Abstract
By the use of x-ray structures and flexible docking, we have developed the first in silico ligand-based view of the structural determinants of the binding of small molecule mimics of gelsolin, natural products bound to actin. Our technique highlights those residues on the actin binding site forming important hydrophobic and hydrogen-bonding interactions with the ligands. Significantly, through the flexible docking of toxin fragments, we have also identified potential residues on the actin binding site that have yet to be exploited. Guided by these observations, we have demonstrated that kabiramide C can be modified to produce a structure with a predicted binding energy increased by 20% while the molecular mass is reduced by 20%, clearly indicating the potential for future elaboration of structures targeting this important component of the cytoskeleton.
INTRODUCTION
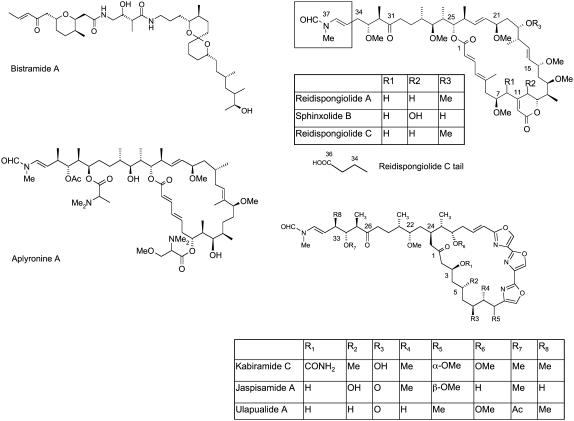
Actin is a major component of the cytoskeleton in eukaryotes, responsible for many important cellular functions such as shape, motility, division, and adhesion (1). Apart from its intrinsic cell biological interest, it therefore makes an attractive target for cancer research. Several natural product toxins, mainly derived from marine life, have been shown to target actin and to display potent antitumor activity (2). Intriguingly, there is increasing evidence these molecules target the same binding site as, and mimic the interactions of, actin binding proteins such as gelsolin, an F-actin capping and severing protein (3–5). Thus, elucidating the structural basis for the binding of these macrolides is of great cross-disciplinary interest (6–9). Several crystal structures of toxins bound to actin have been deposited in the Protein Data Bank in recent years (Fig. 1): kabiramide C (1QZ5) (3), jaspisamide A (1QZ6) (3), ulapualide A (1S22) (10), swinholide A (1YXQ) (11), reidispongiolide A (2ASM) (12), sphinxolide B (2ASO) (12), reidispongiolide C (2ASP) (12), aplyronine A (1WUA) (13), and bistramide A (2FXU) (14). The relatively large size and complexity of these toxins means that a large number of residues has been implicated in their binding, and unified views of the binding requirements of these structures have begun to appear. As the amount of available data grows, an in silico approach to rationalize the available structural data on protein-ligand binding may now be feasible and perhaps even necessary to take advantage of the data to its fullest. In particular, identifying the most important interactions across all the structural classes for which x-ray data exist would aid future synthetic efforts to develop small molecule binders to actin. Surprisingly, however, no such study has been published to date.
FIGURE 1.
Molecular structures of macrolides bistramide A, reidispongiolide A, sphinxolide B, reidispongiolide C (the macrolide tail of reidispongiolide C differs from that of reidispongiolide A and sphinxolide B in the boxed region), kabiramide C, jaspisamide A, ulapualide A (which share a trisoxazole macrolide ring), and aplyronine A that bind to the gelsolin binding site on actin.
Several molecular dynamics (MD) simulations have considered protein-protein interactions and the dynamics of actin in its F- and G-forms (15–19); Yamada and co-workers have derived structure-cytotoxicity relationships for some derivatives of aplyronine A (20–22). However, no in silico study of ligands binding to the hydrophobic cleft of G-actin has been reported. In this work we describe the first such computational investigation, with the aims of identifying the most important interactions in the hydrophobic binding site of G-actin, exploring whether there are interactions that have yet to be identified and could be exploited by new ligands and identifying structural elements that can be modified or removed to produce more efficient ligands (23) than those characterized hitherto.
