Abstract
Archaeal box C/D sRNAs guide the 2′-O-methylation of target nucleotides using both terminal box C/D and internal C′/D′ RNP complexes. In vitro assembly of a catalytically active Methanocaldococcus jannaschii sR8 box C/D RNP provides a model complex to determine those structural features of the guide:target RNA duplex important for sRNA-guided nucleotide methylation. Watson–Crick pairing of guide and target nucleotides was found to be essential for methylation, and mismatched bases within the guide:target RNA duplex also disrupted nucleotide modification. However, dependence upon Watson–Crick base-paired guide:target nucleotides for methylation was compromised in elevated Mg2+ concentrations where mismatched target nucleotides were modified. Nucleotide methylation required that the guide:target duplex consist of an RNA:RNA duplex as a target ribonucleotide within a guide RNA:target DNA duplex that was not methylated. Interestingly, D and D′ target RNAs exhibited different levels of methylation when deoxynucleotides were inserted into the target RNA or when target methylation was carried out in elevated Mg2+ concentrations. These observations suggested that unique structural features of the box C/D and C′/D′ RNPs differentially affect their respective methylation capabilities. The ability of the sR8 box C/D sRNP to methylate target nucleotides positioned within highly structured RNA hairpins suggested that the sRNP can facilitate unwinding of double-stranded target RNAs. Finally, increasing target RNA length to extend beyond those nucleotides that base pair with the sRNA guide sequence significantly increased sRNP turnover and thus nucleotide methylation. This suggests that target RNA interaction with the sRNP core proteins is also important for box C/D sRNP-guided nucleotide methylation.
Keywords: box C/D RNA, sRNA, snoRNA, RNA-guided nucleotide modification, nucleotide methylation
INTRODUCTION
Box C/D RNAs are abundant in both eukaryotic and archaeal organisms, functioning to direct the 2′-O-methylation of numerous nucleotides found in ribosomal RNAs, tRNAs, snRNAs, and even some mRNAs (Tycowski et al. 1998; Cavaille et al. 2000; Clouet d'Orval et al. 2001; Jady and Kiss 2001; Kiss 2001; Bachellerie et al. 2002; Terns and Terns 2002; Decatur and Fournier 2003). Guide sequences within the box C/D RNA base pair to complementary sequences in the target RNA while associated core proteins carry out ribose methylation of the designated nucleotide. The conservation of box C/D guide RNAs in both Eukarya and Archaea strongly suggests that RNA-guided nucleotide modification is an ancient and evolutionarily conserved process (Tran et al. 2004).
All box C/D guide RNAs are defined by conserved boxes C (RUGAUGA) and D (CUGA) located at the 5′ and 3′ termini of the guide RNA, respectively (Tyc and Steitz 1989; Watkins et al. 1996; Xia et al. 1997; Caffarelli et al. 1998). The terminal box elements fold into a unique stem I-bulge-stem II structure where stem II is established by partial base-pairing of boxes C and D. This RNA element is stabilized by tandem-sheared G:A base pairs that hydrogen bond across the asymmetric bulge. This distinct secondary structure is known as a kink-turn, or K-turn, and serves as a platform for core protein binding and RNP assembly (Watkins et al. 2000; Klein et al. 2001).
Internal boxes C′ and D′ are found in many box C/D RNAs and are most typical in the archaeal sRNAs (Kiss-Laszlo et al. 1998; Gaspin et al. 2000; Omer et al. 2000). Archaeal boxes C′ and D′ are highly conserved with respect to the terminal C and D sequences and they also fold into a K-turn structure. The typical C′/D′ K-turn lacks the canonical stem I structure, which is replaced by a small loop, and the C′/D′ K-turn is often designated the “K-loop.” The K-loop also serves as a core protein binding platform (Tran et al. 2003; Nolivos et al. 2005). Thus, terminal box C/D and internal C′/D′ RNPs both direct site-specific nucleotide modification via base-pairing of target sequences to the guide regions located adjacent to the D and D′ boxes (Kiss-Laszlo et al. 1998; Tran et al. 2003).
Archaeal box C/D and C′/D′ RNPs are assembled with three core proteins (Omer et al. 2002; Rashid et al. 2003; Tran et al. 2003). Ribosomal protein L7 specifically recognizes the box C/D K-turn and C′/D′ K-loop motifs and serves to initiate RNP assembly (Kuhn et al. 2002; Nolivos et al. 2005). Box C/D and C′/D′ RNP assembly is completed by the binding of Nop56/58 and then fibrillarin, the methyltransferase enzyme. The ability of Nop56/58 and fibrillarin to dimerize suggests that these two core proteins may bind as a heterodimer (Aittaleb et al. 2003; Tran et al. 2003; Zhang et al. 2006). Nucleotide methylation depends on complete RNP assembly and the presence of all three core proteins. Assembly of both box C/D and C′/D′ RNPs allows the fully assembled sRNP to function as a dual guide RNA, directing 2′-O-methylation of targeted nucleotides from both RNP complexes (Rashid et al. 2003; Tran et al. 2003).
The ability to assemble in vitro archaeal box C/D sRNP complexes has greatly facilitated our understanding of box C/D RNP structure and function (Omer et al. 2002; Rashid et al. 2003; Tran et al. 2003). Recombinant box C/D sRNP core proteins bind the box C/D and C′/D′ motifs in a stepwise fashion and the fully assembled complexes direct nucleotide-specific methylation of target RNAs from both RNP complexes. Investigations have demonstrated that efficient box C/D and C′/D′ RNP-guided nucleotide modification requires that both complexes be juxtaposed in the assembled sRNP (Tran et al. 2003). Also, the highly conserved spacing between RNPs required for nucleotide methylation indicates inter-RNP interactions are important for the nucleotide methylation reaction (Tran et al. 2005).
In vitro assembled archaeal box C/D sRNPs provide model complexes to investigate structure–function relationships of this RNA:protein enzyme. We have utilized the Methanocaldococcus jannaschii sR8 sRNP to examine the interaction of the target RNA substrate with the guide sequences of this RNA:protein enzyme. The ability to mutate target RNAs as well as sRNA guide sequences permitted the determination of those structural features of the guide:target RNA duplex that are important for nucleotide methylation. Initial experiments demonstrated that Watson–Crick base-pairing between the guide and target nucleotide is essential for nucleotide methylation. Establishment of an RNA:RNA duplex between guide sequence and target RNA was also required and the specificity of nucleotide methylation was compromised by elevated magnesium concentrations. Methylation of target nucleotides positioned in highly structured target RNAs suggested an ability of the sRNP complex to facilitate target RNA unfolding for nucleotide modification. Finally, experiments also indicated that target RNA interaction with core proteins contributed significantly to the efficiency of the nucleotide methylation reaction by increasing sRNP turnover.
RESULTS
Watson–Crick pairing of guide and target nucleotides is essential for nucleotide methylation
The folded secondary structure of M. jannaschii sR8, a model archaeal box C/D sRNA, is illustrated in Figure 1. We have established an in vitro archaeal box C/D sRNP assembly system using in vitro transcribed sR8 sRNA and recombinant M. jannaschii core proteins L7, Nop56/58, and fibrillarin (Tran et al. 2003). This reconstituted sRNP is catalytically active and 2′-O-methylates target nucleotides in synthetic target RNAs complementary to both D and D′ guide regions. S-Adenosylmethionine (SAM) is the methyl donor and 3H–CH3 incorporation into the target RNAs measures box C/D and C′/D′ RNP methylation activities.
