complete loss of helicity and that both denaturation processes are approximated by a two-state transition with a single cooperative system (5). These results suggest that the C- and H-states of this protein have a similar structure.
The A-state shows completely disrupted α-helices and an amount of β-strands (18.1%) that is comparable with those in the C- and H-states. Similar amounts of α-helices (2.2%) and β-strands (22%) are present in the U-state, indicating that the A- and U-states comprise similar structures. The existence of β-strands in the U-state has also been observed by other groups. Wang and Shortle (3) suggested that a fragment of SNase with 131 amino acid residues contains ∼10% β-strands when using 4 M urea, and helices α1 and α2 decrease to undetectable and 10%–15% when using 6 M urea, respectively. The FTIR study of From and Bowler revealed the presence of 10.7% and 7.6% residual β-strands when using 4 and 6 M urea, respectively, as well as other components not attributable to random coils (16).
As shown above, the amounts of β-strands in all the denatured states are similar to that in the N-state. However, this does not necessarily mean that the β-sheet structure in the denatured states is the same as that in the N-state, as expected from the large difference in the VUVCD spectra shown in Fig. 1 b. Depending on the degree of increase in backbone flexibility, each denatured state could take a different ensemble of many short peptide segments with the dihedral angles of β-strands, α-helices, and PPII. This ensemble mode would differ from that for myoglobin because SNase contains a considerable amount of β-strands in the N-state, and they may remain partly folded in the denatured form, thereby restricting the flexibility of the peptide chain. From and Bowler reported that a core region of β-strands is folded in the earliest structural events in SNase (16), indicating that the denatured states of SNase might maintain a structure resembling the folding intermediate state.
Conformation of denatured Trx
Trx is a monomeric protein consisting of a five-stranded twisted β-sheet surrounded by four α-helices (35.3% α-helices and 18.0% β-strands). Its hydrophobic core confers high stability against chemical and thermal denaturation (40). As indicated in Table 1 and Fig. 3 c, in the H-state it conserves a considerable amount of α-helices (14.9%) and a similar amount of β-strands (18.2%). This result quantitatively confirms the suggestion of Maier et al. that thermally denatured Trx can be described as a thermodynamically stable intermediate with properties associated with molten globule-like states, such as a pronounced secondary structure and the absence of a rigid tertiary structure (34). The β-strand content (18.8%) in the C-state is the same as that in the H-state, whereas the α-helices are decreased to 9.0%. Since the cold denaturation of Trx is not complete at −9°C (inset of Fig. 1 c), the helix content in a completely cold-denatured state would be smaller than 9.0%. These results indicate that Trx is more significantly unfolded at low temperature than at high temperature, which is opposite to the observations for metMb and SNase. This is probably because Trx is a highly hydrophobic protein and the cold denaturation is mainly due to a weakened hydrophobic interaction at low temperature. Langsetmo et al. showed that this protein folds into two compact forms with the cis or trans isomers of Pro-76, whose differences in stability are more pronounced at 2°C than at 25°C (54). Richardson et al. suggested that two thermodynamic domains in Trx h from C. reinhardtii interact strongly (high cooperativeness) at high temperature, with the interaction being weakened at low temperature, leading to a two-state transition for heat denaturation and a non-two-state transition for cold denaturation (43). Thus, different mechanisms may underlie cold and heat denaturation.
The A-state (pH 1.8) contains large amounts of residual α-helices (25.1%) and β-strands (17.3%), which are comparable to the percentages in the N-state. This suggests that Trx is not largely unfolded in the acidic condition, in contrast to metMb and SNase. The amounts of the secondary structures in the U-state of Trx are comparable to those found in metMb and SNase, but its overall structure is close to that in the C-state, whereas the U-states of metMb and SNase are close to their A-states. These results indicate that the conformation of denatured states depends on the structural characteristics of the native proteins, such as the secondary-structure contents and the presence of a hydrophobic core.
Generalized features of denatured conformations
This study has revealed characteristic differences between the secondary structures of the three investigated proteins induced by four types of denaturation. It should be noted that α-helices and β-strands are assigned with two dihedral angles and three distances between adjacent amide groups (Xtlsstr program) but not with the hydrogen bonds between peptide groups (DSSP program). Within these limitations, the results allow us to derive general features for the conformation of denatured proteins, although the extent of unfolding depends on four types of denaturation: 1) α-helices are broken, 2) β-sheets are broken but β-strands increase, 3) turns are almost unchanged, and 4) PPII and unordered structures increase. It should be emphasized that even denaturation by urea and GdnHCl results in many residual secondary structures in the proteins. The most common feature of denatured proteins is a loss of α-helices, as has also been revealed by other techniques (3,16,51). However, this does not necessarily mean that a particular α-helix in the native protein is completely unfolded by an all-or-none mechanism—some α-helices may remain intact, whereas others are partly unfolded or distorted in the denatured state. An important finding from our VUVCD spectroscopy is that many β-strands are formed in all the denatured proteins, as exemplified by the metMb results (Fig. 3 a).
