Abstract
We present here the parmbsc0 force field, a refinement of the AMBER parm99 force field, where emphasis has been made on the correct representation of the α/γ concerted rotation in nucleic acids (NAs). The modified force field corrects overpopulations of the α/γ = (g+,t) backbone that were seen in long (more than 10 ns) simulations with previous AMBER parameter sets (parm94-99). The force field has been derived by fitting to high-level quantum mechanical data and verified by comparison with very high-level quantum mechanical calculations and by a very extensive comparison between simulations and experimental data. The set of validation simulations includes two of the longest trajectories published to date for the DNA duplex (200 ns each) and the largest variety of NA structures studied to date (15 different NA families and 97 individual structures). The total simulation time used to validate the force field includes near 1 μs of state-of-the-art molecular dynamics simulations in aqueous solution.
INTRODUCTION
Although the first molecular dynamics (MD) simulations of proteins were published in the late 1970s (1,2), the first restrained simulations of nucleic acids (NAs) did not appear until the mid-1980s (3,4), and reliable unrestrained simulations of these molecules were not possible until the mid-1990s, when new force fields were developed and methods for the proper representation of long-range electrostatic effects were incorporated into simulation codes (5–12). This time lag illustrates the technical problems intrinsic to the MD simulation of very charged and flexible polymers, such as NAs, in aqueous solution.
With these difficulties addressed, the last decade has seen a wide use of MD to study a very large number of NAs (13–18) in water for simulation periods in the range 1–50 ns. Most of these simulations have used explicit models of solvent and the particle mesh Ewald method (PME) (5) to account for long-range electrostatic effects. Although others are available, the force fields implemented in AMBER and CHARMM have been the most widely used (6–8,10). In particular, MD simulations using AMBER force fields have been shown to accurately reproduce the structural and dynamic properties of a large variety of canonical and noncanonical NAs in water (13–20). Moreover, they have satisfactorily described complex conformational changes such as the A → B transition in duplex and triplex DNAs (21–27) and have performed well in simulations of DNAs in extreme environments (28–30). Finally, several systematic studies have demonstrated the excellent ability of the standard AMBER force field to reproduce very high-level QM data for hydrogen bond and stacking interactions in the gas phase (31–36). Overall, these studies suggest that the AMBER force field is physically meaningful and retains a proper balance between intramolecular and intermolecular forces.
The latest versions of the AMBER force field, parm94 and parm99 (6,10), were parameterized when “state-of-the-art” simulations were on the 1-ns time scale and QM calculations were limited to small model systems and to moderate levels of theory. Quite surprisingly, both still perform well in simulations in the 10-ns range, which is the normal simulation period at the present time. However, in an extended MD simulation of a DNA duplex, Varnai and Zakrzewska (37) found massive α/γ transitions to the gauche+, trans geometry (away from the g−,g+ state), which introduced severe distortions in DNA in 50-ns trajectories. This effect, which was later found in other simulations by different groups, emerged as a general sequence-independent problem of parm99 or parm94 simulations (see simulations from the Ascona B-DNA consortium, http://humphry.chem.wesleyan.edu:8080/MDDNA, and more extensive simulations by our collaborative groups) (38–40). Fortunately, analysis of data shows that these errors are not very significant in shorter (10 ns or less) simulations and do not invalidate most previous simulations with these force fields, where none or only one of these transitions is evident. However, it is clear that this error needs to be corrected because within a very few years standard MD simulations of NAs will approach 100 ns in length, and in this range of simulation, massive irreversible α/γ transitions disrupt the duplex structure (see results below).
In this article we present a full reparameterization of the α/γ torsional term to derive a new AMBER force field, based on AMBER-parm99, which will be named parmbsc0. This new force field, not only appears to model accurately the structural and dynamic properties of a large variety of NAs over current MD simulation time scales (∼10 ns) but also provides very good representations of these structures in simulations 20 times longer. The extensive use of the Mare Nostrum supercomputer in Barcelona and supercomputing facilities in Brno and the Pittsburgh Supercomputing Center has allowed us to perform a comprehensive and intensive test of the force field (near 1 μs of unrestrained trajectories on 97 different structures, 2 of them 200 ns long), a study that is orders of magnitude greater than those reported in any previous parameterization work and that guarantees that this modified AMBER force field can be safely used to study NAs in a time scale at least one order of magnitude greater than current parm94 and parm99 versions.
METHODS
Reference quantum mechanical calculations
Many studies support the overall very satisfactory performance of the parm99 (or parm94) force field, with the most serious artifact reported so far being the α/γ imbalance occurring in longer B-DNA simulations (see Introduction). Thus, we have adopted a conservative reparameterization protocol in which we have modified the minimum number of parameters required to correct the α/γ conformational transition. In particular, the obvious choice was to reparameterize the α and γ torsional parameters. For this purpose, we chose an extended model (Fig. 1) to explore the potential energy surface (grid spacing 30°) associated with rotations around α and γ torsions. The model was chosen to place the α and γ torsions in a correct chemical environment while maintaining the simplicity needed to reduce potential sources of noise in the quantum mechanical calculations. The system was fully optimized (at both LMP2/6-31+G(d) and B3LYP/6-31+G(d) levels) (41–43) at each point of the potential energy surface except for α and γ (fixed at values of the grid) as well as δ, which was fixed at either B- (δ = 156.5°) or A- (δ = 84.0°) fibber values. As described below the use of these particular δ values for restraint instead of other possible values does not have any impact on the fitted parameters. In summary, four potential energy surfaces were built up to represent the α/γ space for DNA and RNA. As noted below, all these data were merged to improve the statistical quality of the fitting.
FIGURE 1.
Schematic representation of the molecular model used to parameterize α and γ torsions. The atom-type definition is also displayed.
To further test the quality of the force field potential, CCSD(T)/complete basis set calculations (44,45) were performed on the four minima regions. These calculations were carried out by reoptimizing the B3LYP geometry (keeping only δ constant at A- or B-standard values) at the MP2/aug-cc-pVDZ level. Single point calculations were then performed at the MP2/aug-cc-pVXZ (X = D,T) levels extrapolating to infinite basis set with the method developed by Halkier et al. (45). Finally, MP2 → CCSD(T) corrections were included using the 6-31+G(d) basis set.
Force field fitting
With our conservative approach, aimed to retain the beneficial features of the AMBER parm94-99 parameterizations, only torsional parameters involving α and γ torsions were refined, and all other parameters were kept at standard parm99 values (charges for the model system used in the parameterization were determined from standard RESP/6-31G(d) (46) calculations in AMBER). The residual energy (Eq. 1) was fitted to an extended Fourier series (Eq. 2), where the barrier and the phase angle for each periodicity (1, 2, or 3) term were adjusted to obtain the minimum error. Note that the use of the Fourier expansion has no physical foundation and is just a simple empirical correction useful to fit residual QM-classical energies.
In principle, although any dihedral angle(s) can be used to fit a torsion, we chose to follow the standard nomenclature using the O3′-P-O5′-C5′ and O5′-C5′-C4′-C3′ atoms to represent α and γ dihedrals. This differs slightly from the original parm99 force field, where the γ torsion is defined by the O5-C5′-C4′-O4′ atoms using the same set of atom types (OS-CT-CT-OS) as the sugar ring torsion O4′-C4′-C3′-O3′ and all other anomeric torsions. To avoid altering other conformational profiles (such as that of sugar puckering) a new atom type (CI) was introduced and assigned to C5′. Defining a new atom type for the C5′ makes intuitive sense because it is expected that the O5′-C5′-C4′-O4′ anomeric torsion, adjacent to the phosphorus, should be distinct from the standard (OS-CT-CT-OS) anomeric torsion.