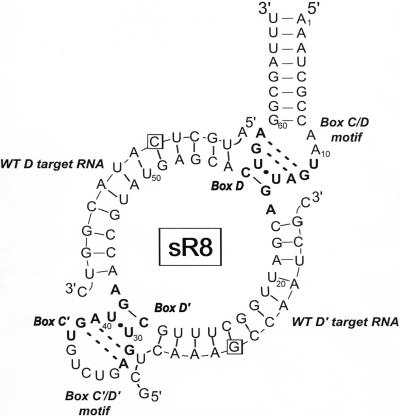
FIGURE 1.
Secondary structure of the M. jannaschii sR8 box C/D sRNA base paired with D and D′ target RNAs. M. jannaschii sR8 box C/D sRNA folded into its secondary structure to form the box C/D and C′/D′ motifs. Boxes C, C′, D, and D′ are indicated in bold. The box C/D and C′/D′ motifs form K-turn structures critical for the binding of archaeal box C/D sRNP core proteins with base-pairing of tandem-sheared G:A pairs indicated by dashed lines. D and D′ sRNA guide regions are base paired with the respective WT D and WT D′ target RNAs used in these studies. The boxed target nucleotide will be 2′-O-methylated during the sRNP-guided nucleotide modification reaction.
Previous in vivo and in vitro systems have assessed the importance of Watson–Crick pairing between guide and target nucleotides for 2′-O-methylation with some data suggesting that methylation of non-Watson–Crick pairs depends upon the specific experimental system used for analysis (Cavaille and Bachellerie 1998; Singh et al. 2004). In light of these conflicting results, analysis of sRNA guide sequence:target RNA interaction began by assessing the importance of Watson–Crick paired guide and target nucleotides for methylation in this system. Synthetic target RNAs of 14 and 16 nucleotides (nt) in length used in these studies were complementary to the sR8 D and D′ guide regions, respectively (Fig. 1). Target RNAs possessing 2′-O-methylated target nucleotides served as negative controls. Mutant D and D′ target RNAs were synthesized to alter guide:target nucleotide base pairing. For A:U Watson–Crick pairs between guide and target nucleotides, point mutations were also made in the sR8 D and D′ guide sequences. Each sR8 sRNP complex tested contained only an altered D or D′ guide:target nucleotide pair, and the nonmutated guide:target nucleotide pair served as an internal control for comparison.
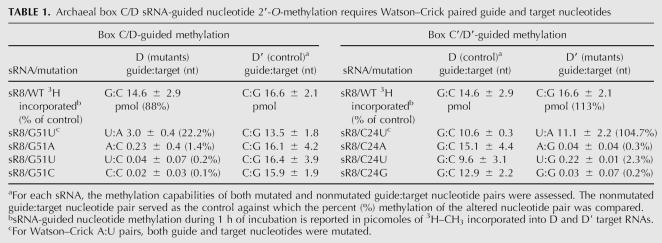
Table 1 summarizes the methylation capabilities of the various guide:target nucleotide base pairs for both D and D′ guide:target RNA duplexes. Watson–Crick G:C base pairs for both D and D′ guide regions exhibited robust target nucleotide methylation. This was also the case for Watson–Crick U:A base pairs although the level of A residue methylation was significantly reduced for the D target U:A pair. This lower level of methylation may be a result of overall guide:target RNA duplex structure. In contrast, a non-Watson–Crick G:U pair (D′ target) or nucleotide mismatches (both D and D′ targets) were severely deficient. Interestingly, the non-Watson–Crick G:U base pair was not significantly methylated despite the hydrogen bonding between guide and target nucleotides. Compensatory mutations of the D and D′ sRNA guide sequences that restored Watson–Crick pairing also restored target nucleotide methylation (data not shown). Collectively, these results clearly demonstrated that, for this in vitro assembled sRNP, Watson–Crick pairing between guide and target nucleotide is critical for nucleotide methylation.
TABLE 1.
Archaeal box C/D sRNA-guided nucleotide 2′-O-methylation requires Watson–Crick paired guide and target nucleotides
Target nucleotide methylation requires a continuous, Watson–Crick paired, guide:target RNA duplex
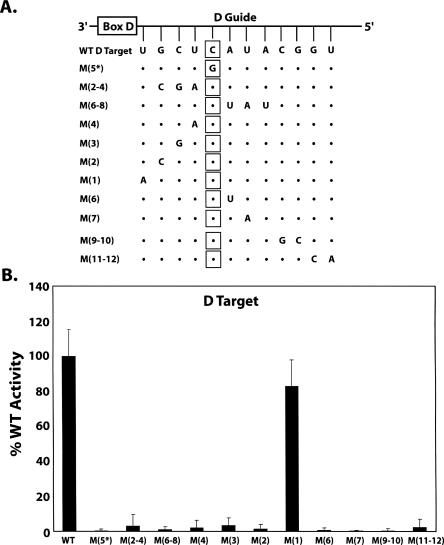
The importance of Watson–Crick base-pairing between the sRNA guide sequence and the target RNA was examined next. Target RNAs were mutated to disrupt Watson–Crick pairing of selected RNA duplex nucleotide pairs (Fig. 2A) and the effect of these mismatches upon D target nucleotide methylation assessed (Fig. 2B). Results clearly demonstrated that Watson–Crick pairing of the guide:target RNA duplex was critical for nucleotide modification. Disruption of three base pairs on either side of the guide:target nucleotide pair abolished methylation of the target nucleotide. Target nucleotide methylation was disrupted even when single nucleotide pairs around the target nucleotide were mismatched. The requirement for base-pairing extended along the entire guide:target RNA duplex. The single exception was the guide:target nucleotide pair immediately adjacent to box D where single mismatches at this position have been reported for some wild-type box C/D RNAs (Kiss-Laszlo et al. 1996; Cavaille and Bachellerie 1998; Gaspin et al. 2000). This may suggest that the guide:target RNA duplex between box D and the target nucleotide is less critical for substrate:enzyme interaction and more important for the correct spatial positioning of the target nucleotide with respect to box D and the assembled sRNP (see Discussion).
FIGURE 2.
Target nucleotide methylation requires a continuous, Watson–Crick paired guide:target RNA duplex. D target RNAs possessing mismatched nucleotides were incubated with in vitro assembled sR8 box C/D sRNP and 2′-O-methylation of the target nucleotide assessed. (A) Schematic presentation of sR8 D guide region with associated box D base paired to wild-type D target RNA (WT D target). Mutated WT D target RNAs are indicated below with mismatched nucleotides designated in capital letters and their distance from box D indicated in parentheses. The target nucleotide is enclosed in squares with dots indicating conserved WT D target RNA ribonucleotides. (B) Incorporation of 3H–CH3 into D target RNAs. The level of target nucleotide methylation for each mismatched target RNA is indicated as the percent of WT D target RNA methylation.
