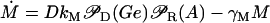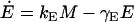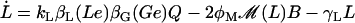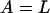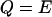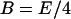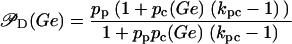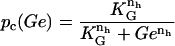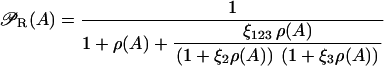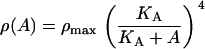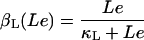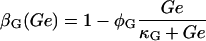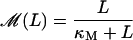Abstract
Multistability is an emergent dynamic property that has been invoked to explain multiple coexisting biological states. In this work, we investigate the origin of bistability in the lac operon. To do this, we develop a mathematical model for the regulatory pathway in this system and compare the model predictions with other experimental results in which a nonmetabolizable inducer was employed. We investigate the effect of lactose metabolism using this model, and show that it greatly modifies the bistable region in the external lactose (Le) versus external glucose (Ge) parameter space. The model also predicts that lactose metabolism can cause bistability to disappear for very low Ge. We have also carried out stochastic numerical simulations of the model for several values of Ge and Le. Our results indicate that bistability can help guarantee that Escherichia coli consumes glucose and lactose in the most efficient possible way. Namely, the lac operon is induced only when there is almost no glucose in the growing medium, but if Le is high, the operon induction level increases abruptly when the levels of glucose in the environment decrease to very low values. We demonstrate that this behavior could not be obtained without bistability if the stability of the induced and uninduced states is to be preserved. Finally, we point out that the present methods and results may be useful to study the emergence of multistability in biological systems other than the lac operon.
INTRODUCTION
At the molecular level, biological systems function using two types of information: genes, which encode the molecular machines that execute the functions of life, and networks of regulatory interactions, specifying how genes are expressed. Substantial progress over the past few decades in biochemistry, molecular biology, and cell physiology has ushered in a new era of regulatory interaction research. Recent analysis has revealed that cell signals do not necessarily propagate linearly. Instead, cellular signaling networks can be used to regulate multiple functions in a context-dependent fashion. Because of the magnitude and complexity of the interactions in the cell, it is often not possible to understand intuitively the systems behavior of these networks. Rather, it has become necessary to develop mathematical models and analyze the behavior of these models, both to develop a systems-level understanding and to obtain experimentally testable predictions. This, together with the fact that DNA micro-arrays, sequencers, and other technologies have begun to generate vast amounts of quantitative biological data, has accelerated the shift away from a purely descriptive biology and toward a predictive one.
Recent computer simulations of partial or whole genetic networks have demonstrated collective behaviors (commonly called systems, or emergent, properties) that were not apparent from examination of only a few isolated interactions alone. Among the various patterns of complex behavior associated with nonlinear kinetics, multistability is noteworthy. Multistability corresponds to a true switch between alternate and coexisting steady states, and so allows a graded signal to be turned into a discontinuous evolution of the system along several different possible pathways. Multistability has certain unique properties not shared by other mechanisms of integrative control. These properties may play an essential role in the dynamics of living cells and organisms. Moreover, multistability has been invoked to explain catastrophic events in ecology (1), mitogen-activated protein kinase cascades in animal cells (2–4), cell cycle regulatory circuits in Xenopus and Saccharomyces cerevisiae (5,6), the generation of switchlike biochemical responses (2,3,7), and the establishment of cell cycle oscillations and mutually exclusive cell cycle phases (6,8), among other biological phenomena. On the other hand, there are also serious doubts that multistability is the dynamic origin of some biological switches (9). Nevertheless, it is generally accepted that two paradigmatic gene-regulatory networks in bacteria, the phage λ switch and the lac operon (at least when induced by nonmetabolizable inducers), do show bistability (10–12). In the former, bistability arises through a mutually inhibitory double-negative-feedback loop, while in the latter, a positive-feedback loop is responsible for the bistability.
The lac operon, the phage-λ switch, and the trp operon, are three of the best known and most widely studied systems in molecular biology. The inducible lac operon in E. coli is the classic example of bistability. It was first noted by Monod and co-workers more than 50 years ago, although it was not fully recognized at the time. The bistable behavior of the lac operon has been the subject of a number of studies. It was first examined in detail by Novick and Weiner (13) and Cohn and Horibata (14). Later experimental studies include those of Maloney and Rotman (15) and Chung and Stephanopoulos (10). Over the last few years, the lac operon dynamics have been analyzed mathematically by Wong et al. (16), Vilar et al. (17), Yildirim and Mackey (18), Santillán and Mackey (19), Yildirim et al. (20), Tian and Burrage (21), and Hoek and Hogeweg (22). This was possible because this system has been experimentally studied for nearly 50 years and there is a wealth of biochemical and molecular information and data on which to draw. Recently, Ozbudak et al. (23) performed a set of ingenious experiments that not only confirm bistability in the lac operon when induced with the nonmetabolizable inducer thiomethylgalactoside (TMG), but also provide new and novel quantitative data that raise questions that may be answered via a modeling approach.
Previous studies have analyzed bistability and its dynamic properties as a systems phenomenon. However, to our knowledge, none of them have dealt with questions like: “How did multistability arise within the context of gene regulatory networks?” or “What evolutionary advantages do bistable regulatory networks have when compared with monostable ones?” In this article, we address these issues from a mathematical modeling approach, basing our examination on the dynamics of the lac operon.
THEORY
Model development
A mathematical model for the lac operon is developed in Appendices A–C. All of the model equations are tabulated in Table 1 and they are briefly explained below. The reader may find it useful to refer to Fig. 1, where the lac operon regulatory pathway is schematically represented.
TABLE 1.
Full set of equations for the model of lactose operon regulatory pathway depicted in Fig. 1
Differential Eqs. 1–3 govern the time evolution of the intracellular concentration of mRNA (M), polypeptide (E), and lactose (L) molecules. Ge and Le, respectively, stand for the extracellular glucose and lactose concentrations. A, Q, and B represent the intracellular allolactose, permease, and β-galactosidase molecule concentrations. The functions 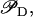
 βL, and βG, respectively, account for the negative effect of external glucose on the initiation rate of transcription (via catabolite repression), the probability that the lactose promoter is not repressed, the positive effect of external lactose on its uptake rate, and the negative effect of external glucose on lactose uptake (inducer exclusion). The expression
βL, and βG, respectively, account for the negative effect of external glucose on the initiation rate of transcription (via catabolite repression), the probability that the lactose promoter is not repressed, the positive effect of external lactose on its uptake rate, and the negative effect of external glucose on lactose uptake (inducer exclusion). The expression 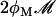 is the rate of lactose metabolism per β-galactosidase. Finally, pc and ρ represent internal auxiliary variables.
is the rate of lactose metabolism per β-galactosidase. Finally, pc and ρ represent internal auxiliary variables.
FIGURE 1.
(A) Schematic representation of the lac operon regulatory mechanisms. This operon comprises genes lacZ, lacY, and lacA. Protein LacZ is a permease that transports external lactose into the cell. Protein LacY polymerases into a homo-tetramer named β-galactosidase. This enzyme transforms internal lactose (Lac) into allolactose (Allo) and galactose (Gal), and further transforms allolactose into galactose. Allolactose can bind to the repressor (R) inhibiting it. When not bound by allolactose, R can bind to a specific site upstream of the operon structural genes and thus avoid transcription initiation. External glucose inhibits production of cAMP which, when bound to protein CRP to form complex CAP, acts as an activator of the lac operon. External glucose also inhibits lactose uptake by permease proteins. (B) Graphical representation of the interactions accounted for by the lac operon mathematical model. The meaning of the variables appearing in this figure is as follows. Ge and Le stand for external glucose and lactose concentrations; M, Q, and B denote mRNA, permease, and β-galactosidase, molecule concentrations, respectively; and L, A, and G correspond to the intracellular lactose, allolactose, and galactose molecule concentrations. All of the processes underlying the lac operon regulatory pathway are represented by rectangles. Inputs (outputs) are denoted with empty arrowheads (circles). Finally, plus and minus signs stand for the effect each input variable has on every output variable.
The model consists of three differential equations (Eqs. 1–3), that respectively account for the temporal evolution of mRNA (M), lacZ polypeptide (E), and internal lactose (L) concentrations.