Docking has been widely and successfully used in virtual screening for drug design (24). It is a computationally cheaper alternative to MD simulations for determining the binding properties of a small molecule to a protein receptor. In this computational technique, the structure of the protein is normally kept rigid, and combinations of rigid body rotations and bond torsions in the ligand are sampled, the search being guided by a heuristic method such as genetic algorithms (GAs) or simulated annealing. To allow for rapid evaluation of protein-ligand affinity, force field-based energy evaluations are often replaced by empirical equations, known as scoring functions, where entropic and solvation effects are accounted for implicitly. Despite being much faster than MD simulations, docking of large flexible molecules remains challenging. To circumvent these difficulties, in this work we ‘scan’ the actin binding site by means of flexible docking of smaller substructures (or ‘fragments’) of the macrolides, in addition to flexible docking of the whole macrolide structures. This maintains the speed of computation of docking, allowing us to compute a much larger configurational space of the ligands and obviating problems with uncoverged simulations.
Nonetheless, by using substructures of macrolides known to bind to actin, we are able to ensure the relevance of the resulting interactions, in contrast to the results from more generalized probes, such as those often used in MD simulations (25). Further, the smaller size of the fragments enables the exploration of regions of the binding site that are not accessible to the full macrolide structures, revealing potential interactions that have yet to be targeted by natural or synthetic methods. Keeping the protein structure rigid is obviously an approximation, but a reasonable one in this case, as we shall show that the relevant region of the binding site does not demonstrate a large degree of flexibility. A more physically rigorous method of exploring the binding of macrolides to actin would be an MD simulation. However, ensuring sufficient convergence of the simulations is challenging, and running individual simulations for each macrolide is likely to be computationally prohibitive. Finally, it is not yet possible to accurately measure binding affinities with the scoring functions used in docking programs (26). However, neither is it feasible to compute binding affinities for the wide range of structures found in macrolides using more rigorous methods such as free energy perturbation (27), which are capable of predicting relative affinities only, and only within a series of structurally similar molecules. Hence, docking is the most efficient and appropriate means to obtain a semiquantitative picture of binding.
METHODS
To model actin, we used the x-ray structure of actin bound with swinholide A (1YXQ). There are no missing atoms in the structure, which aided the preprocessing step, performed with AutoDock Tools (28,29). All the heteroatoms were removed from the structure, and we did not consider the impact of crystallographic water molecules or metal ions. X-ray coordinates of all the ligands considered in this study were available in their bound poses to actin and were therefore aligned with the binding site of 1YXQ by a least squares fit of the C-α atoms in selected residues of the respective actin structures. This was accomplished using the Tcl interface in the visual molecular dynamics (VMD) viewer (30). We used residues 23–25, 116, 133–148, 166–170, and 330–355 for fitting, based on previous reports that had identified these residues as important parts of the gelsolin binding site. The C-α root mean-square deviation (RMSD) for these residues, after alignment, ranged between 0.28 and 0.46 Å, indicating no significant induced fit behavior on binding of any of these molecules. We therefore concluded that the binding site of 1YXQ should provide a suitable receptor for all the ligands we studied. For processing and modifying the ligands, we used the molecular modeling package Ghemical 2.01 (http://www.uku.fi/∼thassine/projects/ghemical/) to add hydrogen atoms and, where necessary, brief minimizations of any manually added atoms to generate sensible bond lengths, constraining the rest of the structure to the x-ray coordinates.
The macrolides shown in Fig. 1 are large, flexible molecules and represent a challenge to current docking methodologies. To ensure convergence of the docking, we ran a series of control dockings using the x-ray structures of selected macrolides, i.e., aplyronine A, bistramide A, kabiramide C, jaspisamide A, reidispongiolide A, and reidispongiolide C. All of these compounds had completely defined atomic positions, with the exception of reidispongiolide C, which has a disordered terminal carboxylate group (but we did not anticipate that this would have a major effect on the results). The tail region of ulapualide A was disordered, swinholide A binds as a dimer, and the crystal structure of sphinxolide B appears to be distorted due to an interaction with a symmetry-related actin molecule. Therefore these structures were not used in our benchmarking. We used AutoDock Tools to determine the number of flexible bonds; this excluded any part of the rings in the molecule, even though they are likely to be flexible. However, our attention was focused on the portion of the molecules that occupy the hydrophobic cleft, which did not include any of the macrolide rings.