However, there is no direct evidence of β-sheets in the denatured apoMb from NMR measurements (51). X-ray solution analyses show the largely extended conformation of denatured proteins (4,52). Therefore, most of the β-strands in denatured proteins, locally possessing the dihedral angles of β-strands, would not form a sheet structure with interpeptide hydrogen bonds, although some β-sheets may remain intact in the denatured state of proteins that are rich in β-structures, such as SNase and Trx. From these considerations, we may speculate that the conformation of denatured proteins is an ensemble of residual α-helices and β-sheets, partly unfolded (or distorted) α-helices and β-strands, PPII, and unordered structures. The populations and positions of these elements would vary with the denaturing conditions (e.g., temperature, pressure, and chemical reagents) and the type of proteins (e.g., amino acid sequence and presence of secondary structures, disulfide bonds, and a hydrophobic core). The β-strands could form intermolecular β-sheets under appropriate conditions, which cause aggregation or amyloid formation (55). In general, some of the denatured states are close to the kinetically observed intermediates (molten globule or burst phase) of protein folding (47,48,56). However, our data are clearly inconsistent with a completely unfolded random coil assumed as a starting conformation for the folding process.
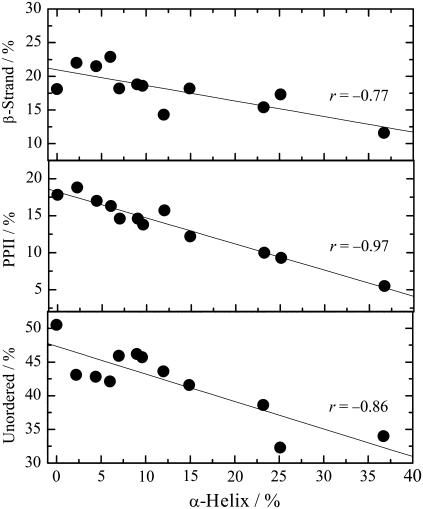
Any correlation between the constituent elements may be useful for understanding the denatured structure and denaturation mechanism of proteins. Fig. 4 plots the percentages of β-strands, PPII, and unordered structures against the α-helix content for all the denatured forms of the three proteins examined. The observed negative slopes confirm that the unfolding of α-helices results in the formation of β-strands, PPII, and unordered structures, with the unfolded residues in α-helices appearing to be almost equally assigned to the other three elements. The increase in unordered structures with decreasing α-helices is reasonable, but the reason for the increases in β-strands and PPII is unclear. The most likely explanation is that extended conformation such as β-strands and PPII are more favorable in the denatured states where intramolecular interactions (particularly hydrogen bonds) are disfavored. A very high correlation between the contents of α-helices and PPII (r = −0.97) suggests that the PPII formed upon denaturation is mainly located in the unfolded α-helix region. Further characterization of β-strands is required to elucidate the conformation of the denatured proteins.
FIGURE 4.
Plots of the β-strands, PPII, and unordered structures contents against α-helix content for the A-, C-, H-, and U-states of metMb, SNase, and Trx.
At present, it appears to be difficult to experimentally detect the ensemble structure of scattered elements using other techniques: NMR is unsuitable for a structure subject to rapid fluctuations, FTIR would require a much higher resolution for assigning the amide bands, and x-ray solution scattering is not sensitive to the fine structure of denatured proteins, although the persistence length and the radius of gyration may be simulated by modeling the composition of the structural elements. An eventual goal of our VUVCD study is to predict not only the contents' and segments' number of the secondary structures, but also their positions on the amino acid sequence. A VUVCD spectroscopy combined with a neural network analysis in progress would provide new insight into the fine structures of both denatured and native proteins.
Acknowledgments
We thank Professors Hirofumi Namatame and Masaki Taniguchi at the HSRC for their technical support of our use of the VUVCD spectrophotometer.
This work was financially supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan (No. 16350088 to K.G.) and by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (No. 15008237 to K.M.).