Messenger RNA (mRNA) is produced via transcription of the lac operon genes, and its concentration decreases because of active degradation and dilution due to cell growth. The value γM represents the degradation plus dilution rate, kM is the maximum transcription rate per promoter, and D is the average number of promoter copies per bacterium. The function 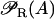 (defined in Eq. 9) accounts for regulation of transcription initiation by active repressors. The fraction of active repressors is proportional to ρ(A) (compare Eq. 10); since repressors are inactivated by allolactose (A), ρ is a decreasing function of A. Furthermore, the rate of transcription initiation decreases as the concentration of active repressors increases. Concomitantly,
(defined in Eq. 9) accounts for regulation of transcription initiation by active repressors. The fraction of active repressors is proportional to ρ(A) (compare Eq. 10); since repressors are inactivated by allolactose (A), ρ is a decreasing function of A. Furthermore, the rate of transcription initiation decreases as the concentration of active repressors increases. Concomitantly, 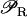 is a decreasing function of ρ, and thus it is an increasing function of A. The function
is a decreasing function of ρ, and thus it is an increasing function of A. The function 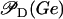 (Eq. 9) denotes the modulation of transcription initiation by external glucose, i.e., through catabolite repression. Production of cyclic AMP (cAMP) is inhibited by extracellular glucose. cAMP further binds the so-called cAMP receptor protein (CRP) to form the CAP complex. Finally, CAP binds a specific site near the lac promoter and enhances transcription initiation. The probability of finding a CAP molecule bound to its corresponding site is given by pc (Eq. 8) and is a decreasing function of Ge. Moreover,
(Eq. 9) denotes the modulation of transcription initiation by external glucose, i.e., through catabolite repression. Production of cyclic AMP (cAMP) is inhibited by extracellular glucose. cAMP further binds the so-called cAMP receptor protein (CRP) to form the CAP complex. Finally, CAP binds a specific site near the lac promoter and enhances transcription initiation. The probability of finding a CAP molecule bound to its corresponding site is given by pc (Eq. 8) and is a decreasing function of Ge. Moreover, 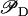 is an increasing function pc and, therefore, a decreasing function of Ge.
is an increasing function pc and, therefore, a decreasing function of Ge.
The translation initiation rate of lacZ transcripts is kE, while γE is the dilution and degradation rate of lacZ polypeptides. Let B and Q, respectively, denote the β-galactosidase and permease concentrations. Given that the corresponding parameters for lacY transcripts and polypeptides attain similar values, that β-galactosidase is a homo-tetramer made up of four identical lacZ polypeptides, and that permease is a lacY monomer, it follows that Q = E and B = E/4 (Eqs. 5 and 6).
Lactose is transported into the bacterium by a catalytic process in which permease protein plays a central role. Thus, the lactose influx rate is assumed to be kLβL(Le)Q, with the function βL(Le) given by Eq. 11. Extracellular glucose negatively affects lactose uptake (so-called inducer exclusion), and this is accounted for by βG(Ge) (Eq. 12), which is a decreasing function of Ge. Once inside the cell, lactose is metabolized by β-galactosidase. Approximately half of the lactose molecules are transformed into allolactose, while the rest enter the catalytic pathway that produces galactose. In our model, 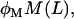 with ℳ defined in Eq. 13, denotes the lactose-to-allolactose metabolism rate, which equals the lactose-to-galactose metabolism rate. Allolactose is further metabolized into galactose by β-galactosidase. From the assumptions that the corresponding metabolism parameters are similar to those of lactose, and that the allolactose production rate is much higher than its degradation plus dilution rate, it follows that A ≈ L (Eq. 4).
with ℳ defined in Eq. 13, denotes the lactose-to-allolactose metabolism rate, which equals the lactose-to-galactose metabolism rate. Allolactose is further metabolized into galactose by β-galactosidase. From the assumptions that the corresponding metabolism parameters are similar to those of lactose, and that the allolactose production rate is much higher than its degradation plus dilution rate, it follows that A ≈ L (Eq. 4).
Parameter estimation
All of the parameters in the model are estimated in Appendix D. Their values are tabulated in Table 2.
TABLE 2.
Value of all of the parameters in the equations of Table 1, as estimated in Appendix D
| μ ≈ 0.02 min−1 | KG ≈ 2.6 μM |
| D ≈ 2 mpb | nh ≈ 1.3 |
| kM ≈ 0.18 min−1 | ξ2 ≈ 0.05 |
| kE ≈ 18.8 min−1 | ξ3 ≈ 0.01 |
| kL ≈ 6.0 × 104 min−1 | ξ123 ≈ 163 |
| γM ≈ 0.48 min−1 | ρmax ≈ 1.3 |
| γE ≈ 0.03 min−1 | KA ≈ 2.92 × 106 mpb |
| γL ≈ 0.02 min−1 | κL ≈ 680 μM |
| kpc ≈ 30 | φG ≈ 0.35 |
| pp ≈ 0.127 | κG ≈ 1.0 μM |
| φM ≈ ∈ [0, 4.0 × 104] min−1 | κM ≈ 7.0 × 105 mpb |
The term mpb indicates molecules per average bacterium.
METHODS
Numerical experiments and analytical studies
We carried out stochastic simulations with the above-described model. This was done by means of Gillespie's Tau-Leap algorithm (24,25), which we implemented in Python (http://www.python.org/download/).
All of the analytical results we used to study the model dynamic behavior are explained in detail in the Appendices.
RESULTS
A mathematical model for the lac operon regulatory pathway was developed as explained in Theory, above. The model equations are tabulated in Table 1. These equations determine the time evolution of variables M, E, and L, which respectively stand for mRNA, lacZ polypeptide, and internal lactose concentrations. Special attention was paid to the estimation of the model parameters from reported experimental data. The model parameters are tabulated in Table 2. Below we describe the results obtained from this model.
The model steady states and their stability are analyzed in Appendix E. As seen there, depending on the values of Le and Ge, there can be up to three steady states. For very low Le values, there is a single stable steady state corresponding to the uninduced state. As Le increases, two more steady states appear via a saddle-node bifurcation. One of these new fixed points is stable, corresponding to the induced state, while the other is a saddle node. With further increases in Le the saddle node and the original stable steady-state approach until they eventually collide and are annihilated via another saddle-node bifurcation. Afterwards, only the induced stable steady state survives.
Ozbudak et al. (23) carried out a series of clever experiments regarding the bistable behavior of the lac operon in E. coli. In these experiments, the Le values at which the bifurcations take place were experimentally determined for various extracellular glucose concentrations, using a nonmetabolizable inducer (TMG). In the present model, the usage of a nonmetabolizable inducer can be modeled by setting φM = 0; recall that this parameter stands for the maximum rate of lactose-to-allolactose and of lactose-to-galactose metabolism.
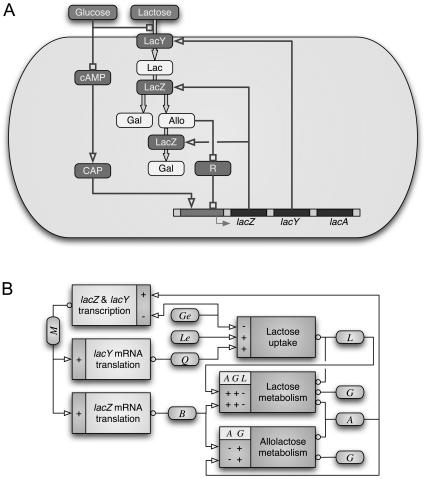
Using the procedure described in Appendix E, we numerically calculated the model Le bifurcation values for several Ge concentrations when a nonmetabolizable inducer is employed. The results are shown in the bifurcation diagram of Fig. 2 A, and compared with the experimental results of Ozbudak et al. (23). The agreement between the experimental data and the model predictions is sufficiently close to give us confidence in the other results we report here.
FIGURE 2.
(A) Bifurcation diagram in the Le versus Ge parameter space, calculated with φM = 0. As discussed in the text, this choice of the parameter φM simulates induction of the lac operon with a nonmetabolizable inducer, like TMG. The dots correspond to the experimental results of Ozbudak et al. (2004). Panels B–D show bifurcation diagrams calculated with the following lactose metabolism rates: φM = 3.6 × 103 min−1, φM = 1.5 × 104 min−1, and φM = 4.0 × 104 min−1, respectively.
