Abstract
The GLD-2 family of poly(A) polymerases add successive AMP monomers to the 3′ end of specific RNAs, forming a poly(A) tail. Here, we identify a new group of GLD-2-related nucleotidyl transferases from Arabidopsis, Schizosaccharomyces pombe, Caenorhabditis elegans, and humans. Like GLD-2, these enzymes are template independent and add nucleotides to the 3′ end of an RNA substrate. However, these new enzymes, which we refer to as poly(U) polymerases, add poly(U) rather than poly(A) to their RNA substrates.
Keywords: poly(U) polymerase, RNA modification, β-nucleotidyl transferase, poly(A)
INTRODUCTION
DNA replication, transcription, and aspects of RNA processing depend on the addition of nucleotide monomers to nucleic acid substrates. Nucleotidyl transferases catalyze these reactions, adding nucleotides to the hydroxyl group of an acceptor substrate. The DNA polymerase β-like nucleotidyl transferases, a superfamily of such enzymes, use many different acceptors, including RNA, DNA, and antibiotics (Holm and Sander 1995; Aravind and Koonin 1999). These transferases play key roles in the processing and control of RNAs.
Despite their wide variation in substrate specificity, the catalytic domains of these enzymes are structurally related and include three carboxylate amino acids essential for catalytic activity (Aravind and Koonin 1999; Keller and Martin 2002). The enzymes appear to generally utilize the same mechanism of nucleotidyl transfer, involving a two-metal-coordinated attack by the 3′-OH of the primer on the α-phosphate of the incoming nucleoside triphosphate (Holm and Sander 1995; Martin and Keller 1996). Certain nucleotidyl transferases copy the sequence of a template, while others are template-independent. Several template-independent enzymes are critical in RNA metabolism, including the poly(A) polymerase (PAP), CCA-adding enzyme, and terminal uridylyltransferase (TUTase). Thus mRNAs, tRNAs, and noncoding RNAs are all substrates of enzymes in this group.
GLD-2 enzymes are template-independent members of the DNA polymerase β-like superfamily and are found throughout Eukarya. They are PAPs and add AMP monomers to the 3′ end of RNAs. The GLD-2 PAPs are distinct from canonical, eukaryotic PAPs in several aspects: they diverge in sequence, lack recognizable RNA-binding motifs, and localize to the cytoplasm (Wang et al. 2002). Most importantly, the GLD-2 enzymes only add poly(A) to selected cytoplasmic mRNAs, while the canonical nuclear PAPs act on most pre-mRNAs (Barnard et al. 2004; Suh et al. 2006; Rouhana and Wickens 2007). Relatives of GLD-2 in Schizosaccharomyces pombe, CID1 and CID13, also have been reported to be cytoplasmic poly(A) polymerases (Read et al. 2002; Saitoh et al. 2002). GLD-2-related enzymes have diverse biological roles, including the regulation of stem cells in Caenorhabditis elegans (Kadyk and Kimble 1998), the meiotic cell cycle in Xenopus (Barnard et al. 2004; Rouhana et al. 2005) and mouse oocytes (Nakanishi et al. 2006), and the S-M checkpoint in S. pombe (Wang et al. 1999, 2000).
Saccharomyces cerevisiae TRF4 and TRF5, two closely related proteins of the superfamily, add poly(A) to a specific subset of nuclear RNAs (Vanacova et al. 2005; Egecioglu et al. 2006; Kadaba et al. 2006) and histone mRNAs (Reis and Campbell 2006). Polyadenylation by TRF4/5 triggers destruction of the RNA by the exosome (Kadaba et al. 2004; LaCava et al. 2005; Egecioglu et al. 2006). Both TRF4/5 and GLD-2 enzymes acquire sequence specificity for particular RNAs by interacting with distinct RNA-binding proteins (Wang et al. 2002; Kadaba et al. 2004; LaCava et al. 2005; Vanacova et al. 2005; Suh et al. 2006).