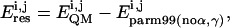 |
(1) |
where i,j stand for a combination of α/γ torsions, and parm99 (no α,γ) means a parm99 calculation with all standard parameters but those involving α/γ set to zero. All energy values are referred to a common structure.
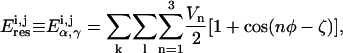 |
(2) |
where k stands for a torsion, α or γ, l stands for the number of dihedral angles (Φ) used to describe this torsion, and ζ is the phase angle.
Multiple different fitting algorithms were explored. A direct nonlinear fitting of the residual plot was initially investigated and discarded because it was very prone to errors as a result of its very high energy points, which, although never sampled in MD simulations, tend toward an excessive weight in the fitting at the expense of minima or flat regions. To overcome these problems, we developed a very flexible Metropolis-Monte Carlo program, where the merit function (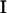 ) is not the variance but the total absolute deviation (reducing the impact of outliers) as in a robust regression. The introduction of weighting factors at each point (
) is not the variance but the total absolute deviation (reducing the impact of outliers) as in a robust regression. The introduction of weighting factors at each point (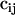 in Eq. 3) allowed us to easily maximize the quality of the fitting in especially important regions or to mix data from different sources, giving each different importance in the fitting (for example, giving more emphasis to data of higher quality). During the initial fittings we realized that very similar values were obtained from DFT and LMP2 reference data and also when models simulating DNA or RNA geometries were considered. Thus, in the final fitting we use this similarity and combined all the data (the weight of LMP2 data was 50% higher than DFT data) into a single set, which was enriched by introducing minima obtained by optimizing (only δ-restrained) the systems in the four distinct minima regions. To guarantee that the minima and the “artifactual α/γ region” (specifically the region that is significantly overpopulated in parm94 and parm99 simulations) were properly represented, points in these regions were assigned an overweight of 100% over the rest. Note that our procedure allows phase angles to be optimized instead of keeping them fixed at 0/180° as usual in AMBER. The relaxation of phase angles provides some improvement in the fitted maps, especially in canonical regions without any increase in the complexity of the Fourier functional used for fitting the residual energy (see Eq. 2).
in Eq. 3) allowed us to easily maximize the quality of the fitting in especially important regions or to mix data from different sources, giving each different importance in the fitting (for example, giving more emphasis to data of higher quality). During the initial fittings we realized that very similar values were obtained from DFT and LMP2 reference data and also when models simulating DNA or RNA geometries were considered. Thus, in the final fitting we use this similarity and combined all the data (the weight of LMP2 data was 50% higher than DFT data) into a single set, which was enriched by introducing minima obtained by optimizing (only δ-restrained) the systems in the four distinct minima regions. To guarantee that the minima and the “artifactual α/γ region” (specifically the region that is significantly overpopulated in parm94 and parm99 simulations) were properly represented, points in these regions were assigned an overweight of 100% over the rest. Note that our procedure allows phase angles to be optimized instead of keeping them fixed at 0/180° as usual in AMBER. The relaxation of phase angles provides some improvement in the fitted maps, especially in canonical regions without any increase in the complexity of the Fourier functional used for fitting the residual energy (see Eq. 2).
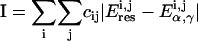 |
(3) |
Molecular dynamics simulations
A complete set of simulations (Table 1) was carried out to validate the parmbsc0 force field. Dickerson's dodecamer (DD) (47) was the first benchmarking model and was simulated in two independent trajectories of 0.2 μs each. Results were compared with equivalent trajectories obtained with parm94 and parm99 force fields. This extended simulation time (to our knowledge, the longest trajectory for DD ever published) is sufficient to reveal structural degradation with old force fields, which was not so evident in standard 5- to 10-ns trajectories. We also performed a number of short (3-ns) MD simulations on the Nottingham database of DNA duplexes (A, B, and Z) for which crystal structures are available and that show in some cases unusual α/γ combinations (see http://holmes.cancres.nottingham.ac.uk/∼charlie/autoDNA/NDB). In addition we carried out a massive analysis of the performance of the new parameter set for different RNAs and for various “exotic” NAs such as triplexes, quadruplexes, and Hoogsteen DNAs, for all of which parm94/99 performs well on the 1-ns time scale. As shown in Table 1, systems were simulated for 10 ns (close to the current “state of the art” length) to verify that the parmbsc0 force field retains the quality of the original AMBER parameterization with regard to other structural and dynamic features than the α/γ behavior. Finally, we tested the ability of the new force field to capture conformational transitions in DNA like the A → B one in duplexes and triplexes and its capability to correct both small (d(GCGC)2) and large (d(CCATGCGCTGAC)·d(GTCAGCGCATGG)) models of canonical duplexes starting from seriously distorted structures populated with multiple α/γ = g+t conformation substates.
TABLE 1.
Summary of MD simulations performed in this article with indication of simulation time and whether or not the simulation follows a conformational transition
| Sequence | Family | Length, ns | Transition |
|---|---|---|---|
| D(CGCGAATTCGCG)2 | B-DNA duplex DD | 2 × 200 | – |
| d(T10·A10-T10) | Triplex PS | 10 | – |
| d(C10·G10-G10) | Triplex APS | 10 | – |
| d(T10·A10-A10) | Triplex APS | 10 | – |
| d(GGGG)4 | G-DNA PS | 10 | – |
| d(GGGG)4 | G-DNA APS | 10 | – |
| d(CGCGCGCGCG)2 | Z-DNA duplex | 10 | – |
| d(CGCGAATTCGCG)2 | A-DNA duplex | 10 | A → B |
| d(T10·A10-T10) | A-DNA triplex | 10 | A → B |
| d(ATATATATATAT)2 | Hoogsteen duplex | 10 | – |
| r(UUCGGGCGCC)·d(GGCGCCCGAA) | DNA·RNA hybrid(PDB code 1FIX) | 10 | – |
| r(CGCGAAUUCGCG)2 | RNA duplex DD | 10 | – |
| r(GCGAGAGUAGG)·r(CCGAUGGUAGU) | RNA duplex(NDB code URL064) | 10 | – |
| r(CGCGGCACCGUCCGCGGAACAAACGG) | RNA pseudoknot(NDB code UR0004) | 10 | – |
| Nottingham dataset | B-DNA | 38 × 3 | – |
| Nottingham dataset | A-DNA | 36 × 3 | A → B |
| Nottingham dataset | Z-DNA | 6 × 3 | – |
| d(CCATGCGCTGAC)·d(GTCAGCGCATGG) | Pathological B-DNA | 25 | Pathol. → B |
APS, antiparallel; PS, parallel-stranded.
Possible transitions are: A, A-DNA conformation; B, B-DNA conformation; Pathol., structure severely distorted due to high number of alpha/gamma (g+/t) substates.
All MD simulations were performed following a similar standard protocol. Crystal or canonical structures were neutralized by a minimum amount of Na+ counterions (K+ for the Nottingham database structures) placed at the points with more electronegative potential, hydrated by TIP3P (48) water molecules, optimized, thermalized, and preequilibrated using our standard equilibration protocol (26,49) doubling the lengths of each individual equilibration substep followed by an extra 1–2 ns (as noted above DD was equilibrated for a longer period of time). All simulations were carried out using periodic boundary conditions and PME (5) under isothermal/isobaric conditions (T = 298 K; P = 1 atm). SHAKE (50) on bonds involving hydrogens was used in conjunction with an integration step of 2 fs.
RESULTS AND DISCUSSION
Fitting to quantum mechanical calculations
The full α/γ potential energy map of the model system (Fig. 1) was determined from B3LYP and LMP2 calculations considering both N- and S-sugar puckerings (Fig. 2). The computed maps were quite similar. Four broad minima appear clearly in the maps: 1), the canonical g−g+ one (located around α ≈ −90°, γ ≈ 60°), 2), the g−g− (α ≈ −90°, γ ≈ −60°), 3), the g+g− (α ≈ 90°, γ −60°), and 4), the g+g+ (α ≈ 60°, γ ≈ 60°). The g+t (α ≈ 100°, γ ≈ −160°), a region that shows some significant overpopulation in our collective set of simulations of DNA duplexes, with the parm94/99 force fields including the ABC-simulations of duplex DNA (38,39), is largely disfavored in the gas phase (Table 2).