Despite the “anchor” portion of the molecules being held rigid, they are all very flexible by the standard of the ‘drug-like’ or ‘fragment-like’ molecules typically used in docking studies, with the number of torsions ranging from 16 in jaspisamide A to 29 in aplyronine A. To find a set of parameters adequate for docking these molecules, we carried out the following protocol: First, as a control experiment to ensure that the actin binding site is “dockable”, we performed a rigid docking of each x-ray structure into the receptor. For docking, we used AutoDock 3.0 (31), employing the Lamarckian GA optimizer. Default parameters were used, except that we increased the population size of the GA to 300 and the number of energy evaluations to 5,000,000 per GA run. After establishing that the rigid dockings produced acceptable results, a series of flexible dockings was carried out. Due to the extremely large search space, we performed several docking runs in parallel, with 20 separate runs, and pooled the results. Previously, we built 49 small molecule fragments of ulapualide A (and minor structural variations) to probe its action as a toxin and as a modulator of gene expression (32). We therefore also used these fragments to ensure the actin binding site was mapped at a sufficient level of detail. This technique should allow a more exhaustive sampling of the configurational space of the docked ligand, compared to using an entire macrolide structure, while maintaining interactions that are relevant between the actin binding site and the molecules under study.
Structures of the fragments are given in the Supplementary Material. Flexible dockings were performed using the same parameters as the rigid docking, except that we carried out 600 runs. The best docked structures for each run were collected, and the atoms with the most negative (i.e., most favorable) van der Waals and electrostatic interactions were extracted and displayed as clusters of points in VMD, where each cluster represented an area of the actin binding site into which docked structures were consistently placed. We selected atoms from each cluster to represent the interaction, and these were overlaid on the protein-ligand complex for visualization. This allowed us to identify parts of the actin structure of particular importance for the binding of the ligands studied, using VMD, MSMS (33), and Persistence of Vision Raytracer (POV-Ray; http://www.povray.org/).
RESULTS
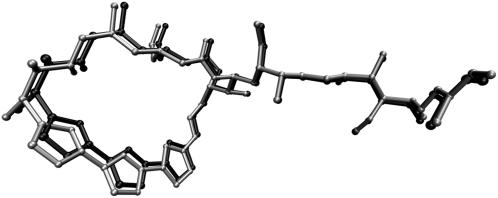
Before exploring the actin binding site via docking, it was necessary to confirm that the actin binding site was dockable using AutoDock. To this end, we rigidly docked the x-ray structures of six macrolides into the x-ray receptor structure of 1YXQ. We used default parameters, as the large size of the ligands, and their lack of flexibility should reduce the search space considerably. For each of the ligands, the 10 docking runs converged to the same structure within an RMSD of 0.50 Å. Compared to the x-ray structures obtained by superposition of the receptor α carbons, the rigidly docked structures differed by no more than 1.07 Å. A typical result is shown in Fig. 2, where the x-ray coordinates of kabiramide C after superposition of the C-α carbon atoms of actin in 1QZ5 and 1YXQ are shown in light shading and those of the x-ray coordinates after rigid docking to the actin structure of 1YXQ are in dark shading. The RMSD between the two structures is 0.71 Å. An RMSD of 2.0 Å is widely considered to match up important interactions between a ligand and receptor (34), and given the 0.46 Å RMSD between the C-α atoms of actin in 1YXQ and 1QZ5, this result suggests that the structures of macrolides can be redocked successfully to the 1YXQ receptor. Therefore we proceeded with flexible docking of each of the seven x-ray structures and the 49 fragments. Subsequently, we examined the distribution of docking scores contributed by each atom to identify a cutoff value for visualization.
FIGURE 2.
Comparison of rigidly docked structure of kabiramide C into the receptor coordinates of actin in 1YXQ (light shading) against the x-ray coordinates in 1QZ5 (dark shading), after alignment of C-α carbons in actin. RMSD between the two kabiramide structures is 0.71 Å, compared to the C-α RMSD of 0.41 Å.