Elevated Mg2+ concentrations promote 2′-O-methylation of mismatched guide:target nucleotide pairs
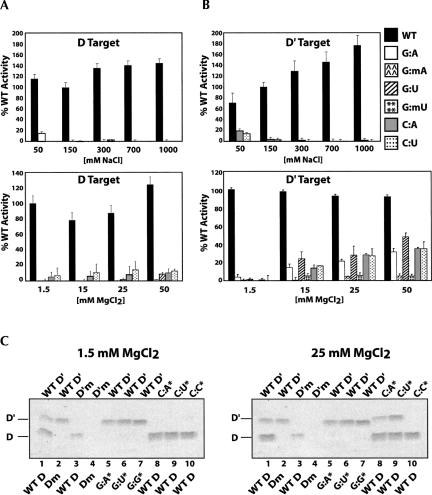
Wild-type D and D′ target RNAs possessing Watson–Crick as well as mismatched guide:target nucleotide pairs were assessed for nucleotide methylation under increasing concentrations of NaCl and MgCl2. A standard NaCl concentration of 150 mM is used for both sRNP assembly and methylation. NaCl concentrations were incrementally increased up to 1 M and the modification activity of both box C/D and C′/D′ RNPs determined (Fig. 3A,B, top panels). Previous work in our laboratory has shown that the sR8–sRNP complex is stable in 1 M NaCl (E. Tran and E.S. Maxwell unpubl.). For wild-type Watson–Crick base pairs, increasing NaCl concentrations had no significant effect upon the methylation levels of the D target nucleotide. Interestingly however, target nucleotide methylation levels increased almost twofold for the D′ target nucleotide at 1 M NaCl concentrations. Methylation for both D and D′ target RNAs was site specific, as control target RNAs possessing a 2′-O-CH3 at the target nucleotide were not methylated (data not shown). For mismatched guide:target nucleotide pairs, there was no methylation of target nucleotides except perhaps for some very low levels of the D′ target at suboptimal NaCl concentrations.
FIGURE 3.
Elevated Mg+2 concentrations promote 2′-O-methylation of mismatched guide and target nucleotides. WT D and WT D′ target RNAs with wild-type or mismatched guide:target nucleotide pairs were incubated with in vitro assembled sR8 sRNP and increasing concentrations of NaCl or MgCl2, and the 2′-O-methylation of target nucleotides was assessed. Specific mismatched guide:target ribonucleotides are indicated at the side with NaCl or MgCl2 concentrations indicated below. Methylation is reported as percent (%) of wild-type, Watson–Crick guide:target nucleotide base pairs. (A) WT D target nucleotide methylation of Watson–Crick and mismatched guide:target nucleotide base pairs in increasing concentrations of NaCl (top panel) and MgCl2 (bottom panel). (B) WT D′ target nucleotide methylation of Watson–Crick and mismatched guide:target nucleotide base pairs in increasing concentrations of NaCl (top panel) and MgCl2 (bottom panel). (C) WT D and WT D′ target RNAs possessing wild-type Watson–Crick or mismatched guide:target nucleotide base pairs were incubated in low (1.5 mM) or high (25 mM) MgCl2 concentrations and nucleotide methylation assessed by polyacrylamide gel electrophoresis of 3H-methylated target RNAs visualized by autoradiography. Electrophoretic migration positions of D′ and D target RNAs are indicated at the side. WT target RNAs, target RNAs possessing a methylated target nucleotide, and target RNAs possessing mutated guide:target nucleotide base pairs (asterisks) are indicated for the D′ target and D target RNAs above and below the individual gel lanes, respectively.
The effect of increasing Mg2+ concentrations upon box C/D and C′/D′ RNP methylation was also examined, as previous work has demonstrated the importance of MgCl2 for sRNP assembly and nucleotide methylation (Galardi et al. 2002; Goody et al. 2004; Hardin and Batey 2006). Increasing MgCl2 concentrations from the standard assay condition from 1.5 to 50 mM had little effect upon methyltransferase activity of Watson–Crick guide:target nucleotide pairs for both the box C/D and C′/D′ RNPs (Fig. 3A,B, bottom panels). However, a substantial level of methylation for mismatched guide:target nucleotide pairs was observed. The methylation of non-Watson–Crick paired target nucleotides was particularly significant for the C′/D′ RNP and approached 40% that of wild-type target RNA at 50 mM MgCl2. These non-Watson–Crick pairs included the hydrogen bonded G:U pair as well as several mismatches. Notably, increased methylation was a general phenomenon for all non-Watson–Crick pairs tested. Also important was the fact that methylation was specific for the fifth or target nucleotide, as mismatched target nucleotides possessing a 2′-O-CH3 exhibited little modification. Preferential methylation of mismatched D′ target nucleotides was confirmed with polyacrylamide gel electrophoresis of selected D and D′ target RNAs possessing target nucleotide mismatches (Fig. 3C). Collectively, these analyses demonstrated a differential response for box C/D versus C′/D′ RNP-guided nucleotide methylation under nonstandard assay conditions, suggesting that distinct structural features of the two different RNPs can affect their catalytic activities.
Target nucleotide methylation requires an RNA:RNA duplex formed between the sRNA guide sequence and the target RNA
Utilization of an in vitro assembled box C/D sRNP methylation system allowed us to examine the importance of forming an RNA:RNA duplex between the guide sequence and target RNA. Initial experiments replaced single ribonucleotides with deoxynucleotides at position 3, 4, 5, 6, or 7 (Fig. 4A). Methylation was disrupted only when the fifth target nucleotide lacked a 2′-OH for methyl addition. These results demonstrated not only site-specific nucleotide methylation but also that insertion of a single deoxynucleotide into the guide:target RNA duplex has no effect upon methylation.
FIGURE 4.
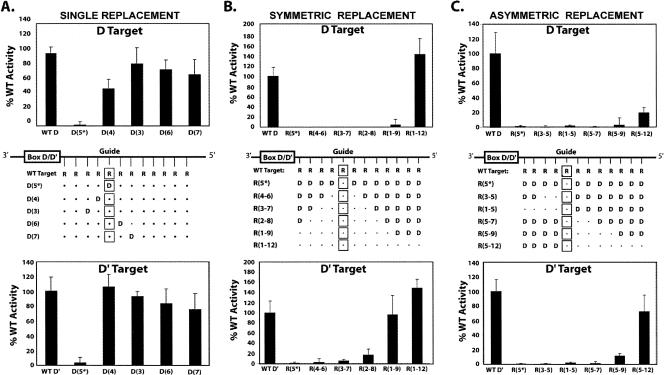
Target nucleotide methylation requires an RNA:RNA duplex formed between the guide sequence and target RNA. WT D and WT D′ target RNAs substituted with deoxynucleotides were incubated with in vitro assembled sR8 box C/D sRNP and 2′-O-methylation of the target nucleotide assessed. Schematic presentation of sR8 D and D′ guide regions base paired with wild-type target RNA (WT D) and chimeric RNA–DNA targets are shown in each panel. Wild-type ribonucleotides are indicated by dots and target nucleotides are enclosed in squares. Substituted deoxynucleotides are indicated by D and their position from box D or D′ is indicated in parentheses. All D and D′ targets excepting D(5*) possessed a ribonucleotide with 2′-OH at the fifth or target nucleotide. Methylation of D and D′ targets is indicated as percent of 3H–CH3 incorporation into WT D or WT D′ target RNAs. (A) Single ribonucleotides of the WT target RNA replaced with deoxynucleotides and target RNA nucleotide methylation assessed. (B) Deoxynucleotide D and D′ target RNAs symmetrically replaced outward from the target nucleotide with increasing numbers of ribonucleotides. (C) Deoxynucleotide D and D′ target RNAs asymmetrically replaced upstream of or downstream from the target nucleotide with increasing numbers of ribonucleotides.