References
- 1.Dill, K. A., and D. Shortle. 1991. Denatured states of proteins. Annu. Rev. Biochem. 60:795–825. [DOI] [PubMed] [Google Scholar]
- 2.Irace, G., E. Bismuto, F. Savy, and G. Colonna. 1986. Unfolding pathway of myoglobin: molecular properties of intermediate forms. Arch. Biochem. Biophys. 244:459–469. [DOI] [PubMed] [Google Scholar]
- 3.Wang, Y., and D. Shortle. 1995. The equilibrium folding pathway of staphylococcal nuclease: identification of the most stable chain-chain interactions by NMR and CD spectroscopy. Biochemistry. 34:15895–15905. [DOI] [PubMed] [Google Scholar]
- 4.Nishii, I., M. Kataoka, F. Tokunaga, and Y. Goto. 1994. Cold denaturation of the molten globule states of apomyoglobin and a profile for protein folding. Biochemistry. 33:4903–4909. [DOI] [PubMed] [Google Scholar]
- 5.Griko, Y. V., P. L. Privalov, J. M. Sturtevant, and S. Yu. Venyaminov. 1988. Cold denaturation of staphylococcal nuclease. Proc. Natl. Acad. Sci. USA. 85:3343–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi, Z., R. W. Woody, and N. R. Kallenbach. 2002. Is polyproline II a major backbone conformation in unfolded proteins? Adv. Protein Chem. 62:163–240. [DOI] [PubMed] [Google Scholar]
- 7.Whittington, S. J., B. W. Chellgren, V. M. Hermann, and T. P. Creamer. 2005. Urea promotes polyproline II helix formation: implications for protein denatured states. Biochemistry. 44:6269–6275. [DOI] [PubMed] [Google Scholar]
- 8.Pappu, R. V., and G. D. Rose. 2002. A simple model for polyproline II structure in unfolded states of alanine-based peptides. Protein Sci. 11:2437–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drozdov, A. N., A. Grossfield, and R. V. Pappu. 2004. Role of solvent in determining conformational preferences of alanine dipeptide in water. J. Am. Chem. Soc. 126:2574–2581. [DOI] [PubMed] [Google Scholar]
- 10.Privalov, P. L. 1990. Cold denaturation of proteins. Crit. Rev. Biochem. Mol. Biol. 25:281–305. [DOI] [PubMed] [Google Scholar]
- 11.Goto, Y., N. Takahashi, and A. Fink. 1990. Mechanism of acid-induced folding of proteins. Biochemistry. 29:3480–3488. [DOI] [PubMed] [Google Scholar]
- 12.Jonas, J., L. Ballard, and D. Nash. 1998. High-resolution, high-pressure NMR studies of proteins. Biophys. J. 75:445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura, A., K. Kimura, and K. Akasaka. 1991. Cold denaturation and heat denaturation of Streptomyces subtilisin inhibitor. 2. 1H NMR studies. Biochemistry. 30:11313–11320. [DOI] [PubMed] [Google Scholar]
- 14.Panick, G., J. A. Vidugiris, R. Malessa, G. Rapp, R. Winter, and C. A. Royer. 1999. Exploring the temperature-pressure phase diagram of staphylococcal nuclease. Biochemistry. 38:4157–4164. [DOI] [PubMed] [Google Scholar]
- 15.Konno, T., M. Kataoka, Y. Kamatari, K. Kanaori, A. Nosaka, and K. Akasaka. 1995. Solution x-ray scattering analysis of cold-, heat-, and urea-denatured states in a protein, Streptomyces subtilisin inhibitor. J. Mol. Biol. 251:95–103. [DOI] [PubMed] [Google Scholar]
- 16.From, N. B., and B. E. Bowler. 1998. Urea denaturation of staphylococcal nuclease monitored by Fourier transform infrared spectroscopy. Biochemistry. 37:1623–1631. [DOI] [PubMed] [Google Scholar]
- 17.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 18.King, S. M., and W. C. Johnson. 1999. Assigning secondary structure from protein coordinate data. Proteins. 35:313–320. [PubMed] [Google Scholar]
- 19.Sreerama, N., and R. W. Woody. 2000. Circular dichroism of peptides and proteins. In Circular Dichroism: Principles and Applications, 2nd ed. N. Berova, K. Nakanishi, and R. W. Woody, editors. Wiley-VCH Press, New York. 55–95.