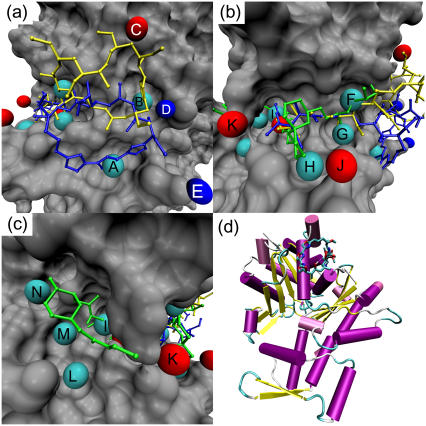
For the molecules we studied, van der Waals interactions displayed a wider range of contributions than electrostatic ones. Given the hydrophobic nature of the binding site, this is not surprising. Therefore, we retained atoms from any structure that contributed >0.6 kcal/mol to the docking score via van der Waals interactions and >0.3 kcal/mol through electrostatic interactions. Examination of the distribution of the atoms showed clear clustering; hence we represented each cluster by a single atom chosen manually from the approximate center of each cluster. The majority of van der Waals interactions were mediated through contacts between the receptor atoms and aliphatic and aromatic carbon atoms in the ligand. Hence, these were classified as hydrophobic interactions. Favorable electrostatic interactions were mediated through oxygen and nitrogen atoms and through hydrogen atoms, the latter attached to oxygen or nitrogen atoms. Therefore, contacts involving the first two atom types are classified as hydrogen-bond acceptors, and the latter as hydrogen-bond donor interactions. The binding site of the natural products (see Fig. 1) can be split into three main sections: the hydrophobic pocket, where most macrolides have a large hydrophobic ‘anchor’ (except for bistramide A); the hydrophobic ‘cleft’, where a hydrophobic tail is intercalated and which is responsible for the depolymerization effects of the molecules; and a region at the other end of the cleft, where only bistramide A has been found to bind. We therefore examined each region in turn for important interactions. The results are displayed in panels a–c of Fig. 3. Panel d shows the binding site of kabiramide C in the context of the entire actin structure.
FIGURE 3.
Predicted important interactions at the gelsolin binding site of actin. Cyan spheres indicate hydrophobic interactions, red spheres hydrogen-bond acceptor interactions, and blue spheres hydrogen-bond donor interactions. (a) The hydrophobic pocket of actin. Kabiramide A (blue) and reidispongiolide A (yellow) are shown for reference. Key: A — hydrophobic interaction at Gly-23; B — hydrophobic interaction at Ser-141, Ala-144, Pro-332, Ser-338; C— hydrogen-bond acceptor interaction with Arg-147; D hydrogen-bond donor interaction with Glu-334; E — hydrogen-bond donor interaction with Asp-25. (b) The hydrophobic cleft of actin. Bistramide A (green), kabiramide A (blue), and reidispongiolide A (yellow) are shown for reference. Key: F — hydrophobic interaction with Arg-147 and Thr-148; G — hydrophobic interaction with Tyr-143 and Leu-346; H — hydrophobic interaction with Thr-351; I — hydrophobic interaction with Ile-135, Val-139, Tyr-169; J — hydrogen-bond acceptor interaction with Thr-351; K — hydrogen-bond acceptor interaction with Tyr-169. (c) The far end of the hydrophobic cleft of actin. Bistramide A is shown for reference. Key: L — hydrophobic interaction with Tyr-133 and Val-370; M — hydrophobic interaction with Val-134; N — hydrophobic interaction with Leu-110, Asn-111. (d) Cartoon representation of actin, with kabiramide C in its binding site at the top of the panel.