FIGURE 2.
Potential energy maps (DFT and LMP2) for α and γ torsions. (A) Furanose forced to be in South conformation. (B) Furanose in North conformation. (C) Difference (absolute values in kcal/mol) between LMP2 and classical α/γ potential energy maps (values are displayed for sugars in South puckering). (Left) Parm99. (Right) Parmbsc0. Energy values in kcal/mol, and angles in degrees.
TABLE 2.
Energies (kcal/mol) relative to the canonical g−g+ minimum computed at different levels of theory
| Geometry | Energy | g− g− | g+ g− | g+ g+ | g+ t(pathol) |
|---|---|---|---|---|---|
| Energy maps | B3LYP/6-31+G(d) | 0.9 | 0.8 | 2.4 | 6.8 |
| LMP26-31+G(d) | 1.3 | 0.7 | 2.2 | 8.0 | |
| Parm99 | 2.5 | 3.5 | 1.9 | 4.3 | |
| parmbsc0 | 1.4 | 2.6 | 2.3 | 8.1 | |
| MP2 / aug-cc-pVDZ | MP2 /complete basis set | 2.0 | 2.4 | 2.7 | – |
| CCSD(T) /complete basis set | 2.1 | 2.4 | 2.8 | – | |
| Parm99 | 2.8 | 4.2 | 2.0 | – | |
| parmbsc0 | 1.8 | 3.2 | 2.0 | – |
Top entries correspond to the energy minima in the QM maps, and those at the bottom to geometries reoptimized at the quoted level of theory. The pathological g+t conformation is not a minimum, and optimization drives geometry out of the region.
Because of the large similarity of the four potential energy surfaces, very similar parameters were obtained in the fitting procedure of each set independently. Accordingly, data from the four maps (B3LYP and LMP2 for both S- and N-sugar puckerings) were mixed together for the parameterization process, which yields a robust α/γ effective potential map that can be used to parameterize these torsions in both DNA and RNA environments. The MC fitting procedure was used to bias the procedure to ensure a particularly good description of those regions sterically accessible for NAs. The final fitted parameters (Table 3) yield an average absolute error (taking the S-LMP2 map as the reference) of only 0.8 kcal/mol. There are only small regions distributed through the map where the error is greater than 2 kcal/mol, and only in the very unstable eclipsed region (α and γ around 120°) are the errors greater than 3 kcal/mol (Fig. 2 and see Supplementary Figs. S1 and S2). In contrast, the differential map obtained using the standard parm99 force field shows sizable errors in the important areas around α ≈ 100°, γ ≈ 100° and throughout the trans region for γ (between −100° and −180°) and, overall, a worse fitting (average absolute error to LMP2 data: 2 kcal/mol).
TABLE 3.
Force field parameters describing the α/γ torsion in parmbsc0 force field
| Torsion | No. of dihedrals | Vn/2 | Phase | Periodicity |
|---|---|---|---|---|
| X-CI-OS-X | 3 | 1.15 | 0 | 3 |
| X-CI-OH-X | 3 | 0.5 | 0 | 3 |
| X-CI-CT-X | 9 | 1.4 | 0 | 3 |
| CT-OS-CT-CI | 1 | 0.383 | 0 | −3 |
| CT-OS-CT-CI | 1 | 0.1 | 180 | 2 |
| H1-CI-CT-OS | 1 | 0.25 | 0 | 1 |
| H1-CI-CT-OH | 1 | 0.25 | 0 | 1 |
| H1-CT-CI-OS | 1 | 0.25 | 0 | 1 |
| H1-CT-CI-OH | 1 | 0.25 | 0 | 1 |
| CI-CT-CT-CT | 1 | 0.18 | 0 | −3 |
| CI-CT-CT-CT | 1 | 0.25 | 180 | −2 |
| CI-CT-CT-CT | 1 | 0.2 | 180 | 1 |
| OS-P-OS-CI | 1 | 0.185181 | 31.79508 | −1 |
| OS-P-OS-CI | 1 | 1.256531 | 351.9596 | −2 |
| OS-P-OS-CI | 1 | 0.354858 | 357.24748 | 3 |
| OH-P-OS-CI | 1 | 0.185181 | 31.79508 | −1 |
| OH-P-OS-CI | 1 | 1.256531 | 351.9596 | −2 |
| OH-P-OS-CI | 1 | 0.354858 | 357.24748 | 3 |
| CT-CT-CI-OS | 1 | 1.17804 | 190.97653 | −1 |
| CT-CT-CI-OS | 1 | 0.092102 | 295.63279 | −2 |
| CT-CT-CI-OS | 1 | 0.96283 | 348.09535 | 3 |
| CT-CT-CI-OH | 1 | 1.17804 | 190.97653 | −1 |
| CT-CT-CI-OH | 1 | 0.092102 | 295.63279 | −2 |
| CT-CT-CI-OH | 1 | 0.96283 | 348.09535 | 3 |
Vn/2 are in kcal/mol, and phase angles in degrees. For atom description see Fig. 1. Van der Waals and bond and angle parameters involving the new CI atom are taken from equivalent ones in parm99. A library file containing all parameters is accessible from http://mmb.pcb.ub.es/PARMBSC0. Note that we use standard nomenclature in AMBER datafile, where a negative value of periodicity means that additional Fourier terms for the dihedral will follow. Values in bold are those that were parameterized here under the restraint imposed by the other parameters transferred from standard parm99.
Geometries in the four minima were reoptimized without restrictions (other than the δ torsion) at the MP2/aug-cc-pVDZ level and used to perform single-point calculations at even higher levels (MP2/CBS and MP2/CBS+corr(CCSD(T)) to verify whether or not the force field provides a reasonable description of the four minima. The results in Table 2 demonstrate the quality of the parmbsc0 force field. Compared with parm99, the largest improvement of parmbsc0 is found in a nonminimum region, α/γ = g+t, which was significantly overstabilized in parm99 (or parm94) calculations, thus explaining the pathological behavior detected in parm94/parm99 MD simulations.
Simulations of Dickerson's dodecamer
No oligonucleotide has been the subject of more studies, both theoretical and experimental, than the DD (d(CGCGAATTGCGC)2). All the experimental data indicate that it is a stable duplex pertaining to the B-family, but with sizable sequence dependence in its helical parameters. Analysis of 12 structures of DD deposited in the PDB (six solved by x-ray crystallography and six by NMR) show that all sugars are in South and South-East regions except some cytidines, which in certain structures sample N regions. All backbones are in the canonical g−g+ α/γ region, without any nucleotide in the g+t region. The major groove width is around 18 Å, and the minor groove oscillates between 10 (NMR) and 12 (x-ray) Å. Hydrogen bonding is well preserved in all experimental structures, though local distortions of linearity appear. The average roll is around 0° (x-ray) or 3° (NMR), and the average twist is 34 ± 3° (NMR) or 35 ± 0.3° (x-ray), with a clear dependence on sequence (stronger in x-ray-derived structures; Table 4).
TABLE 4.