Ozbudak et al. further assert that, when their experiments were repeated with the natural inducer (lactose), they were unable to identify any bistable behavior. This result, and other studies reporting similar conclusions (22), have sparked a lively debate about whether bistability is a property of the lac operon under naturally occurring conditions, or it is only an artifact introduced by the use of nonmetabolizable inducers.
To investigate the effects of lactose metabolism, we recalculated the bifurcation diagram of Fig. 2 A with different values of the parameter φM. Bifurcation diagrams in the Le versus Ge parameter space, calculated with φM = 0, 3.6 × 103, 1.5 × 104, and 4.0 × 104 min−1, are shown in Fig. 2, B–D. From these results we conclude that the main effects of lactose metabolism on the Le versus Ge bifurcation diagrams are
The border between the uninduced monostable and the bistable regions is located at higher Le values as φM increases.
The width of the bistable region increases as both Ge and φM increase.
For very high values of φM (larger than 2.5 × 104 min−1), bistability is not present at very low Ge concentrations, and only appears (via a cusp catastrophe) after Ge exceeds a threshold.
In their experiments, Ozbudak et al. (23) let a bacterial culture grow for a long time (more than four hours) in a medium with constant glucose and TMG concentrations. Then they measured the expression level of gene lacZ in every bacterium and plotted the corresponding histogram. When the Ge and TMG concentrations were set such that the induced and uninduced stable steady states were both available, a bimodal distribution for the lacZ expression level was observed. Otherwise, they obtained unimodal distributions.
Ozbudak et al. later repeated the same experiments using lactose as an inducer, and observed only unimodal distributions, even for saturating values of Le. Thus, they did not find any evidence of bistability with lactose as an inducer. According to the bifurcation diagrams in Fig. 2, these findings can be explained if 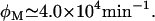 In that case, even when Le is as high as 1000 μM, the system should be in the monostable uninduced region, or close to it, and thus any sign of bistability will be hard to detect. To test this hypothesis we carried out stochastic simulations by means of Gillespie's Tau-Leap algorithm (24,25), which we implemented in Python. In these simulations we set the values of Ge and Le, and let the system evolve for 20,000 min, recording the value of all variables every minute. We performed these numerical experiments with Le = 1000 μM, and Ge = 4, 10, 20, 40, 100, 200, 400, and 1000 μM.
In that case, even when Le is as high as 1000 μM, the system should be in the monostable uninduced region, or close to it, and thus any sign of bistability will be hard to detect. To test this hypothesis we carried out stochastic simulations by means of Gillespie's Tau-Leap algorithm (24,25), which we implemented in Python. In these simulations we set the values of Ge and Le, and let the system evolve for 20,000 min, recording the value of all variables every minute. We performed these numerical experiments with Le = 1000 μM, and Ge = 4, 10, 20, 40, 100, 200, 400, and 1000 μM.
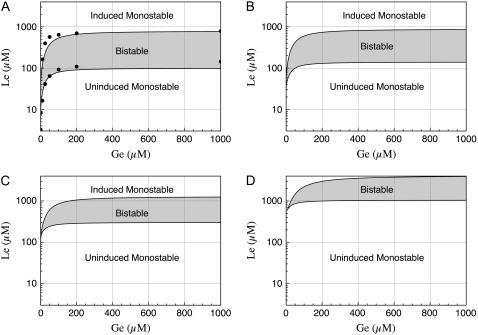
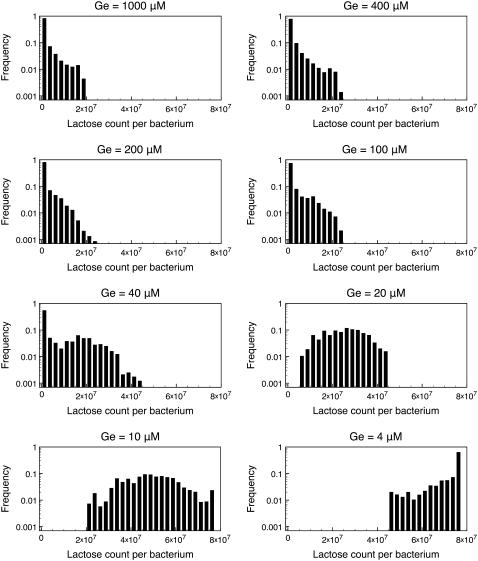
In Fig. 3 we show the histograms of the internal lactose molecule count calculated from these numerical experiments. Notice that:
Unimodal distributions, corresponding to the uninduced steady state, were obtained for Ge = 100, 200, 400, 1000 μM. In all these cases, the number of lactose molecules per average bacterium is small—as compared with the corresponding numbers in the induced state—most of the times, and the frequency of higher molecule counts rapidly decays.
We obtained unimodal distributions as well for Ge = 4, 10, 20 μM. These distributions correspond to the induced steady state, and the number of lactose molecules per average bacterium fluctuates around values larger than 2 × 107 mpb.
Of all the calculated distributions, only that for Ge = 40 μM vaguely resembles a bimodal distribution.
FIGURE 3.
Normalized histograms for the number of lactose molecules per average-size bacterium. This histograms were calculated from the results of stochastic simulations in which we let the system evolve, for 10,000 min, with Le = 1000 μM and Ge = 1000, 400, 200, 100, 40, 20, 10, and 4 μM; see the main text for details. Notice that the lac operon remains in the uninduced state for Ge ≥ 100 μM, and then it jumps to the induced state when Ge ≤ 20 μM. Of all the histograms, only that corresponding to Ge = 40 μM shows a vaguely bimodal distribution characteristic of bistability. These results are in complete accord with the bifurcation diagrams presented in Fig. 2.
Following Santillán and Mackey (19), we analyze the effect of inducer exclusion on the bistable behavior of the lac operon. For this, we construct—by setting βG(Ge) = 1 in Eq. 3—an in silico mutant strain of E. coli in which this regulatory mechanism is absent, and recalculate the bifurcation diagram in the Le versus Ge parameter space. The result is shown in Fig. 4. As seen there, the absence of this mechanism moves the bifurcation region downwards, so the monostable induced region starts at Le values similar to those obtained when TMG is used as inducer (Fig. 2 A). If this mutant strain can be engineered, it may be possible to identify bistability with experiments like those of Ozbudak et al. (23).
FIGURE 4.
Bifurcation diagram, in the Le versus Ge parameter space, for an in silico mutant strain of E. coli in which the inducer-exclusion regulatory mechanism is absent. This bifurcation diagram was calculated with φM = 4.0 × 104 min−1.
DISCUSSION
We have developed a mathematical model of the lac operon that, though not accurate in all detail, captures the essential system behavior with respect to its bistable characteristics. The model reproduced the experimental results of Ozbudak et al. (23), who report the bifurcation diagram for the lac operon in E. coli, when induced with a nonmetabolizable inducer.
As mentioned above, there is currently a debate about whether bistability is a property of the lac operon under naturally occurring conditions. Ozbudak et al. reported that bistability was not observed when the natural inducer, lactose, was employed. Later, van Hoek and Hogeweg (22) simulated the in silico evolution of a large set of lac operons (under conditions of fluctuating external lactose and glucose), and concluded that bistability is present for artificial inducers, but not for lactose. Moreover, Narang and Pilyugin (26) argued, from a modeling study, that E. coli can exhibit diauxic growth when growing in a mixture of sugars, even if bistability is absent. To gain further insight into this matter, we analyzed the influence of lactose metabolism on the system bistable behavior, by varying the parameter φM (the maximum lactose-to-allolactose metabolism rate). Below, we discuss how our results provide a possible explanation to such apparent absence of bistability, and suggest that bistability does not disappear because of lactose metabolism, although it is highly modified.
In terms of external lactose, the bistability region moves upwards in the Le versus Ge parameter space, as φM increases. If φM > 2.5 × 104 min−1, bistability is not present in the limit of very low Ge, and it only appears after Ge surpasses a given threshold, via a cusp catastrophe. We suspect that this last situation is the most likely in wild-type bacteria, because it becomes practically impossible for the operon to switch into the induced state when there is an abundance of glucose. Moreover, there is no resistance, due to bistability, to activate the lac genes when glucose is absent, and so to employ the energy provided by lactose.