The most important interactions in the hydrophobic pocket are shown in Fig. 3 a, with the rigidly docked structures of jaspisamide A (blue) and reidispongiolide A (yellow) for reference. Hydrophobic interactions are represented by cyan spheres, hydrogen-bond acceptor interactions are represented by red spheres, and hydrogen-bond donor sites indicated by blue spheres. For jaspisamide A the hydrophobic interaction between the trisoxazole rings and Gly-23 (labeled A) is highlighted. Conversely, the reidispongiolide A ring occupies hydrophobic interaction B, formed between the lactone group and a pocket formed of Ala-144, Ser-145, Pro-332, Arg-335, Ser-338, and Ile-341. No obvious hydrogen-bond donors exist sufficiently close to the carbonyl group oxygen, so the interaction seems to be entirely hydrophobic. Indeed, replacing the carbonyl group oxygen with an ethylene carbon and repeating the rigid docking of reidispongiolide A had no deleterious effect on the final docked energy; in fact, the energy became more favorable by −0.4 kcal/mol. Additionally, a hydrogen-bond interaction at Arg-147 is highlighted (C). These interactions were noted by Allingham and co-workers (3,12). However, docking also suggested the possibility of hydrogen-bond donor interactions with Asp-25 and Glu-334 (D and E). As no macrolide structure has the necessary donor groups in the right arrangement for hydrogen-bonding to occur, hitherto these residues have not been identified as potentially important.
Fig. 3 b shows the hydrophobic cleft, with the rigidly docked structures of bistramide A, kabiramide C, and reidispongiolide A superimposed. Unsurprisingly, the cleft is dominated by hydrophobic interactions. Important residues for these interactions include Ile-135, Val-139, Tyr-143, Tyr-169, Leu-346, and Thr-351. This region of the cleft has been extensively studied, and these interactions are consistent with those identified by Allingham and co-workers (12). However, we did notice that during flexible docking, more extensive hydrophobic interactions were formed with Thr-351 (H) than in the x-ray structures, perhaps due to the fact that only a methoxy group is substituted at the relevant position (C32 in kabiramide C and C33 in reidispongiolide A). Below we investigate the possibility that replacement or extension of this group with a moderately larger moiety may increase binding affinity. Two hydrogen-bond interactions are also indicated in this region (J and K); these positions would be exposed to solvent and may be useful areas to introduce polar contacts either for direct hydrogen-bonding interactions with Thr-351 or Tyr-169 or mediated through water interactions.
Fig. 3 c shows the enone side chain of bistramide A near Tyr-169. This is at the opposite end of the hydrophobic cleft to where all other previously characterized ligands have been anchored and is therefore of special interest. Kozmin and co-workers' description of the binding of bistramide A notes a high preponderance of polar contacts in this structure and that the enone side chain is disordered and does not play a critical role in the binding of bistramide A (14). Therefore, it is interesting that three hydrophobic interactions are close to the enone structure, indicating substantial potential for further structural elaboration of bistramide-based molecules. These interactions are mediated by Tyr-133 and Val-370 (interaction L), Val-134 (interaction M), Leu-110 (interaction N), and Leu-136 (interaction I). Interaction I is also close to Val-139 and Tyr-169. However, the unfavorable electrostatic interactions are only minor, and the shape of the receptor in this region provides a strongly favorable van der Waals interaction that outweighs the electrostatic interaction by nearly an order of magnitude according to the AutoDock scoring function. That these interactions are possibly valid sites for compounds based on bistramide A is demonstrated by the highest scoring flexibly docked structure, where the C12-C13 rotates the enone and pyran group so they occupy interaction sites M and L. As Kozmin and co-workers identified three polar contacts mediated by water molecules (14), which we have not modeled in our docking study, we can expect that some deviation from the x-ray structure will occur to compensate for these missing interactions. Although we found a hydrogen-bonding interaction near Tyr-169, this is likely to be occupied by solvent; an analysis of the x-ray structure of 2FXU reveals a water molecule close to that position.