Chimeric DNA–RNA oligonucleotides complementary to D and D′ guide sequences were then incubated with in vitro assembled sR8 sRNP complexes and methylation of the target nucleotide assessed. Each of these chimeric oligonucleotides possessed a ribonucleotide at the fifth target nucleotide, thus providing a 2′-OH for methyl addition. Chimeric D and D′ target DNA deoxynucleotides were gradually replaced with ribonucleotides in either symmetric or asymmetric fashion progressing outward from the target nucleotide (Fig. 4A,B, upper panels). In striking contrast to single ribonucleotide replacement, chimeric D and D′ targets were not methylated, and recovery of target nucleotide methylation required the substantial replacement of the deoxynucleotides with ribonucleotides (Fig. 4B,C, lower panels). These results indicated that formation of an RNA:RNA helix is critical for target nucleotide methylation.
Progressive symmetric replacement of deoxynucleotides with ribonucleotides gradually restored methylation activity for both RNP complexes, although the methylation activity of the two RNPs recovered at different rates (Fig. 4B). Only slight methyltransferase activity was observed for the box C/D RNP when nine ribonucleotides replaced the corresponding deoxynucleotides and full recovery of wild-type (WT) methyltransferase activity required the base-pairing of all 12 guide sRNA nucleotides with target ribonucleotides. Significant D′ target nucleotide methylation was observed when 7 nt were substituted for the corresponding deoxynucleotides (∼20% activity) and full activity was restored with the replacement of nine ribonucleotides. Interestingly, target RNAs replacing all 12 deoxynucleotides pairing with the guide sequence with ribonucleotides exhibited somewhat higher methylation levels than the WT D and WT D′ target RNAs. However, the WT D and R(1–12) target RNAs differ in the 5′ and 3′ nucleotides that extend beyond the target RNA sequence complementary to the guide sequence. The WT D and D′ target RNAs possessed ribonucleotides, whereas the D and D′ R(1–12) target RNAs possessed deoxynucleotides. At present, it is not apparent why these different extended nucleotides result in altered methylation levels.
Asymmetric replacement of either upstream or downstream target deoxynucleotides with ribonucleotides was examined next. Complete restoration of the upstream guide RNA:target RNA duplex between the target nucleotide and the D or D′ box did not result in any target nucleotide methylation (Fig. 4C). In contrast, target nucleotide methylation was partially recovered as the downstream RNA:RNA helix of the guide:target duplex was restored. Approximately 20% of wild-type methylation levels were observed for the D target nucleotide when the entire downstream guide:target duplex was converted to an RNA:RNA duplex. The D′ target nucleotide was methylated to ∼70% of wild-type levels when the downstream guide:target duplex was entirely ribonucleotide base pairs. This suggested that establishment of an RNA:RNA helix was more important downstream from the target nucleotide than upstream (see Discussion). The fact that box C/D and C′/D′ RNP nucleotide methylation activities were differentially affected by either symmetric or asymmetric replacement of deoxynucleotides with ribonucleotides again suggested that distinct RNP structural differences might account for the observed differences.
Target nucleotides positioned within highly folded target RNAs are efficiently methylated
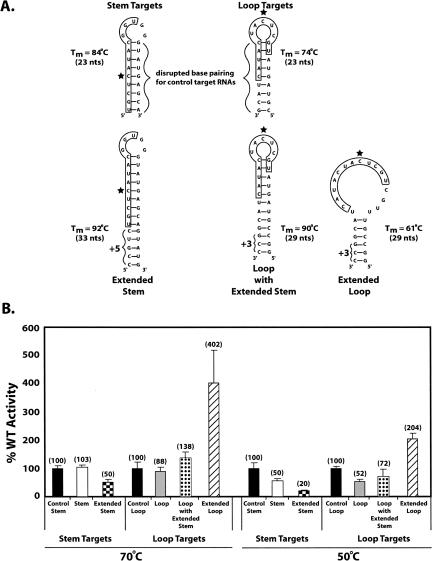
Nucleotides targeted by box C/D RNPs for methylation are often positioned within highly folded or structured regions of the target RNA. As the sequence surrounding the target nucleotide must be single stranded to base pair with the guide region, unfolding this structured target RNA for nucleotide methylation is necessary. To assess the possible ability of the sRNP complex to facilitate unfolding, target RNAs were constructed that positioned the target nucleotide in highly structured regions (Fig. 5A). The first class of target RNAs positioned the target nucleotide in the center of a double-stranded RNA hairpin (stem targets) whereas the second class positioned the target nucleotide in the loop region of a hairpin–loop structure (loop targets). In addition to simple stem and loop target RNAs, some targets possessed extended stem structures or an expanded loop region. The Tm of each target RNA target was determined by UV melting analysis and is indicated in Figure 5A. Control stem and loop target RNAs were also synthesized where the double-stranded stem was disrupted by altering one strand of the complementary sequence. The control RNA for the loop target possessed an altered sequence for base-pairing to the sRNA guide sequence and thus required a new sRNA guide sequence (sR8 GRDtl) to maintain RNA:RNA pairing between the guide sequence and target RNA.
FIGURE 5.
Target nucleotides positioned within highly folded target RNAs are methylated. D target RNAs possessing target nucleotides within highly folded RNA secondary structures were incubated with in vitro assembled sR8 box C/D sRNP and the methylation of target nucleotides assessed. (A) D target RNAs folded into hairpin-loop structures with the target nucleotide positioned within the RNA hairpin (stem targets) or RNA loop region (loop targets). Target RNA sequences base-pairing with the sR8 D guide region are boxed and target nucleotides are indicated with stars. Brackets indicate the RNA hairpin region disrupted in the control stem and loop target RNAs. Target RNA melting temperatures (Tm) are indicated at the side. (B) Incorporation of 3H–CH3 into D target RNAs. Levels of D target nucleotide methylation (various folded hairpin-loop structures indicated by assorted bar coloration) are presented in parentheses as the percent of nonfolded, control D target RNAs (black bars) with methylation incubation temperatures indicated below.
For simple stem and loop target RNAs, both were highly methylated at 70°C and at comparable levels to their unstructured control target RNAs (Fig. 5B). To address the possibility that the standard assay condition of 70°C was sufficiently close to the target RNA melting temperatures to promote transient target RNA hairpin melting, a second assay was conducted at a lower reaction temperature of 50°C. This lower temperature was ∼25°C–35°C below these target RNA's hairpin Tm. Therefore, the stability of the hairpin structures should be significantly increased. While reduction in temperature reduced the overall levels of nucleotide methylation from optimal levels observed at 70°C, both target RNAs were effectively methylated (∼50%) in comparison to their unstructured RNA controls (Fig. 5B). These results suggested that the archaeal sRNP complex can facilitate unfolding of RNA hairpin structures so as to promote guide:target base-pairing and nucleotide methylation.