Selected parameters describing an average Dickerson's dodecamer (DD) from x-ray, NMR, and MD simulations (parm94, parm99, and parmbsc0)
| Parameter | NMR | X-ray | parm94 | parm99 | parmbsc0 |
|---|---|---|---|---|---|
| Average twist | 34 ± 2 | 35 ± 0.1 | 30 ± 2 | 26 ± 4 | 33 ± 1 |
| Average roll | 3 ± 1 | 0 ± 0.5 | 4 ± 2 | 4 ± 2 | 3 ± 2 |
| mG-width | 12 ± 1 | 10 ± 0.2 | 13 ± 2 | 12 ± 1 | 12 ± 1 |
| MG-width | 18 ± 3 | 18 ± 0.3 | 20 ± 1 | 21 ± 1 | 19 ± 1 |
| % S-puckering | ∼100 ± 0 | 89 ± 5 | 75 ± 16 | 96 ± 5 | 93 ± 6 |
| G | ∼100 | 100 | 85 ± 15 | 98 ± 6 | 97 ± 6 |
| C | ∼100 | 64 ± 16 | 80 ± 18 | 96 ± 8 | 88 ± 12 |
| A | ∼100 | 100 | 56 ± 36 | 94 ± 12 | 94 ± 12 |
| T | ∼100 | 100 | 72 ± 27 | 94 ± 12 | 91 ± 14 |
| % g− g+ | 98 ± 4 | 99 ± 2 | 66 ± 17 | 67 ± 8 | 98 ± 4 |
| No. H-bonds | 26 ± 0 | 26 ± 0 | 26 ± 0.3 | 25 ± 1 | 26.0 ± 0.1 |
| % BI in (BI/BII) Eq. | 98 ± 4 | 87 ± 3 | 84 ± 8 | 80 ± 6 | 82 ± 7 |
Rotational parameters are in degrees, and distances in Å. The canonical g−g+ is defined in regions of α 240–360° and γ 0–120°. North is defined by phase angles smaller than 90°.
No detailed NMR analysis of sugar puckering is provided in DD structures deposited in PDB. Accurate estimates for a related sequence suggest an average South population around 81%, with more purines than pyrimidines in the South conformations (see text for details).
DD has been successfully simulated over the 1- to 50-ns time scale using parm94 or parm99 (see Introduction), but longer simulations approaching the 100-ns barrier are scarce (51). Analysis of Fig. 3 shows that at around 60 ns the structure of the duplex is severely distorted because of numerous α/γ transitions to the g+t region (Figs. 3 and 4 and extended data at Supplementary Fig. S3; http://mmb.pcb.ub.es/PARMBSC0). These transitions, similar to those reported in shorter trajectories (see, for example, ABC simulations), are stochastic in nature and irreversible on the 100-ns time scale for both parm94 and parm99 force fields. A few of these transitions could be tolerated in the duplex, but when they accumulate, they result in a considerable departure of the structure from the canonical B-form: lower helical twist (average twist around 30° (parm94) or 26° (parm99); see Table 4), distorted grooves, and even the wrong puckering population. Clearly, the MD simulations presented here, the longest ever published for parm94/99, backed by similar results on a wide variety of NA structures by this group, ranging from DNA minicircles to A-tract DNA to a large set of DNA duplexes, confirm that although reliable results can be obtained in short or medium (<20 ns) simulations, severe artifacts will be found over longer simulations.
FIGURE 3.
MD simulations of Dickerson's dodecamer with parm94 (orange) and parm99 (magenta) force fields. Average values from x-ray (black) and NMR (red) experiments (for the same sequence) are shown as solid straight lines with the associated standard association as dashed lines. (A) Percentage of canonical α/γ torsions. (B) Average twist. (C) RMSd (Å) from crystal data.
FIGURE 4.
Structures of Dickerson's dodecamer obtained by averaging the last 5 ns of the trajectories obtained with parm99 and parmbsc0 force fields. The crystal structure is shown as reference.
Two independent 200-ns MD simulations were performed with the parmbsc0 force field using different starting geometries and velocities. In both cases, the duplex samples the same region of the conformational space, which is close to the experimental structure (see Table 4, Figs. 4 and 5, and complete data at http://mmb.pcb.ub.es/PARMBSC0). The simulated duplex remains within the range of helical parameters expected for a canonical B-form duplex in aqueous solution, showing an improvement in global helical twist representation with respect to parm94 and parm99 force fields. As expected from the regularity of the duplex, the Watson-Crick hydrogen-bonding scheme is fully maintained except for some breathing events at the nucleobases at the ends of the helix (see Supplementary Fig. S4 and trajectory videos at http://mmb.pcb.ub.es/PARMBSC0). The sugar puckers mainly sample the South region, as expected for a B-DNA, but a nonnegligible percentage of N-puckering is observed. In fact, the integration of puckering populations using a two-state model shows ∼9% (T), 12% (C), 7% (A), and 3% (G) of North puckering in the simulations, which are close to the most accurate NMR estimates of the population of N-sugars in B-DNA (14% (T), 24% (C), 5% (A), and 6%(G); see Isaacs and Spielmann (52)). Finally, it is worth noting that the new force field not only provides a good global geometry of the duplex but also reproduces some sequence-dependent variations in the structure (such as the undertwist of d(CG) steps; Fig. 6) or the higher tendency of C to display N-puckerings, which might be important to understand biological properties of DNA (see Table 4). Note that all these details are lost in long parm94 or parm99 simulations (see Table 4 and Supplementary Fig. S5).
FIGURE 5.
Analysis for two independent parmbsc0 trajectories (blue and green) of Dickerson's dodecamer. (A) RMSd (Å) with respect to the corresponding MD-averaged (light) or experimental structure (dark). (B) Percentage of canonical α/γ torsions. (C) Widths of the grooves (Å). (D) Average twist (in degrees). Average experimental values are shown as reference (solid lines) with the associated standard deviations as dashed lines (black, x-ray; red, NMR).
FIGURE 6.
Variation of twist (in degrees) with the sequence in parmbsc0 simulations (the averages of the two simulations are shown here) of Dickerson's dodecamer (light gray line). The average values obtained by taking all NMR (dark gray) or x-ray (black) structures for this sequence are shown as reference. Error bars are displayed at each step.
Additional comparisons were carried out taken as reference values of B-DNA structures in a manually cured subset of the NDB database, where anomalous duplexes (containing drugs, mismatches, etc.) were removed (53). These comparisons need to be taken with some caution because we are now comparing simulated data for a given duplex with experimental data for a large set of different oligos of different lengths and sequences, but in any case, if the force field works well, we should find similarity in the distribution of backbone torsions and helical parameters between MD simulations and the experimental database. Results in Supplementary Fig. S6 confirm that the distribution of torsions sampled by parmbsc0 simulations reproduce very well that found in the experimental databases, and the same is detected for helical parameters (see Supplementary Fig. S7). In both cases, the improvement with respect to parm99 calculations is very clear. Not surprisingly, the greatest improvement in performance between parmbsc0 and parm99 is found for the α and γ torsion and for the closely coupled χ one (see Supplementary Fig. S6). In terms of helical parameters, the greatest improvement of parmbsc0 is found in the helical twist (see Supplementary Fig. S7).
As the standard deviations of the various averages indicate, the parmbsc0 force field does not allow a rigid picture of DNA. On the contrary, the structure is very flexible, and many reversible transitions are found. The most common of these changes is that between BI (around 82%) and BII (18%) forms. This transition, the equilibrium constant of which is well reproduced by parmbsc0 MD simulations (see Table 4), occurs with a very high frequency during the two 200-ns trajectories, indicating that the force field is not rigidifying the structure (Fig. 7). Many α/γ transitions are also detected in the simulations, but all of them are reversible after a few nanoseconds. This finding indicates that the new force field, while maintaining the necessary flexibility, captures properly the α/γ equilibrium (an example of the time evolution of one individual set of α/γ values in Fig. 7, additional examples at Supplementary Fig. S8, and complete data at http://mmb.pcb.ub.es/PARMBSC0), without an artifactual rigidification of the structure.
FIGURE 7.