The fact that Ozbudak et al. (23) did not observe signs of bistability when lactose is used as an inducer suggests that 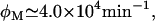 or higher. In that case, even when Le is as high as 1000 μM, the (Ge, Le) point lies most of the time in the monostable uninduced region of the bifurcation diagram (see Fig. 2 D), or it is very close to it, and thus bistability should be very hard to detect. Our stochastic simulations support this assertion given that, of all the experiments we carried out, only that corresponding to Ge = 40 μM and Le = 1000 μM rendered a bimodal like distribution.
or higher. In that case, even when Le is as high as 1000 μM, the (Ge, Le) point lies most of the time in the monostable uninduced region of the bifurcation diagram (see Fig. 2 D), or it is very close to it, and thus bistability should be very hard to detect. Our stochastic simulations support this assertion given that, of all the experiments we carried out, only that corresponding to Ge = 40 μM and Le = 1000 μM rendered a bimodal like distribution.
According to Ozbudak et al., whenever a bimodal distribution is found, it is considered a proof of bistability. More precisely, having a bimodal distribution is a sufficient but not a necessary condition for this behavior: for instance, if we have large standard deviations, a bimodal distribution will hardly be noticed even is bistability is present. Then, our numeric experiments confirm that—if 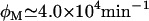 —identifying bistability via expression-level distributions is very difficult. We have also shown that if a mutant strain of E. coli in which inducer exclusion is absent or deficient can be found, it may be possible to identify bistability with the methods of Ozbudak et al., even if the natural, induced lactose is employed.
—identifying bistability via expression-level distributions is very difficult. We have also shown that if a mutant strain of E. coli in which inducer exclusion is absent or deficient can be found, it may be possible to identify bistability with the methods of Ozbudak et al., even if the natural, induced lactose is employed.
E. coli and other bacteria can feed on both lactose and glucose. However, when they grow in a medium rich in both sugars, glucose is utilized before lactose starts being consumed. This phenomenon, known as diauxic growth (27,28), represents an optimal thermodynamic solution given that glucose is a cheaper energy source than lactose since, to take advantage of lactose, the bacteria needs to expend energy in producing the enzymes needed to transport and metabolize this sugar (29).
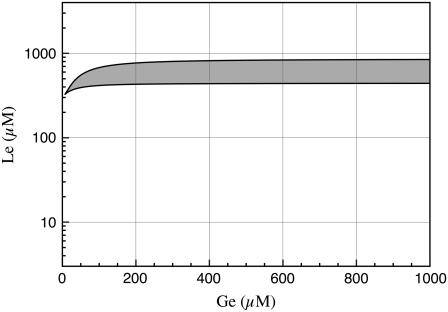
Consider a bacterial culture growing in a medium containing a mixture of glucose and lactose. According to the bifurcation diagram in Fig. 2 D, induction of the lac operon, as glucose is exhausted, can take place in two different ways: if Le < 600 μM, the cells undergo a smooth transition from the uninduced to the induced state; otherwise, the lac operon shifts abruptly from the off- to the on-state when glucose is almost completely depleted.
To better understand the behaviors described in the previous paragraph, we carried out numeric experiments in which the lactose operon is induced by changing the bacterial culture from a medium rich in glucose (in which it has been growing for a long time) to a medium with no glucose, while the lactose concentration is kept constant. Such simulations were performed by numerically solving the model differential equations with the Runge-Kutta method, implemented in Octave's algorithm, lsode. Our results (not shown) indicate that it takes >1000 min for the lac operon to become fully induced when Le = 300 μM, whereas when Le = 1000 μM, the operon achieves a 97% induction level 300 min after the medium shift.
From these results we conclude that, with a very high external lactose concentration, the lac operon can remain fully uninduced until glucose utilization is almost completely given up, because the response time is short in these conditions. That is, the most efficient performance consists of an abrupt change from the off- to the on-state, driven by a small variation on the external glucose concentration, if Le is very high. An analysis of Fig. 2 D reveals that, if Le = 1000 μM, the system goes from the uninduced to the induced monostable regions when Ge decreases from 210 to 25 μM.
We have argued that bistability helps to guarantee an efficient performance of the lac operon in E. coli, when feeding on glucose, lactose, or both sugars. Could a regulatory pathway involving only monostability be equally efficient? The lac operon in E. coli is an inducible operon. This means that it is subject to positive feedback regulation through lactose (allolactose). In our model, this is accounted for by the fact that 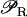 in Eq. 1 is an increasing function of A. Therefore, the higher the intracellular level of lactose, the higher the transcription initiation rate of the lactose operon genes. Depending on the functional form of
in Eq. 1 is an increasing function of A. Therefore, the higher the intracellular level of lactose, the higher the transcription initiation rate of the lactose operon genes. Depending on the functional form of 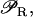 we can have either bistability for some parameter values, or a unique single stable steady state for all the parameters. In general,
we can have either bistability for some parameter values, or a unique single stable steady state for all the parameters. In general, 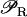 needs to be highly sigmoidal to have bistability.
needs to be highly sigmoidal to have bistability.
Expression of the lac operon is modulated by external glucose through the function 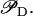 On the other hand, by analyzing the model steady state we can see that as Ge decreases from 210 to 25 μM, the system jumps from the uninduced to the induced states, and the L* steady-state value increases by a factor of 56. Substituting all these values into Eqs. 7 and 8, we conclude that an incremental increase of
On the other hand, by analyzing the model steady state we can see that as Ge decreases from 210 to 25 μM, the system jumps from the uninduced to the induced states, and the L* steady-state value increases by a factor of 56. Substituting all these values into Eqs. 7 and 8, we conclude that an incremental increase of 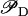 from 0.14 to 0.26 drives the 56-fold increase of L*. That is, the following amplification relation is observed:
from 0.14 to 0.26 drives the 56-fold increase of L*. That is, the following amplification relation is observed:
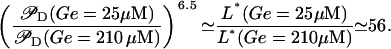 |
It is well known, since the invention of the regenerative circuit in the early decades of the 20th Century, that very large amplifications can destabilize a monostable system subject to positive feedback (30–32), and that the only way to get large amplifications is to approach as much as possible to the stability limit. In Appendix F we analyze the possibility of having large amplifications, with monostable regulation, in the present model of the lac operon. There we show that the stability of the steady state is highly compromised whenever the amplification exponent is >4. Since we predict an amplification exponent of 6.5, larger than four, the stability of our system would be at risk if it were controlled by a monostable pathway. In Appendix F we also demonstrate that bistability (multistability) allows both large amplification and strongly stable steady states. Hence, since a large amplification of the lac operon expression level (driven by a small change in 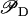 ) is advantageous for E. coli (when Le is very high), we conclude that bistability ensures an efficient consumption of lactose and glucose without jeopardizing the system stability.
) is advantageous for E. coli (when Le is very high), we conclude that bistability ensures an efficient consumption of lactose and glucose without jeopardizing the system stability.
Acknowledgments
We thank P. Swain for invaluable comments and suggestions.
This research was supported by the Natural Sciences and Engineering Research Council (grant No. OGP-0036920, Canada), Mathematics of Information Technology and Complex Systems (Canada), Consejo Nacional de Ciencia y Tecnología (CONACyT, Mexico), Comisión de Operación y Fomento de Actividades Académicas del Instituto Politécnico Nacional (COFAA-IPN, Mexico), and Estímulo al Desempeño de la Investigación del Instituto Politécnico Nacional (EDI-IPN, Mexico), and partially carried out while M.S. was visiting McGill University in 2005.
APPENDIX A: MODEL DEVELOPMENT
In this section, a mathematical model of the lac operon in E. coli is developed. The reader may find it convenient to refer to Fig. 1 of the main text, where the lac operon regulatory mechanisms are schematically represented. The model presented here accounts for the time evolution of the following variables: intracellular mRNA (M), lacZ polypeptide (E), and intracellular lactose (L) concentrations. The dynamics of these variables are modeled by the (balance) differential equations:
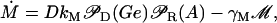 |
(14) |
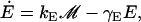 |
(15) |
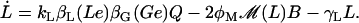 |
(16) |
The meaning of the functions and parameters in these equations is as follows:
A is the intracellular allolactose concentration.
D is the concentration of lac promoter within the bacterium.
Q and B are the permease and β-galactosidase concentrations, respectively.
kM, kE, and kL are, respectively, the lac promoter transcription initiation rate, the lacZ mRNA translation initiation rate, and the maximum lactose uptake rate per permease.
γM, γE, and γL, respectively, stand for the dilution plus degradation rates of M, E, and L.