- 20.Venyaminov, S. Yu., and K. S. Vassilenko. 1994. Determination of protein tertiary structure class from circular dichroism spectra. Anal. Biochem. 222:176–184. [DOI] [PubMed] [Google Scholar]
- 21.Manavalan, P., and W. C. Johnson Jr. 1983. Sensitivity of circular dichroism to protein tertiary structure class. Nature. 305:831–832. [Google Scholar]
- 22.Toumadje, A., S. W. Alcorn, and W. C. Johnson Jr. 1992. Extending CD spectra of proteins to 168 nm improves the analysis for secondary structure. Anal. Biochem. 200:321–331. [DOI] [PubMed] [Google Scholar]
- 23.Venyaminov, S. Yu., I. A. Baikalov, Z. M. Shen, C. S. C. Wu, and J. T. Yang. 1993. Circular dichroic analysis of denatured proteins: inclusion of denatured proteins in the reference set. Anal. Biochem. 214:17–24. [DOI] [PubMed] [Google Scholar]
- 24.Sreerama, N., S. Y. Venyaminov, and R. W. Woody. 2000. Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 287:243–251. [DOI] [PubMed] [Google Scholar]
- 25.Snyder, P. A., and E. M. Rowe. 1980. The first use of synchrotron radiation for vacuum ultraviolet circular dichroism measurements. Nucl. Instrum. Methods Phys. Res. A. 172:345–349. [Google Scholar]
- 26.Sutherland, J. C., A. Emrick, L. L. France, D. C. Monteleone, and J. Trunk. 1992. Circular dichroism user facility at the National Synchrotron Light Source: estimation of protein secondary structure. Biotechniques. 13:588–590. [PubMed] [Google Scholar]
- 27.Jones, G. R., and D. T. Clarke. 2004. Applications of extended ultra-violet circular dichroism spectroscopy in biology and medicine. Faraday Discuss. 126:223–236. [DOI] [PubMed] [Google Scholar]
- 28.Wallace, B. A., F. Wien, A. J. Miles, J. G. Lees, S. V. Hoffmann, P. Evans, G. J. Wistow, and C. Slingsby. 2004. Biomedical applications of synchrotron radiation circular dichroism spectroscopy: identification of mutant proteins associated with disease and development of a reference database for fold motifs. Faraday Discuss. 126:237–243. [DOI] [PubMed] [Google Scholar]
- 29.Ojima, N., K. Sakai, K. Matsuo, T. Matsui, T. Fukazawa, H. Namatame, M. Taniguchi, and K. Gekko. 2001. Vacuum-ultraviolet circular dichroism spectrophotometer using synchrotron radiation: optical system and on-line performance. Chem. Lett. (Jpn.). 30:522–523. [Google Scholar]
- 30.Matsuo, K., K. Sakai, Y. Matsushima, T. Fukuyama, and K. Gekko. 2003. Optical cell with a temperature-control unit for a vacuum-ultraviolet circular dichroism spectrophotometer. Anal. Sci. 19:129–132. [DOI] [PubMed] [Google Scholar]
- 31.Matsuo, K., R. Yonehara, and K. Gekko. 2004. Secondary-structure analysis of proteins by vacuum-ultraviolet circular dichroism spectroscopy. J. Biochem. (Tokyo). 135:405–411. [DOI] [PubMed] [Google Scholar]
- 32.Matsuo, K., R. Yonehara, and K. Gekko. 2005. Improved estimation of the secondary structures of proteins by vacuum-ultraviolet circular dichroism spectroscopy. J. Biochem. (Tokyo). 138:79–88. [DOI] [PubMed] [Google Scholar]
- 33.Jennings, P. A., and P. E. Wright. 1993. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 262:892–896. [DOI] [PubMed] [Google Scholar]
- 34.Maier, C. S., M. I. Schimerlik, and M. L. Deinzer. 1999. Thermal denaturation of Escherichia coli thioredoxin studied by hydrogen/deuterium exchange and electrospray ionization mass spectrometry: monitoring a two-state protein unfolding transition. Biochemistry. 38:1136–1143. [DOI] [PubMed] [Google Scholar]
- 35.Shortle, D., and A. K. Meeker. 1989. Residual structure in large fragments of staphylococcal nuclease: effects of amino acid substitutions. Biochemistry. 28:936–944. [DOI] [PubMed] [Google Scholar]
- 36.Crumpton, M. J., and A. Polson. 1965. A comparison of the conformation of sperm whale metmyoglobin with that of apomyoglobin. J. Mol. Biol. 11:722–729. [DOI] [PubMed] [Google Scholar]
- 37.Holmgren, A., and P. Reichard. 1967. Thioredoxin 2: cleavage with cyanogen bromide. Eur. J. Biochem. 2:187–196. [DOI] [PubMed] [Google Scholar]
- 38.Sreerama, N., and R. W. Woody. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287:252–260. [DOI] [PubMed] [Google Scholar]
- 39.Fink, A. L., L. J. Calciano, Y. Goto, M. Nishimura, and S. A. Swedberg. 1993. Characterization of the stable, acid-induced, molten globule-like state of staphylococcal nuclease. Protein Sci. 2:1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santoro, M. M., and D. W. Bolen. 1992. A test of the linear extrapolation of unfolding free energy changes over an extended denaturant concentration range. Biochemistry. 31:4901–4907. [DOI] [PubMed] [Google Scholar]
- 41.Koslowski, A., N. Sreerama, and R. W. Woody. 2000. Theoretical approach to electronic optical activity. In Circular Dichroism: Principles and Applications, 2nd ed. N. Berova, K. Nakanishi, and R. W. Woody, editors. Wiley-VCH Press, New York. 55–95.