(Top) Percentage of canonical (BI) conformation during two parmbsc0 simulations (blue and green) of Dickerson's dodecamer. Average experimental values are shown as reference in solid lines with the standard deviations in dashed lines (black x-ray and red NMR). (Bottom) Example of variation of α/γ torsions in a given basepair step along time. Squares correspond to α and triangles to γ. Colors: green/yellow correspond to one nucleotide, and blue/magenta the complementary one.
Is the parmbsc0 force field applicable to different sequences?
We have compared the performance of parm99 and parmbsc0 in relatively short (3-ns) simulations of 38 different duplexes taken from the Nottingham database. Although these simulations are too short to fully sample the conformational space of these systems, they make it possible to evaluate the performance of the reparameterized force fields in the important initial process of relaxing and equilibrating experimental (NMR or x-ray crystallographic) starting structures. Results in Fig. 8 show that the new force field behaves very well in all cases studied, providing stable trajectories, with RMSd from the respective MD-averaged conformations around 1.2 Å, and around 1.6–2.8 Å from experimental structures, the largest deviations being found in all the cases for the longest duplexes. Analysis of the 36 trajectories did not reveal any artifactual behavior or local distortions that might indicate potential sequence-dependent errors in the simulations (see Supplementary Fig. S9). We can then safely recommend the use of parmbsc0 to study any B-DNA duplex.
FIGURE 8.
Distribution of RMSd (Å) in parmbsc0 simulations of Nottingham's database of B-DNAs (light from MD-averaged, and dark from experimental structures).
Can parmbsc0 repair incorrect structures?
Previous simulations show that parmbsc0 leads to a correct representation of the α/γ configurational space in very long simulations started from a crystal structure without anomalous α/γ conformations. In fact, transitions to minor α/γ conformers are not avoided but are reversible in the nanosecond time scale (see Fig. 7 bottom), suggesting that the force field is robust enough to recover from starting structures containing a few isolated anomalous conformations. It is, however, unclear what will happen when the MD simulation starts from a very severely distorted conformation containing many α/γ transitions. To evaluate this point, we performed a series of simulations of the DNA duplex (d(CCATGCGCTGAC)·d(GTCAGCGCATGG)), which should exist in the B-form in solution, starting with the very corrupted structure obtained previously by Varnai and Zakrzewska (37) at the end of their 50-ns-long parm99 force field simulation. The duplex initially contained 13 anomalous α/γ conformers, which severely distorted the backbone. However, the parmbsc0 simulation that started with this structure corrected the anomalous conformations within 25 ns in three distinct simulations (with different initial conditions), leading to samplings close to those expected for a canonical B-helix (see Fig. 9). Extension of this trajectory to 100 ns simulation time (data not shown) confirms that the duplex is maintained in the canonical region. Similar simulations performed with shorter oligonucleotides (such as d(GCGC)2; data not shown) confirm the ability of the parmbsc0 force field to correct erroneous NA conformations. In summary, we can conclude, based on our validation on a large set of NA structures, that the parmbsc0 force field can safely be used to study canonical B-DNAs over long temporal scales and is robust enough to recognize and repair large structural errors while still preserving the essential flexibility of the duplex, not artificially penalizing α/γ transitions as required for a correct representation of distorted NAs (for example, those in complexes with proteins).
FIGURE 9.
Evolution of the percentage of canonical α/γ pairs in a parmbsc0 simulation started from severely distorted duplex. The general shape of the helix in the beginning of the simulation (after 50-ns parm99 trajectory) is compared with that obtained at the end of the parmbsc0 simulation.
Can parmbsc0 be used to represent RNAs?
Considering the changes introduced in parmbsc0 with respect to parm99 and the similarity of α/γ potential energy maps for N- and S-sugars, we expected that the new force field should be well suited to RNA simulations. To verify this point, we performed 10-ns MD simulations for three different RNA systems: 1), the RNA version of DD, 2), an RNA duplex (taken from NDB entry URL064) containing several mismatches, and 3), an RNA pseudoknot (NDB UR0004) containing a wide variety of unusual hydrogen bond schemes including those involving nucleobases in anomalous ionization states. In all cases the trajectories were stable and remain close to the known experimental conformations (Table 5; see Supplementary Figs. S10, S11; and http://mmb.pcb.ub.es/PARMBSC0). The pattern of canonical A-U and G-C Watson-Crick hydrogen bonds is fully preserved, whereas noncanonical pairs are slightly more labile: two of them are partially lost for URL064, whereas only one linking a nucleobase and a sugar is lost in the pseudoknot simulation. All the simulations sample the A- region with 100% North puckerings in the riboses for the canonical DD-RNA and URL064. The pseudoknot presents three sugars in the South conformation during the entire trajectory, in agreement with the corresponding x-ray structure. In summary, parmbsc0 is able to reproduce with good quality the structure of RNAs, including those with mismatches or unusual pairing schemes. Further testing of RNA would be vital because of the complexity of RNA structures (54,55), but the generally good performance of parm94/99 to represent A-RNA crystal structures (in simulations reaching 100 ns) makes us confident of the performance of parmbsc0.
TABLE 5.
Geometric descriptors for different RNA simulations
| System | RMSd(avg) | RMSd(exp) | % North | % hb can | % hb noncan |
|---|---|---|---|---|---|
| DD-RNA | 1.3 | 1.7 | 100 / 100* | 99 | — |
| RNA URL064 | 1.0 | 1.8 | 100 / 100 | 100 | 77 |
| RNA Pseudoknott | 1.2 | 2.2 / 1.7† | 87 / 87 | 99 | 92 |
End bases are excluded from the study and the percentage of maintenance of hydrogen bonds is presented into blocks: canonical Watson-Crick pairs and noncanonical pairs.
The second number corresponds to values found in the crystal.
The second value corresponds to the RMSd when a flexible bulge (containing five residues) is omitted in the calculations.
Can parmbsc0 be used to represent unusual DNA conformations?
One of the most impressive features of the parm94/parm99 force fields is their ability to properly model unusual DNAs (see Introduction), which were not considered or even discovered at the time of the original parameterization. This indicates that the parameterization process based on the study of small model systems used there was quite successful, and any improvement to these force fields should maintain their ability to represent structures very far away from the canonical B-DNA duplex. For this purpose we have analyzed 1), Z-DNA, 2), parallel and antiparallel triplexes based on different purine or pyrimidine motifs, 3), parallel and antiparallel quadruplexes, and 4), antiparallel Hoogsteen duplexes. In all the cases, 10-ns MD simulations were run, extending the length of previous parm99- or parm94-based MD simulations on these systems (see reviews on previous AMBER MD simulations (10–16)). The trajectories were stable, and sampled configurations were very close to the experimental ones, as demonstrated in terms of RMSd, helical properties, and sugar puckerings (Table 6,see Supplementary Figures S12–S18, and data at http://mmb.pcb.ub.es/PARMBSC0). The force field reproduces almost exactly the conformation of both parallel and antiparallel DNA quadruplexes (see Table 6). Similarly, parmbsc0 gives good results for a variety of triplexes, which are found to pertain to the B-family in all cases, in agreement with previous MD simulations and NMR experiments (26,56, and references therein). Interestingly, the B-form for the triplex is also found when the trajectory is started from an A- triplex conformation (26), confirming that the force field can correct (at least some) erroneous starting conformations (see above and below).
TABLE 6.