Ge and Le denote the external glucose and lactose concentrations.
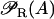 is the probability that promoter P1 is not repressed, while
is the probability that promoter P1 is not repressed, while 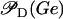 takes into account the effect of external glucose on the probability of having an mRNA polymerase bound to this promoter (catabolite repression).
takes into account the effect of external glucose on the probability of having an mRNA polymerase bound to this promoter (catabolite repression).βL(Le) and βG(Ge) are functions that account for the modulation of lactose uptake as functions of the external lactose and glucose (inducer exclusion) concentrations, respectively.
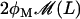 represents the rate of lactose metabolization per β-galactosidase molecule.
represents the rate of lactose metabolization per β-galactosidase molecule.
Probability of having a polymerase bound to the lac promoter, including catabolite repression
Let P and C, respectively, denote the mRNA polymerase and CAP concentrations. A polymerase can bind by itself to the lac promoter, P1. However, the affinity of this reaction is increased when a CAP molecule is bound to its corresponding binding site in the DNA chain. By taking this cooperative behavior into account, as well as the results in Appendices B and C, the probability PD can be calculated as
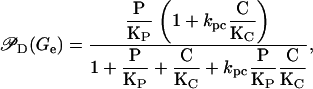 |
(17) |
where KP and KC are the respective dissociation constants of the polymerase-promoter and CAP-DNA complexes, and kpc > 1 is a constant that accounts for the cooperative interaction between the promoter and the CAP binding site. Equation 17 can be rewritten as
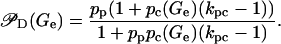 |
(18) |
In this equation, pp and pc(Ge), respectively, denote the probabilities that a polymerase is bound to the promoter in the absence of CAP, and that a CAP molecule is bound to its binding site in the absence of mRNA polymerase. The probability pp is constant and given by
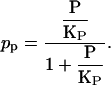 |
However,
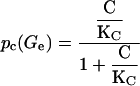 |
is a function of the external glucose concentration. Experimentally pc(Ge) must be a decreasing function of Ge since external glucose inhibits the synthesis of cAMP, which in turns implies a decrease in C and thus in pc(Ge). Here we assume that the functional form for pc(Ge) is given by
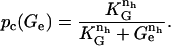 |
(19) |
Probability that the promoter P1 is not repressed
The lac operon has three different operator regions denoted by O1, O2, and O3 all of which can bind active repressor and are involved in transcriptional regulation. A repressor bound to O1 avoids transcription initiation. Conversely, a repressor bound to either O2 or O3 does not seem to affect transcription. Nevertheless, DNA can fold in such a way that a single repressor simultaneously binds two operators in all possible combinations. These complexes are more stable than that of a repressor bound to a single operator, and all of them inhibit transcription initiation (33). From the results in Appendix B, the probability that the lac operon promoter is not repressed can then be calculated as
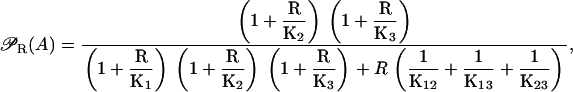 |
(20) |
where R is the concentration of active repressors, Ki (i = 1, 2, 3) is the dissociation constant of the R-Oi complex, and Kij (i, j = 1, 2, 3, i < j) is the dissociation constant of the Oi-R-Oj complex.
Define ρ(A) = R/K1, ξi = K1/Ki (i = 2, 3), and 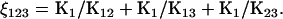 With these definitions, Eq. 20 can be rewritten as
With these definitions, Eq. 20 can be rewritten as
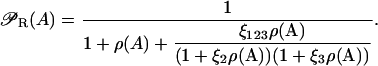 |
(21) |
Repressor molecules are tetramers and each of their subunits can be bound by an allolactose molecule, inactivating the repressor. The concentration of active repressors as a function of the allolactose concentration is given by Eq. 19,
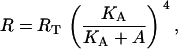 |
where RT is the total repressor concentration, and KA is the allolactose-repressor subunit complex dissociation rate. Since ρ(A) = R/K1, it follows that
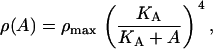 |
(22) |
where ρmax = RT/K1.
Inducer uptake rate
From the results of Ozbudak et al. (23), in the absence of external glucose the dependence of the normalized inducer uptake rate, per β-permease molecule, on the external inducer concentration is given by
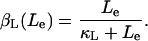 |
(23) |
The results of Ozbudak et al. (23) also allow us to model the decrease in the inducer uptake rate caused by external glucose by
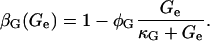 |
(24) |
Lactose metabolism
After being transported into the bacterium, lactose is metabolized by β-galactosidase. Approximately half of the lactose molecules are transformed into allolactose, while the rest are directly used to produce galactose. The lactose-to-allolactose metabolism rate, which as explained above equals the lactose-to-galactose metabolism rate, can be modeled as a Michaelis-Menten function, typical of catalytic reactions
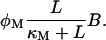 |
Therefore, the total rate of lactose metabolized per single β-galactosidase is
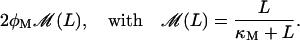 |
(25) |
Allolactose metabolism
Allolactose and lactose are both metabolized into galactose by the same enzyme β-galactosidase. Under the assumption that the allolactose production and metabolism rates are much faster than the cell growth rate, we may assume that these two metabolic processes balance each other almost instantaneously so
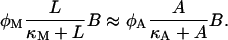 |
On the other hand, allolactose is an isomer of lactose, and therefore we can expect that the parameters related to the metabolism kinetics of both sugars attain similar values: φM ≈ φA and κM ≈ κA. Then
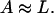 |
(26) |
Permease and β-galactosidase concentrations
The translation initiation and degradation rates of lacZ and lacY are slightly different. Here, we assume they are the same for the sake of simplicity. From this and the fact that β-galactosidase is a homotetramer, made up of four lacZ polypeptides, while permease consists of a single lacY polypeptide, it follows that
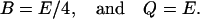 |
(27) |
APPENDIX B: BINDING-STATE PROBABILITY DISTRIBUTION FOR A MOLECULE WITH MULTIPLE, INDEPENDENT, BINDING SITES
Consider a molecule D with specific, independent, binding sites for N different molecules Bi, i = 1…N. Denote the binding state of molecule D by (n1, n2, …nN) = ({ni}), where ni = 1 if a molecule Bi its bound to its corresponding site in molecule D, and ni = 0 otherwise. In what follows, we demonstrate by induction that the probability of any given binding state is
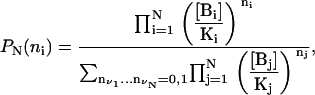 |
where [Bi] is the concentration of chemical species Bi, and Ki is the dissociation constant of the D:Bi complex.
Case N = 1
The reaction leading to the formation of complex D:B1 is
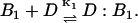 |
At equilibrium, the concentrations of the chemical species involved in this reaction satisfy the relation
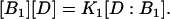 |
(28) |
Assume a constant total concentration for species D, i.e.,
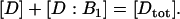 |
(29) |
Then, from Eqs. 28 and 29, the fractions of free and bound B1 sites are respectively given by
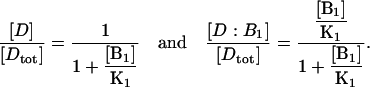 |
From these results, the binding-state probability distribution for the current system can be written as
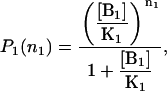 |
(30) |
where n1 = 0 if the B1 binding site is empty, and n1 = 1 otherwise.
Case N = 2
From the N = 1 case, the probability distribution for the B1 binding site in the absence of species B2 is
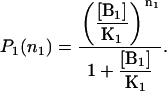 |
Also, the probability distribution for the B2 binding site in the absence of species B1 is
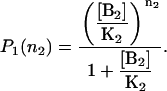 |
If both sites are independent, their joint binding-state probability distribution is
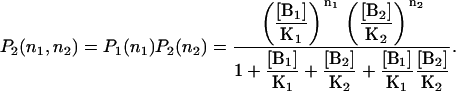 |
(31) |
General case
Take a D molecule with N independent binding sites and assume that the binding-state probability distribution for the first N–1 sites is given, in the absence of species BN, by
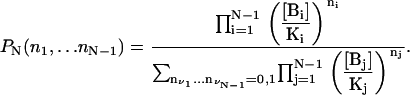 |
In the absence of species B1, …BN–1, the probability of the BN binding site is
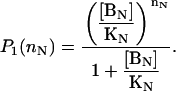 |
Since all sites are independent from each other, the binding-state probability distribution can be calculated as
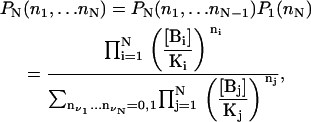 |
(32) |
which proves our original assertion.