- 42.Sreerama, N., and R. W. Woody. 2004. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 383:318–351. [DOI] [PubMed] [Google Scholar]
- 43.Richardson III, J. M., S. D. Lemaire, J. P. Jacquot, and G. I. Makhatadze. 2000. Difference in the mechanisms of the cold and heat induced unfolding of thioredoxin h from Chlamydomonas reinhardtii: spectroscopic and calorimetric studies. Biochemistry. 39:11154–11162. [DOI] [PubMed] [Google Scholar]
- 44.Young, M. A., and E. S. Pysh. 1975. Vacuum ultraviolet circular dichroism of poly(L-proline) I and II. J. Am. Chem. Soc. 97:5100–5103. [DOI] [PubMed] [Google Scholar]
- 45.Woody, R. W. 1992. Circular dichroism and conformation of unordered peptides. Adv. Biophys. Chem. 2:77–79. [Google Scholar]
- 46.Nölting, B., R. Golbik, A. S. Soler-Gonzalez, and A. R. Fersht. 1997. Circular dichroism of denatured barstar suggests residual structure. Biochemistry. 36:9899–9905. [DOI] [PubMed] [Google Scholar]
- 47.Meersman, F., L. Smeller, and K. Heremans. 2002. Comparative Fourier transform infrared spectroscopy study of cold-, pressure-, and heat-induced unfolding and aggregation of myoglobin. Biophys. J. 82:2635–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilmanshin, R., M. Gulotta, R. B. Dyer, and R. H. Callender. 2001. Structures of apomyoglobin's various acid-destabilized forms. Biochemistry. 40:5127–5136. [DOI] [PubMed] [Google Scholar]
- 49.Chi, Z., and S. A. Asher. 1999. Ultraviolet resonance Raman examination of horse apomyoglobin acid unfolding intermediates. Biochemistry. 38:8196–8203. [DOI] [PubMed] [Google Scholar]
- 50.Vassilenko, K. S., and V. N. Uversky. 2002. Native-like secondary structure of molten globules. Biochim. Biophys. Acta. 1594:168–177. [DOI] [PubMed] [Google Scholar]
- 51.Yao, J., J. Chung, D. Eliezer, P. E. Wright, and H. J. Dyson. 2001. NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding. Biochemistry. 40:3561–3571. [DOI] [PubMed] [Google Scholar]
- 52.Nishii, I., M. Kataoka, and Y. Goto. 1995. Thermodynamic stability of the molten globule states of apomyoglobin. J. Mol. Biol. 250:223–238. [DOI] [PubMed] [Google Scholar]
- 53.Kohn, J. E., I. S. Millett, J. Jacob, B. Zagrovic, T. M. Dillon, N. Cingel, R. S. Dothager, S. Seifert, P. Thiyagarajan, T. R. Sosnick, M. Z. Hasan, V. S. Pande, I. Ruczinski, S. Doniach, and K. W. Plaxco. 2004. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA. 101:12491–12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langsetmo, K., J. Fuchs, and C. Woodward. 1989. Escherichia coli thioredoxin folds into two compact forms of different stability to urea denaturation. Biochemistry. 28:3211–3220. [DOI] [PubMed] [Google Scholar]
- 55.Fandrich, M., M. A. Fletcher, and C. M. Dobson. 2001. Amyloid fibrils from muscle myoglobin. Nature. 410:165–166. [DOI] [PubMed] [Google Scholar]
- 56.Georgescu, R. E., J. H. Li, M. E. Goldberg, M. L. Tasayco, and A. F. Chaffotte. 1998. Proline isomerization-independent accumulation of an early intermediate and heterogeneity of the folding pathways of a mixed alpha/beta protein, Escherichia coli thioredoxin. Biochemistry. 37:10286–10297. [DOI] [PubMed] [Google Scholar]