Extending the stem structure (extended stem and loop with extended stem) or increasing the loop size(extended loop) had expected as well as unexpected consequences upon methylation levels. At 70°C, the extended stem target was reduced in methlylation as anticipated and methylation was further reduced when the reaction temperature was lowered to 50°C. Nevertheless, even at 50°C, the extended stem RNA was substantially methylated to ∼20% of the control target RNA. On the other hand, extending the stem of the loop target RNA (loop with extended stem) unexpectedly enhanced methylation with respect to the simple loop target at both 70°C and 50°C. Surprisingly, target RNA methylation was dramatically increased with respect to the control target RNA when the loop was enlarged from 5 (loop) to 17 (extended loop) nt. These results are difficult to rationalize when only considering the target RNA's folded structure. Notably, determination of some target RNA Tm values (extended stem and loop with extended stem) suggested alternative folded structures as evidenced by biphasic melting curves (data not shown). These alternative structures as well as the extended target RNA sequence (see below) may help to explain the enhanced methylation levels observed for some of these target RNAs.
Extending target RNAs beyond the guide:target RNA duplex significantly enhances methylation activity by increasing enzyme turnover
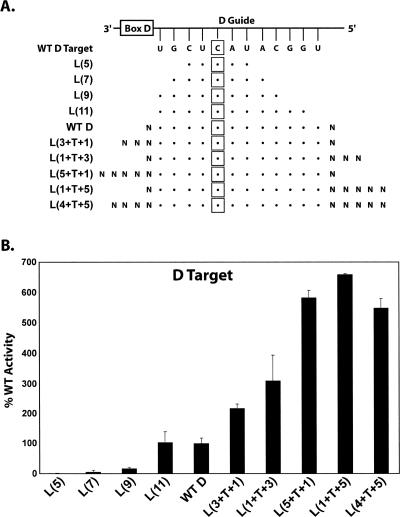
Our studies primarily focused upon those structural features of the guide:target RNA duplex itself that are important for nucleotide methylation. However, concluding experiments investigated the required length of the RNA target for nucleotide methylation and, more specifically, the effect of extending the target RNA beyond those nucleotides that base pair with the sRNA guide sequence (Fig. 6). Initial experiments determined that a minimal D target RNA length of 9 nt was required to obtain low levels of nucleotide methylation. Extension of the target RNA to 11 nt essentially restored nucleotide modification comparable to that of a target RNA pairing with the entire sRNA guide sequence. Strikingly, however, increasing the target RNA to extend beyond the sRNA guide sequence dramatically increased the level of D target RNA methylation. Extension of the RNA 3 nt beyond the 5′ (3 + T + 1) or 3′ (1 + T + 3) end of the target sequence pairing with the sRNA guide sequence increased nucleotide methylation between two- and threefold. Extending either end 5 nt (5 + T + 1 or 1 + T + 5) increased methylation five- to sixfold. The effect of addition at each terminus was not additive, as extending both termini by 5 nt (5 + T + 5) gave the same increase in methylation as did extension on either end. These results suggested that the target RNA binding to the sRNP utilizes nucleotides beyond that RNA sequence hydrogen bonded to the sRNA guide sequence, and this binding most likely involves interactions with sRNP core proteins.
FIGURE 6.
Target RNA methylation increases when the target RNAs extend beyond the guide:target duplex. D target RNAs of increasing length were incubated with in vitro assembled sR8 box C/D sRNP and the methylation of target nucleotides assessed. (A) Schematic presentation of sR8 D guide region base paired with target RNAs of increasing length. Target nucleotides base paired with the guide sequence are indicated by dots with target RNA nucleotides extending beyond the guide:target RNA duplex indicated by N. Target nucleotides are enclosed in squares. Target RNAs are designated at the side with 5′ and 3′ nucleotides extended beyond the guide:target RNA indicated in the parentheses with respect to the 12 target nucleotides (T) base paired to the guide sequence. (B) Incorporation of 3H–CH3 into D target RNAs of increasing length. Levels of target nucleotide methylation are indicated as the percent of the 14 nt control WT D target RNA.
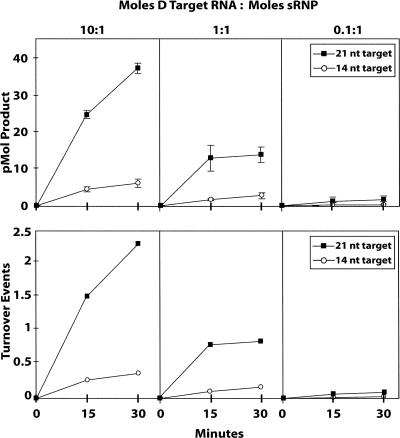
The effect of target RNA length upon the kinetics of nucleotide methylation was then examined using both short and extended D target RNAs at three different D target RNA to sRNP molar ratios. As the ratio of target RNA to assembled sRNP was increased, the picomoles of target RNA methylated were also increased (Fig. 7A). This is consistent with elevated concentrations of the substrate target RNA driving the reaction toward nucleotide methylation. As anticipated, the extended D target RNA of 21 nt (Fig. 6, target RNA L[4 + T + 5]) was methylated at significantly higher levels with respect to the shorter 14 nt WT D target RNA (Fig. 6, WT D target) at all three target RNA-to-sRNP ratios. Enzyme or sRNP turnover number was then calculated from these data (Fig. 7B). For the shorter D target RNA at all three target RNA:sRNP ratios, the turnover number was calculated to be less than one turnover per 30 min. These low values indicated relatively inefficient sRNP-guided nucleotide methylation activity. However, increasing the D target RNA length increased the turnover number at all three target RNA:sRNP ratios. At a target RNA:sRNP ratio of 10:1, a molar ratio comparable to the standard reaction conditions used in this study, a turnover of ∼2.5 catalytic events per sRNP per 30 min was observed. This turnover number is similar to that estimated by Omer et al. (2002) for an in vitro assembled Sulfolobus solfataricus box C/D sRNP complex methylating an extended target RNA. Enhanced turnover for the extended target RNA suggested either more efficient binding of the target RNA to the sRNP (target RNA interaction with the sRNP core proteins?) or more efficient target RNA dissociation from the sRNP complex upon nucleotide modification (see Discussion).
FIGURE 7.
Extended target RNAs significantly enhance nucleotide methylation activity by increasing catalytic turnover. Short WT D (14 nt) and extended D target RNAs (21 nt) were incubated with in vitro assembled sR8 box C/D sRNP and nucleotide methylation assessed at three different molar ratios of D target RNA to sRNP. Upper panels indicate picomoles of short WT (circles) and extended (squares) D target RNAs methylated over 30-min incubation periods. Lower panels indicate sRNP turnover for short WT (circles) and extended (squares) D target RNAs calculated from the average picomoles of product produced per 30-min incubation period. Molar ratios of target RNA to sRNP are indicated at the top.