Average geometric descriptors of different anomalous DNAs obtained by (at least) 10 ns of MD simulations with AMBER parmbsc0 force field
| Descriptor | G-DNA(ps) | G-DNA (aps) | Triplex d(A-A·T) aps | Triplex d(G-G·C) aps | Triplex d(T-A·T)ps | Z-DNA | aps-Hoogsteen |
|---|---|---|---|---|---|---|---|
| rmsd(experimental) | 1.0 | 1.2 | 2.3 | 2.5 | 3.4 | 1.04 | 1.3 |
| rmsd(average) | 0.8 | 0.7 | 1.0 | 1.3 | 0.9 | 0.76 | 1.2 |
| %South | 90 (100) | 97(100) | 73.8 (72) | 71.5 (72) | 74.5 (72) | 50‡(50) | 93 (100) |
| %H-bond* | 97 | 96 | 93 | 93 | 99 | 100 | 98 |
| m-groove† | 12.7 (12.3) | 13.1 (12.7) | 11.4 (12.2) | 12.4 (12.2) | 11.2 (13.0) | 9.8 (9.25) | 11.8 (9–11) |
| M-groove | — | — | — | — | — | — | 21.9 (∼20) |
| mM-groove | — | — | 9.7 (∼9) | 10.5 (∼9) | 9.2 (∼8) | — | — |
| MM-groove | — | — | 15.2 (∼15) | 13.6 (∼15) | 17.1 (∼15) | — | — |
| C1′-C1′ interstrand | 11.4 (11.4) | 11.4 (11.3) | 10.5 (10.4) | 10.7 (10.4) | 10.6 (10.5) | 10.8 (10.7) | 8.6 (8.2) |
| Average twist | 30 (31) | n.a | 29 (30) | 28 (30) | 29 (31) | −30 (−27.) | 32 (35) |
Translational parameters are in angstroms, and rotations in degrees. Values in parentheses correspond to experimental values (PDB entries: 352D and 156D for ps and aps G-DNA (loops excluded in the calculations); 135D and 149D for aps and ps triplexes (backbone atoms), Arnott's values for Z-DNA and 1GQU for the aps Hoogsteen duplex.
Percentage of hydrogen bond from experimental value of from optical canonical pairings if experimental structure is unknown. For G-DNA both canonical and bifurcated hydrogen bonds were considered.
For definition of grooves in triplexes see [23]; all groove widths are reported as P-P distances.
Both in experiment and simulations, North puckerings correspond to residues in syn conformation, and South to those in the anti one.
Especially remarkable is the performance of the parmbsc0 force field in describing non-B-form duplex DNAs such as the Hoogsteen duplex or Z-DNA, where either the pattern of hydrogen bond or the glycosidic bond orientation is unusual. In both cases, simulations are within thermal noise of the experimental structures (see Table 6). The unusual Hoogsteen duplex (57,58) appears very stable during our simulation, maintaining its hydrogen-bond pattern and helical structure extremely well. The same is found for Z-DNA, where the correct balance of N/S puckerings is preserved, showing that the force field does not have any artifactual tendency to increase S-puckerings. This later point was verified by running eight additional (3-ns) simulations for Z-DNAs of known crystal structure (taken from the Nottingham database), and in all cases, parmbsc0 accurately reproduces the experimental structure. Overall, all these tests confirm that the parmbsc0 force field can be safely used to study unusual DNA structures.
Can parmbsc0 represent DNA·RNA hybrids?
High-quality NMR data (59–61) and previous MD simulations (62,63) have demonstrated that in solution DNA·RNA hybrids tend to an intermediate A/B conformation: although the general shape is close to the A- form, other characteristics such as the sugar conformation of the 2′-deoxyriboses or the geometry of the grooves are not far from those of a B-helix (59–63). The most unusual characteristic of the hybrid is its strand asymmetry, which makes its representation by force fields specially challenging. Fortunately, even in this difficult case, parmbsc0 behaves well, not only in general geometric parameters but also in the fine structural details. Thus, MD is able to capture the asymmetry in sugar puckering between riboses (all in North conformation) and 2′-deoxyriboses (70% in South conformation), a result similar to that found in accurate NMR experiments (between 66% and 78% S-puckering in a related sequence; see Soliva et al. (56)). The inversion in the width of minor and major grooves with respect to the canonical A form is also perfectly reproduced in MD simulations (Table 7), as is the average twist, closer to A- than to B- form (see also Supplementary Fig. S19 and visit the URL site http://mmb.pcb.ub.es/PARMBSC0).
TABLE 7.
Geometric descriptors of a DNA·RNA hybrid obtained by MD simulations with parmbsc0
| RMSd(A) | RMSd(B) | RMSd(NMR) | RMSd(cryst) | %N(ribose) |
|---|---|---|---|---|
| 2.9 | 4.9 | 1.9 | 2.4 | 100 |
| %S (2′-deoxy) | %Hbond | Twist | mG width | MG width |
| 70 / 66–78* | 99 / 100 | 30 / 32 | 15 / 15 | 20 / 20 |
The values after the slash correspond to those obtained experimentally in aqueous solution by NMR techniques (pdb code: 1efs).
Values correspond to those obtained by González et al. (60) in accurate measures of sugar puckering in a related sequence. Such analysis was not performed for 1efs, where all sugars were assumed to be in the South conformation.
Can parmbsc0 model DNA conformational transitions?
One of the big successes of the latest generation of force fields is their ability to drive structures from incorrect to correct conformations, simulating well-known conformational transitions. This is the case of the spontaneous A → B transition in duplex DNA in aqueous solution (18–22), the A → A/B transition in DNA·RNA hybrids (62), and the A(t) → B(t) transition found in parallel triplexes (26). As noted above, we found spontaneous A(t) → B(t) transitions in all triplexes, and the subtle A → A/B conformational change in hybrids (see Supplementary Fig. S17). To verify that the classical A → B transition was found in duplex DNA with the parmbsc0 force field, a 10-ns unrestricted MD simulation of DD starting in the A-conformation was run. The trajectory transforms in the nanosecond time scale from the canonical A-form to another structure very close to the canonical B-form (see Fig. 10 and videos at http://mmb.pcb.ub.es/PARMBSC0). To examine the sequence dependence of this transition, a similar study (trajectories were 3 ns long) was performed for the 36 A-DNA structures in Nottingham's database. In all the cases the duplexes jump to the B-form in just 3 ns, confirming that the force field can capture fast conformational transition in DNAs (see Supplementary Fig. S20).
FIGURE 10.
Evolution of a parmbsc0 simulation of DD starting from a canonical A-type conformation. (Top) Comparison of RMSd (Å) from B (black) and A (light gray) forms. (Bottom) Percentage of South puckering along simulation time. Note that time scale starts after the equilibration.
CONCLUSIONS
We present here a reparameterization of the parm99 AMBER force field for NA simulations. Our effort has been limited to improving the representation of the α/γ conformational space, which seems to be poorly represented in very long DNA MD simulations with current AMBER force fields. After a careful parameterization process based solely on B3LYP and LMP2 calculations (as opposed to the iterative refinement based on MD simulations of previous works) (10), validated by CCSD(T)/CBS calculations, fitted parameters have been tested by the most extended set of simulations published to date. Two very long MD simulations (two times longer than any previously published trajectory) of the Dickerson dodecamer show the excellent ability of the new force field to represent canonical DNAs over time scales that led to severe artifacts in previous AMBER simulations. The new force field is able to correct severe errors in structures and to drive known conformational transitions in water. Furthermore, it provides reasonable 3-ns samplings for 87 experimentally determined duplexes of different sequences and conformations, and it represents both canonical and noncanonical RNA structures. Finally, the new force field was able to model very well a large range of anomalous DNA structures. In summary, the parmbsc0 force field maintains all the impressive characteristics of the parm94/99 force fields that have made them the most widely used in the NA modeling field while solving the most obvious shortcomings manifested in very long B-DNA MD simulations performed with previous parm94/99 force fields.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
We are grateful to Dr. Peter Varnai for kindly providing us the coordinates of his simulation of distorted B-DNA duplex and to Prof. F. Javier Luque for valuable comments and critical reading of this article. We also thank the technical personnel of the Barcelona Supercomputer Center, especially those managing Mare Nostrum for making the massive simulations reported here possible.