APPENDIX C: COOPERATIVITY BETWEEN BINDING SITES
We can see from Eq. 32 that the probability of finding ni = 0, 1 molecules Bi bound to its corresponding binding site is proportional to 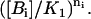 The denominator in the fraction of Eq. 32 plays the role of a normalizing constant.
The denominator in the fraction of Eq. 32 plays the role of a normalizing constant.
Assume that two given sites, say l and m, have a cooperative interaction in the sense that the probability of finding the two of them bound by their respective molecules is larger than the product of the individual probabilities. From this and the considerations of Appendix B, the binding-state probability distribution accounting for cooperativity between sites l and m is
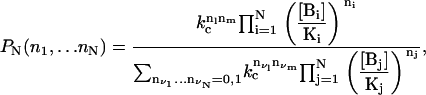 |
(33) |
where kc > 1 measures the strength of the cooperativity.
APPENDIX D: PARAMETER ESTIMATION
Growth rate, μ
The growth rate of a bacterial culture depends strongly on the growth medium. Typically, the mass doubling time varies from 20 to more than 40 min (34). For the purpose of this study, we take a doubling time of 30 min, which corresponds to the growth rate μ ≈ 0.02 min−1.
Lac promoter concentration
According to Bremmer and Dennis (34), there are ∼2.5 genome equivalents per average E. coli cell at the growth rate determined by μ. For the purpose of this work, we take 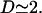
Production rates
Malan et al. (35) measured the transcription initiation rate at the lac promoter and report kM ≈ 0.18 min−1.
From Kennell and Riezman (36), translation of the lacZ mRNA starts every 3.2 s. According to Beckwith (37), the production rate of lac permease is smaller than that of β-galactosidase monomers even though, as Kennell and Riezman (36) report, there are similar levels of both mRNA species. This suggests that lacY mRNA values are translated at a lower rate than is lacZ mRNA. Nevertheless, to our knowledge, there are no reported measurements of the lacY mRNA translation initiation rate. Thus, we assume it is equal to that of lacZ: 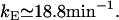
According to Chung and Stephanopoulos (10), the inducer uptake rate per lac permease is 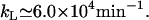
Degradation rates
Kennell and Riezman (36) measured a lacZ mRNA half-life of 1.5 min. That is, its degradation rate is 0.46 min−1, and the corresponding dilution plus degradation rate is 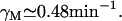
According to Kennell and Riezman (36), the lac permease degradation rate is 0.01 min−1. Thus, its degradation plus dilution rate is 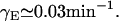
Here we assume that the lactose degradation rate is negligible and so its degradation plus dilution rate is simply 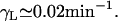
Catabolite repression parameters
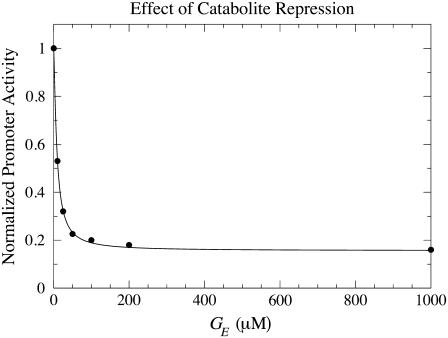
Malan et al. (35) measured the polymerase-lac promoter affinity in the presence and absence of cAMP. From their data, we estimate 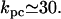
The parameters pp, KG, and nh were estimated by fitting Eq. 5 of the main text (with pc as given by Eq. 6) to the experimental data reported in Ozbudak et al. (23). See Fig. 5. The values we obtained are 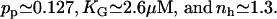
FIGURE 5.
Effect of external glucose, due to catabolite repression, on the activity of the lac promoter. The dots stand for the experimental data of Ozbudak et al. (23), while the solid line was drawn from Eq. 5, with pc as given by Eq. 6, with the estimated parameter values.
Repression parameters
Oehler et al. (33) studied how the three operators in the lac operon cooperate in repression. According to their results, when only O1 and either O2 and O3 are present, the repression level is reduced to 53.85% and 33.85% that of the wild-type operon, respectively. Moreover, when O2 and O3 are destroyed, repression is reduced to 1.38% that of the wild-type operon. This allowed us to estimate the parameters ξ2, ξ3, and ξ123 as 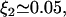
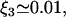 and
and 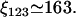
For this, we took into consideration that eliminating O2 (O3) is equivalent to making 1/K2 = 1/K12 = 1/K23 = 0 (1/K2 = K13 = 1/K23 = 0) in Eq. 7 of the main text. Parameters ξ2, ξ3, and ξ123 are modified accordingly in Eq. 8 of the main text.
The parameters ρ max and KA were estimated by fitting the curves in the model bifurcation diagram to the experimental results of Ozbudak et al. (23) (see Fig. 2 A of the main text) obtaining the values 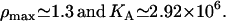
Inducer uptake parameters
From the experimental data of Ozbudak et al. (23), the inducer uptake rate per active permease, as a function of external inducer concentration, can be fitted by a Michaelis-Menten function with a half-saturation concentration of 680 μM. That is, 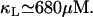
Ozbudak et al. (23) measured the inducer-uptake-rate decrease per active permease as a function of external glucose concentration. We found (plot not shown) that these data are well fit by Eq. 11 of the main text with 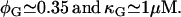
Lactose metabolism parameters
Lactose metabolism rate and saturation constant, φM and κM. We estimate these parameters from the data reported in Martínew-Bilbao et al. (38) as φM ≈ 3.60 × 103 min−1, and κM ≈ 7.0 × 105 mpb, where mpb stands for molecules per average bacterium.
APPENDIX E: STEADY-STATE STABILITY AND BIFURCATION DIAGRAMS
Stability analysis
The model equations (Eqs. 1–3) in the main text can be rewritten as
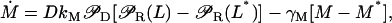 |
(34) |
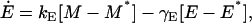 |
(35) |
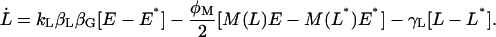 |
(36) |
The steady-state values M*, E*, and L* are, respectively, given by
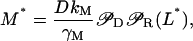 |
(37) |
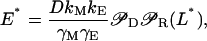 |
(38) |
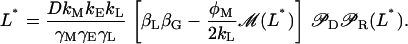 |
(39) |
It is easy to show numerically that given the parameter values tabulated in Table 2 and Ge > 0, Eq. 39 may have up to three roots, and therefore that the model has up to three steady states.
Notice that, except for 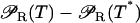 and
and 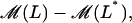 all terms in the right-hand side of Eqs. 34–36 are linear with respect to ℳ−ℳ*, E −E*, and L−L*. Since we are only interested in analyzing the system dynamic behavior in a small neighborhood around the steady states, the following linear approximation is employed:
all terms in the right-hand side of Eqs. 34–36 are linear with respect to ℳ−ℳ*, E −E*, and L−L*. Since we are only interested in analyzing the system dynamic behavior in a small neighborhood around the steady states, the following linear approximation is employed:
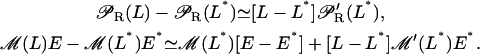 |
With this approximation, the full model can be reduced (around the steady state) to the following system of linear differential equations,
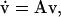 |
(40) |
where
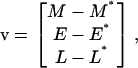 |
and
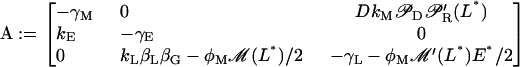 |
The general solution of the linear system (Eq. 40) is 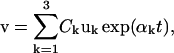 where Ck are undetermined constants, while αk and uk are the respective eigenvalues and eigenvectors of matrix A. Therefore, if all eigenvalues αk have negative real parts, the corresponding steady state is stable. Conversely, the steady state is unstable if at least one eigenvalue has a positive real part.
where Ck are undetermined constants, while αk and uk are the respective eigenvalues and eigenvectors of matrix A. Therefore, if all eigenvalues αk have negative real parts, the corresponding steady state is stable. Conversely, the steady state is unstable if at least one eigenvalue has a positive real part.