DISCUSSION
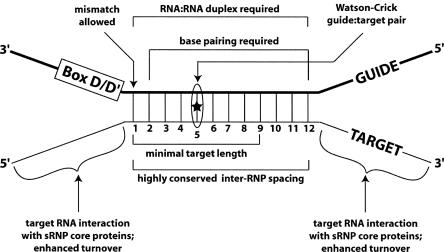
The in vitro assembled archaeal box C/D sRNP provides a model complex for the investigation of box C/D RNA-guided nucleotide modification. The experiments presented here defined those structural features of the guide:target RNA duplex important for sRNA-guided nucleotide 2′-O-methylation. Results demonstrated the importance of a continuously Watson–Crick paired, RNA:RNA duplex for guide sequence:target RNA interaction and nucleotide methylation. A minimal guide:target RNA duplex was also required and increased nucleotide methylation resulted upon extending the target RNA beyond the sRNA guide sequence. These extended target RNAs likely interact with the sRNP core proteins to enhance catalytic turnover of the complex. The ability to methylate target nucleotides within highly folded target RNAs suggested an ability of this RNP complex to facilitate RNA unfolding. A summary of guide:target RNA interactions and structural features important for box C/D sRNA-guided nucleotide modification is presented in Figure 8.
FIGURE 8.
Summary of the structural features of the guide:target RNA duplex important for nucleotide methylation.
Efficient methylation required Watson–Crick pairing of the guide and target nucleotides. Notably, methylation of the D target A:U pair was considerably reduced compared to all other Watson–Crick pairs surveyed. Previous work has suggested that G:C content affects the strength of guide:target RNA interaction and hence methylation efficiency (Cavaille and Bachellerie 1998). However, it is unlikely in this case that reduced methylation is related to base-pairing strength, but rather to an alteration in guide RNA structure. For this A:U pair, the sRNA guide sequence was mutated to establish the A:U pair. Recent work in our laboratory has demonstrated that individual guide region sequences can have significant impact on the overall folding of an sRNA (Gagnon et al. 2006). Thus, we suspect that this sequence change affected the guide region structure and, in turn, interaction with the target RNA and methylation efficiency of this particular target nucleotide. Also notable was the lack of methylation of the non-Watson–Crick G:U pair despite hydrogen bonding between the guide and target nucleotides. Previous in vivo analysis has indicated very low levels of target nucleotide methylation for a G:U pair when positioned within an elongated, G:C-rich, guide:target RNA duplex (Cavaille and Bachellerie 1998). The very slight elevation of methlylation for the G:U pair over that of mismatched guide:nucleotide pairs in our analysis could reflect this minor amount of nucleotide methylation. However, a G:U pair has never been identified at the guide:target nucleotide position for either eukaryotic or archaeal box C/D RNAs, suggesting that the hydrogen bonding of this non-Watson–Crick pair is not sufficient to guide nucleotide methylation in vivo (Cavaille and Bachellerie 1998).
Most striking is the recent demonstration of mismatched target nucleotide methylation for the in vitro assembled box C/D RNP of Haloferax volcanii pre-tRNATrp (Singh et al. 2004). This RNP substantially methylates a variety of mismatched guide:target nucleotide pairs as well as the non-Watson–Crick G:U pair. However, this complex is unique in character in that this intron-encoded box C/D sRNA is positioned in cis with respect to its pre-tRNATrp target nucleotides (Clouet d'Orval et al. 2001; Singh et al. 2004). However, methylation occurs in trans guided by intron-encoded box C/D and C′/D′ RNPs of either the unspliced pre-tRNATrp precursor or the excised circular intron (Singh et al. 2004). The structural constraints imposed by positioning this box C/D sRNP within the pre-tRNATrp intron or excised circular intron coupled with shorter guide region lengths may help explain the unusual methylation characteristics of this in vitro assembled box C/D sRNP.
The stringent requirement for a Watson–Crick guide:target nucleotide pair is compromised with increased concentrations of MgCl2. The effect of Mg2+ ions upon RNA structure is well known (Misra et al. 2003) and suggests that the interaction of the target RNA with the guide sequence is sufficiently altered to allow modification of these mismatched target nucleotides. Recent work has reported similar results for the in vitro assembled Pyrococcus horikoshii box C/D sRNP (Hardin and Batey 2006). Most notable was the methylation of mismatched target nucleotides for the D′ target RNA. Alteration of C′/D′ RNP capabilities for D′ target methylation was also observed with elevated NaCl. We suspect that that the box C/D and C′/D′ complexes are sufficiently distinct in RNP structure despite the fact that both RNP are assembled with the same three core proteins binding a K-turn motif (Omer et al. 2002; Rashid et al. 2003; Tran et al. 2003). Differences in the K-turn structure itself for the box C/D (K-turn) and C′/D′ (K-loop) motifs (Nolivos et al. 2005) may account for the differential activities of the two complexes under altered methylation conditions.
The requirement for Watson–Crick pairing along the entire guide:target RNA duplex is also quite stringent. Mismatches both adjacent to and removed from the guide:target nucleotide pair effectively disrupted target nucleotide methylation. The one exception was the guide:target pair immediately adjacent to the D box sequence. These observations are similar to those originally reported by Cavaille and coworkers for eukaryotic box C/D snoRNA-guided methylation in vivo (Cavaille et al. 1996; Cavaille and Bachellerie 1998). This end of guide sequence:target RNA duplex is adjacent to box D and the assembled sRNP complex. Perhaps this terminal nucleotide interacts with core proteins as do those 5′ nucleotides of the extended target RNA. Non-Watson–Crick pairs and mismatches have also been described in native guide:target RNA duplexes. However, almost all examples are for eukaryotic snoRNAs and the large majority are non-Watson–Crick G:U base pairs, but never at the guide:target nucleotide pair. Only several examples of truly mismatched, nonhydrogen bonded pairs have been described and none at the target nucleotide (Cavaille and Bachellerie 1998). The increased stringency of Watson–Crick pairing observed for this in vitro assembled archaeal sRNP may reflect the minimal nature of the complex and the absence of additional cellular proteins that could play ancillary roles during in vivo methylation.
Utilization of an in vitro assembled sRNP complex made it possible to examine box C/D sRNA-guided methylation of DNA targets. While replacement of single ribonucleotides with deoxynucleotides around the target nucleotide had no effect upon nucleotide methylation, utilization of a DNA target resulted in complete disruption of methylation. This suggested that the overall conformation of the guide:target duplex is likely important for methylation. A DNA target hydrogen bonded to an sRNA guide sequence would not adopt an A-helix configuration like that of an RNA:RNA duplex (Fedoroff et al. 1993). It is thus likely that an RNA:DNA helix disturbs correct target nucleotide positioning at the fibrillarin catalytic site or perhaps interaction with fibrillarin and/or Nop56/58.