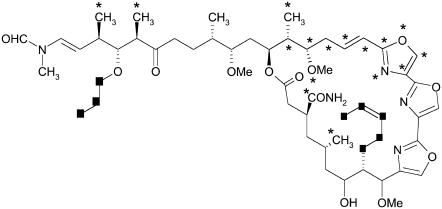
To illustrate the potential use of these interaction maps, we modified the structure of kabiramide C in silico to produce a ligand that bound more “efficiently” (i.e., the docking score per heavy atom was larger) than the original structure. We did not consider synthetic feasibility, nor entropic penalties; we merely wished to demonstrate that the kabiramide C scaffold can be reduced without reducing its binding energy substantially and a small number of targeted elaborations can further improve the strength of binding. From an examination of Fig. 3, we first concentrated on the hydrophobic pocket, where the macrocycle of kabiramide C binds, and targeted the hydrophobic interaction at Pro-332 (marked as interaction B in Fig. 3 a), where the lactone of reidispongiolide A is located. To achieve this we replaced the methyl group at C8 with a pent-2-enyl group. Additionally, hydrophobic interaction site H in Fig. 3 b was identified as a further interaction in the hydrophobic cleft. Here we made a modification at C32, replacing the methoxy group with a propoxy group to increase contact with Thr-351. Elsewhere we identified a portion of the ring of kabiramide C as contributing only weakly to the activity. We experimented with deleting portions of the ring, including one of the trisoxazole rings, and some pendant groups to the main scaffold where our analysis had not indicated any critically important interactions. The deletions and additions to the main scaffold of kabiramide C are marked by stars and squares, respectively, in Fig. 4.
FIGURE 4.
Modified structure of kabiramide C to probe identified interactions in the actin binding site. Parts of the structure marked with stars were deleted; parts marked with squares were added.
Repeating the docking, which allowed flexibility to the newly added groups, successfully placed them at the targeted interactions. Thus, we increased the best docking score by 3.2 kcal/mol over kabiramide C, an increase of 20% and a decrease in Ki of two orders of magnitude, although this neglects the influence of entropy and the inherent error in the AutoDock scoring function (on the order of 2 kcal/mol). At the same time, the molecular mass has been reduced from 943 Da in kabiramide C to 748 Da, a reduction of 20%. Although this is still larger than the value recommended in Lipinski's famous Rule of Five (35) for maximizing the probability of oral bioavailability of a molecules (the threshold is 500), this is clearly a step in the right direction. This demonstrates the ability of our method to indicate the important regions for binding. Clearly, there is substantial scope for development of ligands that bind to actin. The addition of the propoxy group was also of interest, because ulapualide A has an acetate substitution here and it had been considered that this was the cause of the disorder observed in the tail portion of ulapualide A (10). We observed that the addition of an acetate group to kabiramide C reduced the docking score compared to just the alkyl chain, even though the carbon atoms were placed in equivalent positions. This effect could be traced to unfavorable steric interaction between the carbonyl oxygen atom and the sulfur atom of Met-355. This observation provides some support for the hypothesis that the acetate group is the cause of the disorder in ulapualide A.
DISCUSSION
We conclude that flexible docking of ligand fragments is a valuable tool to rationalize and unify structural data across several ligands. Docking-based interaction maps provide a simple visual confirmation of several observations made from examination of individual or small numbers of x-ray structures, as well as extending the analysis to potential interaction sites yet to be exploited. To some extent, structural bioinformatics tools such as LPC/CSU (ligand-protein contacts and contacts of structural units) (36) can reveal similar insights by analyzing ligand-protein atom contacts to highlight regions of particular complementarity or, conversely, regions where there is scope to increase the degree of burial or change the type of interaction. However, this analysis cannot be combined across several ligands and visualized as easily as the docking-based method outlined here. A docking solution also explicitly accounts for conformational effects of modifying the ligand (albeit accounting for protein flexibility remains challenging) and can also indicate promising regions of the binding site where no ligand has hitherto been observed to occupy. For actin in particular, we have been able to focus on a small number of residues, such as the hydrophobic interaction between the trisoxazole ring of the kabiramide C and related compounds at Gly-23, and the hydrophobic interaction between the macrolactone ring of reidispongiolide A at Pro-332. This provides impetus for future synthetic work, to target those structural features that are most vital for binding, and represents the first steps to a pharmacophore for the actin binding site.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
We are grateful for the use of the High Performance Computing facility at the University of Nottingham and the Engineering and Physical Sciences Research Council for support (GR/S75765/01).