The eigenvalues of A are the roots of its characteristic polynomial, det(sI – A). It follows after some algebra that det(sI – A) = P(s) – Q3, with
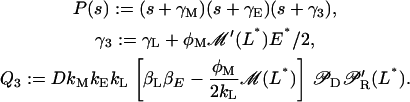 |
Notice that γ3 > 0 because γL > 0 and M is an increasing function; moreover γM > γE > 0. Hence, the polynomial P(s) is positive and strictly increasing for every real s ≥ 0. Assume now that Q3 > P(0), then, P(s) – Q3 has one positive real root since P(0) – Q3 < 0 and P(s) is strictly increasing for s > 0.
On the other hand, we assert that all roots of P(s) – Q3 have negative real part whenever Q3 < P(0). It follows from the work of Strelitz (39) that all roots have negative real part if and only if the following pair of inequalities hold simultaneously:
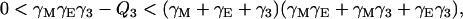 |
and we obviously have that
 |
Therefore, the linear system 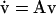 is stable (unstable) whenever Q3 < P(0) (Q3 > P(0)). Furthermore, since the steady-state values of L* are the roots of the equation
is stable (unstable) whenever Q3 < P(0) (Q3 > P(0)). Furthermore, since the steady-state values of L* are the roots of the equation 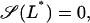 with
with
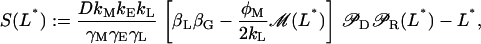 |
(41) |
we can easily calculate that a given steady state is locally stable (unstable) whenever the derivative
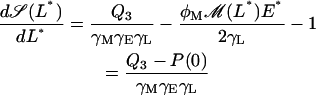 |
(42) |
is strictly negative (positive).
Calculation of the bifurcation points and the bifurcation diagram
Given Ge, the values of L* and Le at which a saddle node bifurcation occurs can be calculated by simultaneously solving 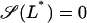 and
and 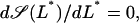 with
with 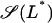 as defined in Eq. 41. The bifurcation diagram in the Le versus Ge parameter space can be calculated by repeating the above procedure for several values of Ge.
as defined in Eq. 41. The bifurcation diagram in the Le versus Ge parameter space can be calculated by repeating the above procedure for several values of Ge.
APPENDIX F: POSITIVE FEEDBACK, STABILITY AND HYSTERESIS
Consider the following simplified model of a biological switch with nonlinear feedback,
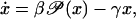 |
(43) |
where γ > 0 is a degradation parameter, β > 0 is an external control parameter, and the smooth function 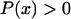 is nonlinear with respect to x. The steady states x* of Eq. 43 are the solutions to
is nonlinear with respect to x. The steady states x* of Eq. 43 are the solutions to
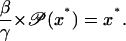 |
(44) |
It is easy to prove that the dynamic system given by Eq. 43 is locally stable at x* if and only if
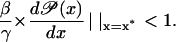 |
(45) |
We choose system Eq. 43 because its dynamic behavior is representative of (and similar to) the behavior of higher dimensional systems. For example, consider the following two-dimensional system, with positive parameters βk > 0 and γk > 0:
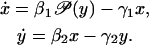 |
The steady states x* and y* are the solutions of
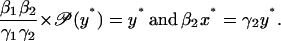 |
These equations are equivalent to Eq. 44 with β = β1β2 and γ = γ1γ2. Furthermore, the two dimensional system is locally stable at x* and y* if and only if both eigenvalues of the matrix
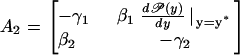 |
have strictly negative real parts. Simple algebraic calculations allows us to prove that both eigenvalues of A2 have strictly negative real part if and only if
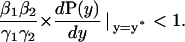 |
Notice that this last result is equivalent to Eq. 45, after setting the degradation parameter γ = γ1γ2 and the external control parameter β = β1β2. Similar results holds for higher dimensions.
Consider the model equations (Eqs. 1–3) with the external glucose Ge values in the interval [25, 210] μM, the external lactose Le = 103 μM and φM = 4 × 104 min−1. It is easy to show, under the previous conditions, that the steady state L* achieves the minimum and maximum values of 4.5 × 105 and 2.53 × 107 molecules per average bacterium, respectively. Therefore,
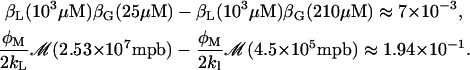 |
The above results indicate that the term βL(Le)βG(Ge) inside the square brackets of Eq. 41 is approximately constant and equal to 0.39. From this, and taking into consideration that the steady states L* are given by the roots 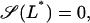 with S defined in Eq. 41, it follows that
with S defined in Eq. 41, it follows that
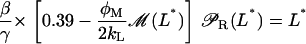 |
(46) |
with the control and degradation parameters defined as
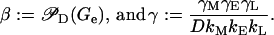 |
(47) |
On the other hand, we have from Eq. 42 that the steady-state L* is locally stable if and only if
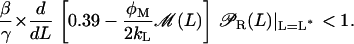 |
(48) |
To finish, notice that Eqs. 46–48 are completely equivalent to Eqs. 44 and 45.
Why hysteresis?
In this section we argue that hysteresis (multistability) is the only dynamic behavior compatible with positive feedback, robust stability, and high amplification factors.
Positive feedback is obtained by demanding that 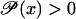 is strictly increasing with respect to x, and robust stability is introduced through a new safety parameter 0 < π < 1 in Eq. 45,
is strictly increasing with respect to x, and robust stability is introduced through a new safety parameter 0 < π < 1 in Eq. 45,
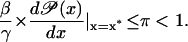 |
(49) |
The parameters in Eq. 43 vary largely due to changes in the experimental variables like temperature, pH, salinity, etc. Hence, robust stability is obtained by setting the safety bound 0 < π < 1 in Eq. 49; a simple estimation yields π ≤ 3/4. If the derivative 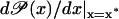 is very close to γ/β, any fluctuation could make
is very close to γ/β, any fluctuation could make 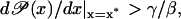 and Eq. 43 will become unstable.
and Eq. 43 will become unstable.
Assume that Eq. 43 is monostable, and thus that Eq. 44 has one single solution (steady state) x*(β) for every external control β > 0. Assume also that this steady state is locally stable and satisfies the robust stability inequality (Eq. 49). Finally, suppose that the external control variable β > 0 lies in the closed interval a ≤ β ≤ b. Notice that x*(b) > x*(a), since 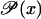 is strictly increasing with respect to x. We shall investigate now what the relation between b/a and x*(b)/x*(a) is.
is strictly increasing with respect to x. We shall investigate now what the relation between b/a and x*(b)/x*(a) is.
Rewrite Eq. 44 under the assumption that the steady-state x*(β) is a function β > 0
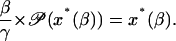 |
Differentiate this equation with respect to β to give
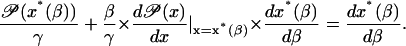 |
Rearranging terms and using the bound in Eq. 49 yields
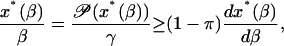 |
then
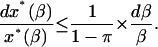 |
Finally, integrating in the interval a ≤ β ≤ b, yields
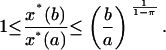 |
(50) |
It follows from the previous results that the hypotheses of monostability, positive feedback, and robust stability, with a safety parameter of π ≤ 3/4, yield an amplification exponent in Eq. 50 of at most 1/(1 – π) ≤ 4. This amplification exponent is quite small if we expect large amplification, so we have to make sacrifices. Presumably, robust stability is not a condition to be sacrificed.
A solution for obtaining large amplification exponents in Eq. 50 is to have a sigmoid function 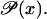 This allows the derivative
This allows the derivative 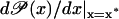 to be almost zero for two steady states, x*, located outside some interval [x1, x2], with x2 > x1 > 0. Conversely
to be almost zero for two steady states, x*, located outside some interval [x1, x2], with x2 > x1 > 0. Conversely 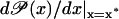 can be ≫1 for a third steady state x* ∈ [x1, x2]. The large value of the derivative inside the interval [x1, x2] permits very large amplification exponents. On the other hand, even when the steady state x* ∈ [x1, x2] is unstable, the system is still stable because the large steady state, x* > x2, and the small one, x* < x1, are both stable.
can be ≫1 for a third steady state x* ∈ [x1, x2]. The large value of the derivative inside the interval [x1, x2] permits very large amplification exponents. On the other hand, even when the steady state x* ∈ [x1, x2] is unstable, the system is still stable because the large steady state, x* > x2, and the small one, x* < x1, are both stable.