Symmetric replacement of deoxynucleotides with ribonucleotides resulted in differential effects for the recovery of D and D′ target methylation. Whereas D target methylation required replacement of all deoxynucleotides, the D′ target recovered with shorter ribonucleotide regions. Asymmetric restoration of either upstream or downstream restoration of ribonucleotides revealed that each region affected methylation activity differently. Restoration of the upstream RNA:RNA duplex did not result in methylation of either D or D′ targets. In contrast, nucleotide methylation was gradually restored as downstream deoxynucleotides were replaced with ribonucleotides. Taken together, these results may suggest different roles for the upstream and downstream regions of the guide:target RNA duplex. We believe that the primary function of the upstream guide:target RNA duplex between box D and the target nucleotide is to simply measure, and thus specifically position, the fifth target nucleotide of the target RNA with respect to the fibrillarin catalytic site on the assembled box C/D RNP. Consistent with this suggestion is the observation that base-pairing of the guide:target nucleotide pair adjacent to the D box is not required although those nucleotides are necessary for determining the fifth nucleotide for modification (Kiss-Laszlo et al. 1996; Cavaille and Bachellerie 1998; Gaspin et al. 2000). The gradual restoration of methylation activity with establishment of a downstream RNA:RNA duplex may well indicate that this region is particularly important for stabilizing target RNA substrate binding to the sRNP enzyme. Finally, the different methylation capabilities of the box C/D and C′/D′ RNPs observed in both symmetric and asymmetric replacement experiments are again consistent with the idea of distinct structural features of the two complexes.
In vivo, box C/D RNA-guided methylation frequently requires unfolding of the target RNA for base-pairing with the guide sequence. The ability to methylate target nucleotides within folded target RNAs suggests an ability of the archaeal box C/D sRNP to facilitate RNA unfolding. However, it appears that more is happening for the larger structured targets than simply unfolding double-stranded RNA hairpins. The loop target with extended stem structure was more effectively methylated at both higher and lower reaction temperatures than the simple loop target despite the fact that the extended stem structure raised the Tm of this folded RNA ∼15°C. As the size of a target RNA increases, those nucleotides beyond the region base-pairing to the sRNA guide sequence could be enhancing interaction of the target RNA with the sRNP core proteins, and in turn enhancing catalytic turnover. Also noteworthy is the melting profile of some of the larger, folded target RNAs used in these studies. While the extended stems stabilize the target RNA as evidenced by the Tm of their most stable structure, UV melting suggested alternative folded structures that could significantly affect guide sequence:target RNA interactions (data not shown). Guide sequence:target RNA interactions could also be facilitated by initial interaction of single-stranded regions of each RNA followed by unzippering of the remaining target RNA hairpins. Such a phenomenon is observed when the “kissing loops” of two hairpin-loop RNAs interact to ultimately unfold each RNA and produce a stable, intermolecular RNA duplex.
The box C/D core proteins could ultimately facilitate such unfolding functions although L7, Nop56/58, and fibrillarin do not possess Walker A and Walker B motifs characteristic of RNA helicases (Wang et al. 2000; Aittaleb et al. 2003; Suryadi et al. 2005; Turner et al. 2005; Cordin et al. 2006). Consistent with the suggestion that the target RNA interacts with the core proteins is the observation that extending the target RNA sequence dramatically increases the efficiency of nucleotide methylation via enhancing catalytic turnover of the sRNP. It is likely that these extra target nucleotides are interacting with fibrillarin and/or Nop56/58 to strengthen and stabilize target RNA-sRNP interactions. Indeed, preliminary experiments using target RNAs with either 5′ or 3′ extensions containing 4-thio-uridine have shown that the target RNA can be UV cross-linked to the Nop56/58 core protein (K. Gagnon and E.S. Maxwell, unpubl.). Ultimately, the molecular details of target RNA interaction with the sRNA guide sequence and the associated sRNP core proteins most likely will be revealed with future crystal structures of the fully assembled box C/D sRNP and bound target RNA.
MATERIALS AND METHODS
Construction and synthesis of wild-type and mutant sR8 RNAs
Wild-type and mutant sR8 DNA oligonucleotides were purchased from Integrated DNA Technologies. The sequences for wild-type sR8 and sR8 GR Dtl were as follows:
sR8 WT:
AAATCGCCAATGATGACGATTGGCTTTGCTGAGTCTGTGATGAACCGTATGAGCACTGAGGCGATTT
sR8 GR Dtl:
AAATCGCCAATGATGACGATTGGCTTTGCTGAGTCTGTGATGAGTATGATGAGCACTGAGGCGATTT
Point mutations in sR8 guide regions were introduced during PCR amplification using the full-length, wild-type sR8 template and primers containing the mutated nucleotide. T7 polymerase promoter sequences were added to sR8 and sR8 guide mutants during PCR amplification. RNA was transcribed in vitro using the AmpliScribe T7-Flash transcription kit (EPICENTRE) according to the manufacturer's protocols. Following transcription and DNase I treatment, all RNAs were gel purified.
Primer pairs for wild-type sR8 and sR8 guide mutants
Wild-type sR8 (primers 1 + 2), and sR8 guide mismatch primers sR8 C24G (primers 2 + 3), sR8 C24A (primers 2 + 4), sR8 C24U (primers 2 + 5), sR8 G51C (primers 1 + 6), sR8 G51A (primers 1 + 7), sR8 G51U (primers 1 + 8) are indicated below.
(1) CTAATACGACTCACTATAGGCCAAATCGCCAATGATGACGATTG
(2) AATCGCCTCAGTGCTCATACGG
(3) CTAATACGACTCACTATAGGCCAAATCGCCAATGATGACGATTGGGTTTG
(4) CTAATACGACTCACTATAGGCCAAATCGCCAATGATGACGATTGGATTTG
(5) CTAATACGACTCACTATAGGCCAAATCGCCAATGATGACGATTGGTTTTG
(6) AAATCGCCTCAGTGCTGATACGGTTC
(7) AAATCGCCTCAGTGCTTATACGGTTC
(8) AAATCGCCTCAGTGCTAATACGGTTC
sR8 targets
Wild-type, mutant, chimeric, and hairpin target RNAs were purchased from Integrated DNA Technologies. Wild-type and methylated control RNA targets are shown below. Additional RNA oligonucleotides used as mutant, chimeric, and hairpin target RNAs are described in the text and figures.
 |
Cloning, expression, and purification of proteins
Cloning and expression of recombinant, archaeal, box C/D sRNP core proteins L7, Nop56/58, and fibrillarin were accomplished as previously described (Tran et al. 2003). His-tagged L7 and fibrillarin were purified by nickel-affinity chromatography using “His-bind” Resin (Novagen) according to the manufacturer's protocols. Untagged Nop56/58 was purified by ion exchange chromatography.
RNP assembly and in vitro methylation
sRNP complexes were assembled by incubating 40 pmol of guide sRNA with 20 pmol of L7, 32 pmol of Nop56/58, and 32 pmol of fibrillarin in assembly buffer containing 20 mM HEPES (pH 7.0), 150 mM NaCl, 0.75 mM dithiothreitol, 1.5 mM MgCl2, 0.1 mM EDTA, and 10% glycerol at 70°C for 10 min. Following sRNP assembly, 360 pmol of target RNA(s) were added along with 360 pmol SAM (S-adenosyl-l-methionine dihydrogen sulfate; Calbiochem) and 1.7 μCi of [3H] SAM (63 Ci/mmol; Amersham) for a final reaction volume of 55 μL. These reactions were incubated at 68°C and 20 μL aliquots were blotted on 3MM Whatman filters at 0 and 60 min. RNA deposited on dry filters was precipitated by soaking in 10% TCA for 15 min at 4°C. Filters were then washed three times in 5% TCA for 15 min at room temperature before air drying. 3H-SAM incorporation into RNA was determined by scintillation counting. Assays were performed in triplicate, standard errors calculated, and results reported as percent activity of wild-type RNA. For electrophoretic analysis of methylated target RNAs, modified RNAs were phenol-chloroform extracted and precipitated with 2% lithium perchlorate in acetone. Isolated RNAs were resolved on denaturing 14% polyacrylamide gels and visualized by autoradiography using BioMax intensifying screens on X-ray film (Kodak).