This work has been supported by the Spanish Ministry of Education and Science (BIO2006-01602) and Fundación La Caixa. Further support was obtained by grants LC06030 and LC512 and by Research Project Z4 055 905 by Ministry of Education of the Czech Republic. The Nottingham database simulations were made possible by the UK National Grid Service and the University of Nottingham's High Performance Computing Resource. We acknowledge additional computer power provided by Brno and Pittsburgh Supercomputer Centers. A.P. and I.M. are fellows of the Catalan and Spanish Ministries of Education and Science, respectively.
A. Pérez and I. Marchán contributed equally to this work.
References
- 1.McCammon, J. A. 1976. Models for Protein Dynamics. H. J. C. Berendsen, editor. CECAM - Universite de Paris IX, Orsay (France).
- 2.McCammon, J. A., B. R. Gelin, and M. Karplus. 1977. Dynamics of folded proteins. Nature. 267:585–590. [DOI] [PubMed] [Google Scholar]
- 3.Levitt, M. 1983. Computer simulation of DNA double-helix dynamics. Cold Spring Harb. Symp. Quant. Biol. 47:251–262. [DOI] [PubMed] [Google Scholar]
- 4.Tidor, B., K. K. Irikura, B. R. Brooks, and M. Karplus. 1983. Dynamics of DNA oligomers. J. Biomol. Struct. Dyn. 1:231–252. [DOI] [PubMed] [Google Scholar]
- 5.Darden, T., D. York, and L. G. Pedersen. 1993. Particle mesh Ewald: An N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 6.Cornell, W. D., P. Cieplak, C. I. Baily, I. R. Gould, K. M. Merz, Jr., D. C. Ferguson, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179–5197. [Google Scholar]
- 7.MacKerell, A. D., J. Wiorkiewicz-Kuczera, and M. Karplus. 1995. An all-atom empirical energy function for the simulation of nucleic acids. J. Am. Chem. Soc. 117:11946–11975. [Google Scholar]
- 8.Foloppe, N., and A. D. Mackerell. 2000. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 21:86–104. [Google Scholar]
- 9.Langley, D. R. 1998. Molecular dynamic simulations of environment and sequence dependent DNA conformations: the development of the BMS nucleic acid force field and comparison with experimental results. J. Biomol. Struct. Dyn. 16:487–509. [DOI] [PubMed] [Google Scholar]
- 10.Cheatham, T. E., III, P. Cieplak, and P. A. Kollman. 1999. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 16:845–862. [DOI] [PubMed] [Google Scholar]
- 11.Cheatham, T. E., III, J. L. Miller, T. Fox, T. A. Darden, and P. A. Kollman. 1995. Molecular dynamics simulations on solvated biomolecular systems: the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J. Am. Chem. Soc. 117:4193–4194. [Google Scholar]
- 12.York, D. M., W. Yang, H. Lee, T. Darden, and L. G. Pedersen. 1995. Toward the accurate modeling of DNA: the importance of long-range electrostatics. J. Am. Chem. Soc. 117:5001–5002. [Google Scholar]
- 13.Beveridge, D. L., and K. J. McConnell. 2000. Nucleic acids: theory and computer simulation, Y2K. Curr. Opin. Struct. Biol. 10:182–196. [DOI] [PubMed] [Google Scholar]
- 14.Cheatham, T. E., III, and P. A. Kollman. 2000. Molecular dynamics simulation of nucleic acids. Annu. Rev. Struct. Dyn. 51:435–471. [DOI] [PubMed] [Google Scholar]
- 15.Cheatham, T. E., III 2004. Simulation and modeling of nucleic acid structure, dynamics and interactions. Curr. Opin. Struct. Biol. 14:360–367. [DOI] [PubMed] [Google Scholar]
- 16.Giudice, E., and R. Lavery. 2002. Simulations of nucleic acids and their complexes. Acc. Chem. Res. 35:350–357. [DOI] [PubMed] [Google Scholar]
- 17.Orozco, M., A. Pérez, A. Noy, and F. J. Luque. 2003. Theoretical methods for the simulation of nucleic acids. Chem. Soc. Rev. 32:350–364. [DOI] [PubMed] [Google Scholar]
- 18.Orozco, M., M. Rueda, J. R. Blas, E. Cubero, F. J. Luque, and C. A. Laughton. 2004. Encyclopedia of Computational Chemistry. http://www.mrw.interscience.wiley.com/ecc/articles/cn0080/bibliography-fs.html.
- 19.Pérez, A., J. R. Blas, M. Rueda, J. M. López-Bes, X. de la Cruz, and M. Orozco. 2005. Exploring the essential dynamics of B.DNA. J. Chem. Theor. Comput. 1:790–800. [DOI] [PubMed] [Google Scholar]
- 20.Cheatham III, T. E., and M. A. Young. 2000. Molecular dynamics simulation of nucleic acids: successes, limitations, and promise. Biopolymers. 56:232–256. [DOI] [PubMed] [Google Scholar]
- 21.Cheatham III, T. E., and P. A. Kollman. 1996. Observation of the A-DNA to B-DNA transition during unrestrained molecular dynamics in aqueous solution. J. Mol. Biol. 259:434–444. [DOI] [PubMed] [Google Scholar]
- 22.Soliva, R., F. J. Luque, C. Alhambra, and M. Orozco. 1999. Role of sugar re-puckering in the transition of A and B forms of DNA in solution. A molecular dynamics study. J. Biomol. Struct. Dyn. 17:89–99. [DOI] [PubMed] [Google Scholar]
- 23.Cheatham III, T. E., M. F. Crowley, T. Fox, and P. A. Kollman. 1997. A molecular level picture of the stabilization of A-DNA in mixed ethanol-water solutions. Proc. Natl. Acad. Sci. USA. 94:9626–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConnell, K. J., and D. L. Beveridge. 2000. DNA structure: what's in charge? J. Mol. Biol. 304:803–820. [DOI] [PubMed] [Google Scholar]
- 25.Sprous, D., M. A. Young, and D. L. Beveridge. 1998. Molecular dynamics studies of the conformational preferences of a DNA double helix in water and an ethanol/water mixture: Theoretical considerations of the A double left right arrow B transition. J. Phys. Chem. B. 102:4658–4667. [Google Scholar]
- 26.Shields, G. C., C. A. Laughton, and M. Orozco. 1997. Molecular dynamics simulations of the d(T·A·T) triple helix. J. Am. Chem. Soc. 119:7463–7469. [Google Scholar]
- 27.Cheatham III, T. E., and P. A. Kollman. 1997. Insight into the stabilization of A-DNA by specific ion association: spontaneous B-DNA to A-DNA transitions observed in molecular dynamics simulations of d[ACCCGCGGGT]2 in the presence of hexaamminecobalt(III). Structure (London). 5:1297–1311. [DOI] [PubMed] [Google Scholar]
- 28.Rueda, M., S. G. Kalko, F. J. Luque, and M. Orozco. 2003. The structure and dynamics of DNA in the gas phase. J. Am. Chem. Soc. 125:8007–8014. [DOI] [PubMed] [Google Scholar]
- 29.Rueda, M., F. J. Luque, and M. Orozco. 2005. Nature of minor-groove binders-DNA complexes in the gas phase. J. Am. Chem. Soc. 127:11690–11698. [DOI] [PubMed] [Google Scholar]
- 30.Rueda, M., F. J. Luque, and M. Orozco. 2006. G-quadruplexes can mantain their structure in the gas phase. J. Am. Chem. Soc. 128:3608–3619. [DOI] [PubMed] [Google Scholar]
- 31.Hobza, P., M. Kabelac, J. Sponer, P. Mejzlik, and J. Vondrasek. 1997. Performance of empirical potentials (AMBER, CFF95, CVFF, CHARMM, OPLS, POLTEV), semiempirical quantum chemical methods (AM1, MNDO / M, PM3), and ab initio Hartree-Fock method for interaction of DNA bases: comparison with nonempirical beyond Hartree-Fock results. J. Comp. Chem. 18:1136–1150. [Google Scholar]