In conclusion, only if the function 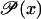 is sigmoid, and the system equation (Eq. 43) shows bistability, can we simultaneously have very large amplifications and robust stability.
is sigmoid, and the system equation (Eq. 43) shows bistability, can we simultaneously have very large amplifications and robust stability.
References
- 1.Rietkerk, M., S. F. Dekker, P. C. de Ruiter, and J. van de Koppel. 2004. Self-organized patchiness and catastrophic shifts in ecosystems. Science. 305:1926–1929. [DOI] [PubMed] [Google Scholar]
- 2.Ferrell, J. E., Jr., and E. M. Machleder. 1998. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 280:895–898. [DOI] [PubMed] [Google Scholar]
- 3.Bagowski, C. P., and J. E. Ferrell. 2001. Bistability in the JNK cascade. Curr. Biol. 11:1176–1182. [DOI] [PubMed] [Google Scholar]
- 4.Bhalla, U. S., P. T. Ram, and R. Iyengar. 2002. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 297:1018–1023. [DOI] [PubMed] [Google Scholar]
- 5.Cross, F. R., V. Archambault, M. Miller, and M. Klovstad. 2002. Testing a mathematical model of the yeast cell cycle. Mol. Biol. Cell. 13:52–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomerening, J. R., E. D. Sontag, and J. E. Ferrell, Jr. 2003. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat. Cell Biol. 5:346–351. [DOI] [PubMed] [Google Scholar]
- 7.Bagowski, C., J. Besser, C. R. Frey, and J. E. J. J. Ferrell, Jr. 2003. The JNK cascade as a biochemical switch in mammalian cells: ultrasensitive and all-or-none responses. Curr. Biol. 13:315–320. [DOI] [PubMed] [Google Scholar]
- 8.Sha, W., J. Moore, K. Chen, A. D. Lassaletta, C. S. Yi, J. J. Tyson, and J. C. Sible. 2003. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proc. Natl. Acad. Sci. USA. 100:975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slepchenko, B. M., and M. Terasaki. 2004. Bio-switches: what makes them robust? Curr. Opin. Genet. Dev. 14:428–434. [DOI] [PubMed] [Google Scholar]
- 10.Chung, J. D., and G. Stephanopoulos. 1996. On physiological multiplicity and population heterogeneity of biological systems. Chem. Eng. Sci. 51:1509–1521. [Google Scholar]
- 11.Ferrell, J. E. 2002. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback, and bistability. Curr. Opin. Chem. Biol. 6:140–148. [DOI] [PubMed] [Google Scholar]
- 12.Angeli, D., J. E. Ferrell, Jr., and E. D. Sontag. 2004. Detection of multistability, bifurcations, and hysteresis in a large class of biological positive-feedback systems. Proc. Natl. Acad. Sci. USA. 101:1822–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novick, A., and M. Weiner. 1957. Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. USA. 43:553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohn, M., and K. Horibata. 1959. Analysis of the differentiation and of the heterogeneity within a population of Escherichia coli undergoing induced beta-galactosidase synthesis. J. Bacteriol. 78:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maloney, L., and B. Rotman. 1973. Distribution of suboptimally induces −D-galactosidase in Escherichia coli. The enzyme content of individual cells. J. Mol. Biol. 73:77–91. [DOI] [PubMed] [Google Scholar]
- 16.Wong, P., S. Gladney, and J. D. Keasling. 1997. Mathematical model of the lac operon: inducer exclusion, catabolite repression, and diauxic growth on glucose and lactose. Biotechnol. Prog. 13:132–143. [DOI] [PubMed] [Google Scholar]
- 17.Vilar, J. M., C. C. Guet, and S. Leibler. 2003. Modeling network dynamics: the lac operon, a case study. J. Cell Biol. 161:471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildirim, N., and M. C. Mackey. 2003. Feedback regulation in the lactose operon: a mathematical modeling study and comparison with experimental data. Biophys. J. 84:2841–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santillán, M., and M. C. Mackey. 2004. Influence of catabolite repression and inducer exclusion on the bistable behavior of the lac operon. Biophys. J. 86:1282–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yildirim, N., M. Santillán, D. Horike, and M. C. Mackey. 2004. Dynamics and bistability in a reduced model of the lac operon. Chaos. 14:279–292. [DOI] [PubMed] [Google Scholar]
- 21.Tian, T., and K. Burrage. 2005. A mathematical model for genetic regulation of the lactose operon. In Lecture Notes in Computer Science, Vol. 3841. O. Gervasi, M. L. Gavrilova, V. Kumar, A. Laganà, H. P. Lee, Y. Mun, D. Taniar, and C. J. K. Tan, editors. Springer-Verlag, Berlin, Heidelberg.
- 22.van Hoek, M. J., and P. Hogeweg. 2006. In silico evolved lac operons exhibit bistability for artificial inducers, but not for lactose. Biophys. J. 91:2833–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozbudak, E. M., M. Thattai, H. N. Lim, B. I. Shraiman, and A. van Oudenaarden. 2004. Multistability in the lactose utilization network of Escherichia coli. Nature. 427:737–740. [DOI] [PubMed] [Google Scholar]
- 24.Gillespie, D. T. 2001. Approximate accelerated stochastic simulation of chemically reacting systems. J. Chem. Phys. 115:1716–1733. [Google Scholar]
- 25.Gillespie, D. T., and L. R. Petzold. 2003. Improved leap-size selection for accelerated stochastic simulation. J. Chem. Phys. 119:8229–8234. [Google Scholar]
- 26.Narang, A., and S. S. Pilyugin. 2007. Bacterial gene regulation in diauxic and non-diauxic growth. J. Theor. Biol. 244:326–348. [DOI] [PubMed] [Google Scholar]
- 27.Roseman, S., and N. D. Meadow. 1990. Signal transduction by the bacterial phosphotransferase system. J. Biol. Chem. 265:2993–2996. [PubMed] [Google Scholar]
- 28.Narang, A. 1998. The dynamical analogy between microbial growth on mixtures of substrates and population growth of competing species. Biotechnol. Bioeng. 59:116–121. [DOI] [PubMed] [Google Scholar]
- 29.Dekel, E., and U. Alon. 2005. Optimality and evolutionary tuning of the expression level of a protein. Nature. 436:588–592. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong, E. H. 1922. Some recent developments of regenerative circuits. Proc. Inst. Radio Eng. 10:244–260. [Google Scholar]
- 31.Britain, J. E. 2004. Electrical Engineering Hall of Fame—Edwin H. Armstrong. Proc. IEEE. 92:575–578. [Google Scholar]
- 32.Garnsback, H. 1924. The crystodyne principle. Radio News. 431:294–295. [Google Scholar]
- 33.Oehler, S., E. R. Eismann, H. Krämer, and B. Müller-Hill. 1990. The three operators of lac operon cooperate in repression. EMBO J. 9:973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bremmer, H., and P. P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell by growth rate. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, Vol. 2. F. C. Neidhart, R. Curtis, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger, editors. American Society of Microbiologists, Washington, DC.
- 35.Malan, T. P., A. Kolb, H. Buc, and W. R. McClure. 1984. Mechanism of CRP-cAMP activation of lac operon transcription initiation activation of the P1 promoter. J. Mol. Biol. 180:881–909. [DOI] [PubMed] [Google Scholar]
- 36.Kennell, D., and H. Riezman. 1977. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J. Mol. Biol. 114:1–21. [DOI] [PubMed] [Google Scholar]
- 37.Beckwith, J. 1987. The lactose operon. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, Vol. 2. F. C. Neidhart, J. L. Ingraham, K. B. Low, B. Magasanik, and H. E. Umbarger, editors. American Society of Microbiologists, Washington, DC.
- 38.Martínez-Bilbao, M., R. E. Holdsworth, R. A. Edwards, and R. E. Huber. 1991. A highly reactive β-galactosidase (Escherichia coli) resulting from a substitution of an aspartic acid for Gly-794. J. Biol. Chem. 266:4979–4986. [PubMed] [Google Scholar]
- 39.Strelitz, S. 1977. On the Routh-Hurwitz problem. Am. Math. Mon. 84:542–544. [Google Scholar]