References
- 1.Pollard, T. D., L. Blanchoin, and R. D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. [DOI] [PubMed] [Google Scholar]
- 2.Allingham, J. S., V. A. Klenchin, and I. Rayment. 2006. Actin-targeting natural products: structures, properties and mechanisms of action. Cell. Mol. Life Sci. 63:2119–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klenchin, V. A., J. S. Allingham, R. King, J. Tanaka, G. Marriott, and I. Rayment. 2003. Trisoxazole macrolide toxins mimic the binding of actin-capping proteins to actin. Nat. Struct. Biol. 10:1058–1063. [DOI] [PubMed] [Google Scholar]
- 4.Klenchin, V. A., S. Y. Khaitlina, and I. Rayment. 2006. Crystal structure of polymerization-competent actin. J. Mol. Biol. 362:140–150. [DOI] [PubMed] [Google Scholar]
- 5.Dominguez, R. 2004. Actin-binding proteins—a unifying hypothesis. Trends Biochem. Sci. 29:572–578. [DOI] [PubMed] [Google Scholar]
- 6.Spector, I., F. Braet, N. R. Shochet, and M. R. Bubb. 1999. New anti-actin drugs in the study of the organization and function of the actin cytoskeleton. Microsc. Res. Tech. 47:18–37. [DOI] [PubMed] [Google Scholar]
- 7.Yeung, K. S., and I. Paterson. 2002. Actin-binding marine macrolides: total synthesis and biological importance. Angew. Chem. Int. Ed. Engl. 41:4632–4653. [DOI] [PubMed] [Google Scholar]
- 8.Fenteany, G., and S. T. Zhu. 2003. Small-molecule inhibitors of actin dynamics and cell motility. Curr. Top. Med. Chem. 3:593–616. [DOI] [PubMed] [Google Scholar]
- 9.Yeung, K. S., and I. Paterson. 2005. Advances in the total synthesis of biologically important marine macrolides. Chem. Rev. 105:4237–4313. [DOI] [PubMed] [Google Scholar]
- 10.Allingham, J. S., J. Tanaka, G. Marriott, and I. Rayment. 2004. Absolute stereochemistry of ulapualide A. Org. Lett. 6:597–599. [DOI] [PubMed] [Google Scholar]
- 11.Klenchin, V. A., R. King, J. Tanaka, G. Marriott, and I. Rayment. 2005. Structural basis of swinholide A binding to actin. Chem. Biol. 12:287–291. [DOI] [PubMed] [Google Scholar]
- 12.Allingham, J. S., A. Zampella, M. V. D'Auria, and I. Rayment. 2005. Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity. Proc. Natl. Acad. Sci. USA. 102:14527–14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirata, K., S. Muraoka, K. Suenaga, T. Kuroda, K. Kato, H. Tanaka, M. Yamamoto, M. Takata, K. Yamada, and H. Kigoshi. 2006. Structure basis for antitumor effect of aplyronine A. J. Mol. Biol. 356:945–954. [DOI] [PubMed] [Google Scholar]
- 14.Rizvi, S. A., V. Tereshko, A. A. Kossiakoff, and S. A. Kozmin. 2006. Structure of bistramide A-actin complex at a 1.35 Å resolution. J. Am. Chem. Soc. 128:3882–3883. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y. M., M. Scolari, W. Im, and H. J. Woo. 2006. Protein-protein interactions in actin-myosin binding and structural effects of R405Q mutation: a molecular dynamics study. Proteins. 64:156–166. [DOI] [PubMed] [Google Scholar]
- 16.Guaqueta, C., L. K. Sanders, G. C. L. Wong, and E. Luijten. 2006. The effect of salt on self-assembled actin-lysozyme complexes. Biophys. J. 90:4630–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minehardt, T. J., P. A. Kollman, R. Cooke, and E. Pate. 2006. The open nucleotide pocket of the profilin/actin x-ray structure is unstable and closes in the absence of profilin. Biophys. J. 90:2445–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu, J. W., and G. A. Voth. 2005. Allostery of actin filaments: molecular dynamics simulations and coarse-grained analysis. Proc. Natl. Acad. Sci. USA. 102:13111–13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wriggers, W., J. X. Tang, T. Azuma, P. W. Marks, and P. A. Janmey. 1998. Cofilin and gelsolin segment-1: molecular dynamics simulation and biochemical analysis predict a similar actin binding mode. J. Mol. Biol. 282:921–932. [DOI] [PubMed] [Google Scholar]