- 32.Alhambra, C., F. J. Luque, F. Gago, and M. Orozco. 1997. Ab initio study of stacking interactions in A- and B-DNA. J. Phys. Chem. B. 101:3846–3853. [Google Scholar]
- 33.Pérez, A., J. Sponer, P. Jurecka, P. Hobza, F. J. Luque, and M. Orozco. 2005. Are the RNA(A·U) hydrogen bonds stronger than the DNA(A·T) ones? Chem. Eur. J. 11:5062–5066. [DOI] [PubMed] [Google Scholar]
- 34.Sponer, J., P. Jurecka, I. Marchan, F. J. Luque, M. Orozco, and P. Hobza. 2006. Nature of base stacking. Reference quantum chemical stacking energies in ten unique B-DNA base pair steps. Chem. Eur. J. 12:2854–2865. [DOI] [PubMed] [Google Scholar]
- 35.Sponer, J., P. Jurecka, and P. Hobza. 2004. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J. Am. Chem. Soc. 126:10142–10151. [DOI] [PubMed] [Google Scholar]
- 36.Sponer, J. E., N. Spackova, J. Leszczynski, and J. Sponer. 2005. Principles of RNA base pairing: structures and energies of the trans Watson-Crick/sugar edge base pairs. J. Phys. Chem. B. 109:11399–11410. [DOI] [PubMed] [Google Scholar]
- 37.Varnai, P., and K. Zakrzewska. 2004. DNA and its counterions: a molecular dynamics study. Nucleic Acids Res. 32:4269–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beveridge, D. L., G. Barreiro, K. S. Byun, D. A. Case, T. E. Cheatham III, S. B. Dixit, E. Giudice, F. Lankas, R. Lavery, J. H. Maddocks, R. Osman, E. Seibert, H. Sklenar, G. Stoll, K. M. Thayer, P. Varnai, and M. A. Young. 2004. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. I. Research design and results on d(CpG) steps. Biophys. J. 87:3799–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixit, S. B., D. L. Beveridge, D. A. Case, T. E. Cheatham 3rd, E. Giudice, F. Lankas, R. Lavery, J. H. Maddocks, R. Osman, H. Sklenar, K. M. Thayer, and P. Varnai. 2005. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. II: Sequence context effects on the dynamical structures of the 10 unique dinucleotide steps. Biophys. J. 89:3721–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixit, S. B., and D. L. Beveridge. 2006. Structural bioinformatics of DNA: a web-based tool for the analysis of molecular dynamics results and structure prediction. Bioinformatics. 22:1007–1009. [DOI] [PubMed] [Google Scholar]
- 41.Saebø, S., and P. Pulay. 1993. Local treatment of electron correlation. Annu. Rev. Phys. Chem. 44:213–236. [Google Scholar]
- 42.Becke, A. D. 1993. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98:5648–5652. [Google Scholar]
- 43.Lee, C., W. Yang, and R. G. Parr. 1988. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 37:785–789. [DOI] [PubMed] [Google Scholar]
- 44.Barlett, R. J. 1995. Modern Electronic Structure Theory. Part I. D. R. Yarkony, editor. World Science. Singapore.
- 45.Halkier, A., T. Helgaker, P. Jorgensen, W. Klopper, H. Koch, J. Olsen, and A. K. Wilson. 1998. Basis-set convergence in correlated calculations on Ne, N2 and H2O. Chem. Phys. Lett. 286:243–252. [Google Scholar]
- 46.Bayly, C. I., P. Cieplak, W. D. Cornell, and P. A. Kollman. 1993. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges. J. Chem. Phys. 97:10269–10280. [Google Scholar]
- 47.Dickerson, R. E., and H. L. Ng. 2001. DNA structure from A to B. Proc. Natl. Acad. Sci. USA. 98:6986–6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 49.Shields, G. C., C. A. Laughton, and M. Orozco. 1998. Molecular dynamics simulation of a PNA.DNA.PNA triple helix in aqueous solution. J. Am. Chem. Soc. 120:5895–5904. [Google Scholar]
- 50.Ryckaert, J. P., G. Ciccotti, and H. J. C. Berendsen. 1977. Numerical-integration of cartesian equations of motion of a system with constraints - molecular-dynamics of N-alkanes. J. Comp. Phys. 23:327–341. [Google Scholar]
- 51.Ponomarev, S. Y., K. M. Thayer, and D. L. Beveridge. 2004. Ion motions in molecular dynamics simulations on DNA. Proc. Natl. Acad. Sci. USA. 101:14771–14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isaacs, R. J., and H. P. Spielmann. 2001. NMR evidence for mechanical coupling of phosphate BI-BII transitions with deoxyribose conformational exchange in DNA. J. Mol. Biol. 311:149–160. [DOI] [PubMed] [Google Scholar]
- 53.Pérez, A., A. Noy, F. Lankas, F. J. Luque, and M. Orozco. 2004. The relative flexibility of DNA and RNA: Database analysis. Nucleic Acids Res. 32:6144–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krasovska, M. V., J. Sefcikova, K. Reblova, B. Schneider, N. G. Walter, and J. Sponer. 2006. Cations and hydration in catalytic RNA: Molecular dynamics of the hepatitis delta virus ribozyme. Biophys. J. 91:626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spackova, N., and J. Sponer. 2006. Molecular dynamics simulations of sarcin-rich rRNA motif. Nucleic Acids Res. 34:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soliva, R., E. Sherer, F. J. Luque, C. A. Laughton, and M. Orozco. 2000. DNA-triplex stabilizing properties of 8-aminoguanine. J. Am. Chem. Soc. 122:5997–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cubero, E., N. G. Abrescia, J. A. Subirana, F. J. Luque, C. A. Laughton, and M. Orozco. 2003. J. Am. Chem. Soc. 125:14603–14612. [DOI] [PubMed] [Google Scholar]
- 58.Abrescia, N. G., A. Thompson, T. Huynh-Dinh, and J. A. Subirana. 2002. Proc. Natl. Acad. Sci. USA. 99:2806–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lane, A. N., S. Ebel, and T. Brown. 1993. NMR assignments and solution conformation of the DNA.RNA hybrid duplex d(GTGAACTT)· r(AAGUUCAC). Eur. J. Biochem. 215:297–306. [DOI] [PubMed] [Google Scholar]
- 60.Gonzalez, C., W. Stec, M. A. Reynolds, and T. L. James. 1995. Structure and dynamics of a DNA.RNA hybrid duplex with a chiral phosphorothioate moiety: NMR and molecular dynamics with conventional and time-averaged restraints. Biochemistry. 34:4969–4982. [DOI] [PubMed] [Google Scholar]
- 61.Gyi, J. I., D. Gao, G. L. Conn, J. O. Trent, T. Brown, and A. N. Lane. 2003. The solution structure of a DNA.RNA duplex containing 5-propynyl U and C; comparison with 5-Me modifications. Nucleic Acids Res. 31:2683–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noy, A., A. Pérez, M. Márquez, F. J. Luque, and M. Orozco. 2005. Structure, recognition properties and flexibility of the DNA·RNA hybrid. J. Am. Chem. Soc. 127:4901–4920. [DOI] [PubMed] [Google Scholar]
- 63.Cheatham, T. E., III, and P. A. Kollman. 1997. Molecular dynamics simulations highlight the structural differences among DNA:DNA, RNA:RNA, and DNA. RNA Hybrid Duplexes. J. Am. Chem. Soc. 119:4805–4825. [Google Scholar]