ACKNOWLEDGMENTS
We thank Keith Gagnon and Xinxin Zhang for assistance with protein purification and providing sR8 templates and Clay Clark for advice on sRNP kinetic analysis. This work was funded by NSF Grant MCB 0543741 (E.S.M.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.517307.
REFERENCES
- Aittaleb, M., Rashid, R., Chen, Q., Palmer, J.R., Daniels, C.J., Li, H. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 2003;10:256–263. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- Bachellerie, J.P., Cavaille, J., Huttenhofer, A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Caffarelli, E., Losito, M., Giorgi, C., Fatica, A., Bozzoni, I. In vivo identification of nuclear factors interacting with conserved elements of box C/D small nucleolar RNAs. Mol. Cell. Biol. 1998;18:1023–1028. doi: 10.1128/mcb.18.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille, J., Bachellerie, J.P. SnoRNA-guided ribose methylation of rRNA: Structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res. 1998;26:1576–1587. doi: 10.1093/nar/26.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille, J., Nicoloso, M., Bachellerie, J.P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- Cavaille, J., Buiting, K., Kiefmann, M., Lalande, M., Brannan, C.I., Horsthemke, B., Bachellerie, J.P., Brosius, J., Huttenhofer, A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet d'Orval, B., Bortolin, M.-L., Gaspin, C., Bachellerie, J.-P. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 2001;29:4518–4529. doi: 10.1093/nar/29.22.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin, O., Banroques, J., Tanner, N.K., Linder, P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Decatur, W., Fournier, M.J. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- Fedoroff, O.Yu., Salazar, M., Reid, B.R. Structure of a DNA:RNA hybrid duplex. Why RNase H does not cleave pure RNA. J. Mol. Biol. 1993;233:509–523. doi: 10.1006/jmbi.1993.1528. [DOI] [PubMed] [Google Scholar]
- Gagnon, K.T., Zhang, X., Agris, P.F., Maxwell, E.S. Assembly of the archaeal box C/D sRNP can occur via alternative pathways and requires temperature-facilitated sRNA remodeling. J. Mol. Biol. 2006;362:1025–1042. doi: 10.1016/j.jmb.2006.07.091. [DOI] [PubMed] [Google Scholar]
- Galardi, S., Fatica, A., Bachi, A., Scaloni, A., Presutti, C., Bozzoni, I. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol. Cell. Biol. 2002;22:6663–6668. doi: 10.1128/MCB.22.19.6663-6668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspin, C., Cavaille, J., Erauso, G., Bachellerie, J.P. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: Lessons from the Pyrococcus genomes. J. Mol. Biol. 2000;297:895–906. doi: 10.1006/jmbi.2000.3593. [DOI] [PubMed] [Google Scholar]
- Goody, T., Melcher, S.E., Norman, D.G., Lilley, D.M.J. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 2004;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin, J.W., Batey, R.T. The bipartite architecture of the sRNA in an archaeal box C/D complex is a primary determinant of specificity. Nuc. Acids Res. 2006;34:5039–5051. doi: 10.1093/nar/gkl644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady, B., Kiss, T. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 2001;20:541–551. doi: 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., Bachellerie, J.P., Caizergues-Ferrer, M., Kiss, T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., Kiss, T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, D.J., Schmeing, T., Moore, P., Steitz, T. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, J., Tran, E., Maxwell, E.S. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5 kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002;30:931–941. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, V.K., Shiman, R., Draper, D.E. A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers. 2003;69:118–136. doi: 10.1002/bip.10353. [DOI] [PubMed] [Google Scholar]
- Nolivos, S., Carpousis, A.J., Clouet-d'Orval, B. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 2005;33:6507–6514. doi: 10.1093/nar/gki962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer, A.D., Lowe, T., Russell, A., Ebhardt, H., Eddy, S., Dennis, P. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- Omer, A.D., Ziesche, S., Ebhardt, H., Dennis, P. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. 2002;99:5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid, R., Aittaleb, M., Chen, Q., Spiegel, K., Demeler, B., Li, H. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J. Mol. Biol. 2003;33:295–306. doi: 10.1016/j.jmb.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Singh, S., Gurha, P., Tran, E.J., Maxwell, E.S., Gupta, R. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J. Biol. Chem. 2004;279:47661–47671. doi: 10.1074/jbc.M408868200. [DOI] [PubMed] [Google Scholar]
- Suryadi, J., Tran, E.J., Maxwell, E.S., Brown, B.A. The crystal structure of the Methanocaldococcus jannaschii multifunctional L7Ae RNA-binding protein reveals an induced-fit interaction with the box C/D RNAs. Biochemistry. 2005;44:9657–9672. doi: 10.1021/bi050568q. [DOI] [PubMed] [Google Scholar]
- Terns, M.P., Terns, R.M. Small nucleolar RNAs: Versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- Tran, E.J., Zhang, X., Maxwell, E.S. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ sRNP core proteins. EMBO J. 2003;22:3930–3940. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, E., Brown, J., Maxwell, E.S. Evolutionary origins of the RNA-guided nucleotide-modification complexes: From the primitive translation apparatus? Trends Biochem. Sci. 2004;29:343–350. doi: 10.1016/j.tibs.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Tran, E.J., Zhang, X., Lackey, L., Maxwell, E.S. Conserved spacing between the box C/D and C′/D′ RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA. 2005;11:285–293. doi: 10.1261/rna.7223405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, B., Melcher, S.E., Wilson, T.J., Norman, D.G., Lilley, D.M. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 2005;11:1192–1200. doi: 10.1261/rna.2680605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc, K., Steitz, J. U3, U8, and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski, K., You, Z., Graham, P., Steitz, J. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- Wang, H., Boisvert, D., Kim, K.K., Kim, R., Kim, S.H. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J. 2000;19:317–323. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, N., Leverett, R., Xia, L., Andrews, M., Maxwell, E.S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- Watkins, N., Segault, V., Charpentier, B., Nottrott, S., Fabrizio, P., Bachi, A., Wilm, M., Rosbash, M., Branlant, C., Luhrmann, R. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- Xia, L., Watkins, N., Maxwell, E.S. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA. 1997;3:17–26. [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Champion, E.A., Tran, E., Brown, B., Baserga, S., Maxwell, E.S. The coiled-coiled domain of the Nop56/58 core protein is dispensable for sRNP assembly but is critical for archaeal box C/D sRNP-guided nucleotide methylation. RNA. 2006;12:1092–1103. doi: 10.1261/rna.2230106. [DOI] [PMC free article] [PubMed] [Google Scholar]