- 20.Kigoshi, H., K. Suenaga, M. Takagi, A. Akao, K. Kanematsu, N. Kamei, Y. Okugawa, and K. Yamada. 2002. Cytotoxicity and actin-depolymerizing activity of aplyronine A, a potent antitumor macrolide of marine origin, and its analogs. Tetrahedron. 58:1075–1102. [Google Scholar]
- 21.Suenaga, K., N. Kamei, Y. Okugawa, M. Takagi, A. Akao, H. Kigoshi, and K. Yamada. 1997. Cytotoxicity and actin depolymerizing activity of aplyronine A, a potent antitumor macrolide of marine origin, and the natural and artificial analogs. Bioorg. Med. Chem. Lett. 7:269–274. [Google Scholar]
- 22.Kigoshi, H., K. Suenaga, T. Mutou, T. Ishigaki, T. Atsumi, H. Ishiwata, A. Sakakura, T. Ogawa, M. Ojika, and K. Yamada. 1996. Aplyronine A, a potent antitumor substance of marine origin, aplyronines B and C, and artificial analogues: total synthesis and structure-cytotoxicity relationships. J. Org. Chem. 61:5326–5351. [Google Scholar]
- 23.Hopkins, A. L., C. R. Groom, and A. Alex. 2004. Ligand efficiency: a useful metric for lead selection. Drug Discov. Today. 9:430–431. [DOI] [PubMed] [Google Scholar]
- 24.Alvarez, J., and B. Shoichet. 2005. Virtual Screening in Drug Discovery. CRC Press, Boca Raton, FL.
- 25.Meagher, K. L., and H. A. Carlson. 2004. Incorporating protein flexibility in structure-based drug discovery: using HIV-1 protease as a test case. J. Am. Chem. Soc. 126:13276–13281. [DOI] [PubMed] [Google Scholar]
- 26.Warren, G. L., C. W. Andrews, A. M. Capelli, B. Clarke, J. LaLonde, M. H. Lambert, M. Lindvall, N. Nevins, S. F. Semus, S. Senger, G. Tedesco, I. D. Wall, J. M. Woolven, C. E. Peishoff, and M. S. Head. 2006. A critical assessment of docking programs and scoring functions. J. Med. Chem. 49:5912–5931. [DOI] [PubMed] [Google Scholar]
- 27.Zwanzig, R. W. 1954. High-temperature equation of state by a perturbation method. 1. Nonpolar gases. J. Chem. Phys. 22:1420–1426. [Google Scholar]
- 28.Sanner, M. F. 1999. Python: a programming language for software integration and development. J. Mol. Graph. Model. 17:57–61. [PubMed] [Google Scholar]
- 29.Sanner, M. F., D. Stoffler, and A. J. Olson. 2002. ViPEr, a Visual Programming Environment for Python. Proc. Inter. Python Conf., 10th, Alexandria, VA. 103–115.
- 30.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- 31.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19:1639–1662. [Google Scholar]
- 32.Vincent, E., J. Saxton, C. Baker-Glenn, I. Moal, J. D. Hirst, G. Pattenden, and P. E. Shaw. 2007. Effects of ulapualide A and synthetic macrolide analogues on actin dynamics and gene regulation. Cell. Mol. Life Sci. 64:487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanner, M. F., A. J. Olson, and J. C. Spehner. 1996. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 38:305–320. [DOI] [PubMed] [Google Scholar]
- 34.Cole, J. C., C. W. Murray, J. W. M. Nissink, R. D. Taylor, and R. Taylor. 2005. Comparing protein-ligand docking programs is difficult. Proteins. 60:325–332. [DOI] [PubMed] [Google Scholar]
- 35.Lipinski, C. A., F. Lombardo, B. W. Dominy, and P. J. Feeney. 1997. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23:3–25. [DOI] [PubMed] [Google Scholar]
- 36.Sobolev, V., A. Sorokine, J. Prilusky, E. E. Abola, and M. Edelman. 1999. Automated analysis of interatomic contacts in proteins. Bioinformatics. 15:327–332. [DOI] [PubMed] [Google Scholar]