Here, we identify a new group of GLD-2-related nucleotidyl transferases from Arabidopsis, S. pombe, C. elegans, and humans. These poly(U) polymerases (PUPs) are template-independent and similar in sequence to GLD-2. However, they add poly(U), rather than poly(A), to the 3′ end of the RNA.
RESULTS
Proteins that do not stimulate translation but add polymeric tails
In experiments designed to identify new poly(A) polymerases related to GLD-2, we tethered candidate proteins to the 3′ UTR of a reporter mRNA and then assayed that mRNA's translation. Candidate proteins were fused to MS2 coat protein and brought to a reporter mRNA using MS2 binding sites in its 3′ UTR. In these studies, Xenopus oocytes were first injected with mRNAs encoding the MS2 fusion proteins and incubated for 6 h to allow the proteins to accumulate. Luciferase mRNAs containing three MS2 binding sites were then injected. As an internal control, β-galactosidase mRNAs lacking MS2 binding sites were coinjected (Fig. 1A). In such assays, tethered poly(A) polymerase adds poly(A) to the reporter mRNA, increasing its translation (Gray et al. 2000; Dickson et al. 2001).
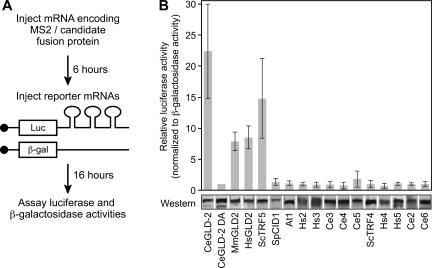
FIGURE 1.
Tethered GLD-2 family members. (A) Protocol. mRNA encoding an MS2 fusion protein with the candidate polypeptide was injected into oocytes. The fusion protein was expressed during a 6-h incubation, and then two reporter mRNAs were coinjected. The luciferase mRNA contained MS2 binding sites; the β-galactosidase lacked MS2 sites, and was used as a control. (B) Results. (Top) Relative stimulation of translation by each protein, expressed as the ratio of luciferase to β-galactosidase activities. Values were normalized to CeGLD-2 D608A, a catalytically inactive form of the enzyme (Kwak et al. 2004). Error bars were derived from more than three experiments. (Bottom) Chimeric fusion proteins detected by Western blotting using anti-HA tag antibody. Lysates equivalent to three oocytes were loaded into each lane.
We tested 16 different sequences derived from six species, including Arabidopsis, C. elegans, S. cerevisiae, S. pombe, mouse, and human. As expected, several stimulated luciferase activity, including the previously identified GLD-2 enzymes of C. elegans, mouse, and humans, and the TRF5 poly(A) polymerase of S. cerevisiae (Fig. 1B, bar graph; Kwak et al. 2004). An active site mutation in C. elegans GLD-2 (D608A), which disrupts its catalytic activity, prevented stimulation. All the proteins tested were expressed at comparable levels (Fig. 1B, Western).
The CID1 protein of S. pombe, previously identified as a cytoplasmic poly(A) polymerase (Read et al. 2002), did not enhance translation of the reporter mRNA (Fig. 1B, bar graph), even though the protein was expressed at normal levels (Fig. 1B, Western). Several other open reading frames (ORFs) behaved similarly, failing to stimulate translation despite being present at normal abundance (Fig. 1B).
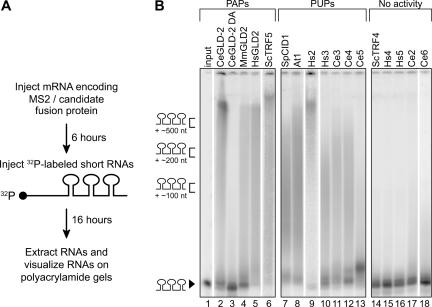
To investigate the biochemical activity of the proteins that lacked apparent polyadenylation activity, we assayed their ability to add residues to the 3′ end of the RNA. To do so, we injected short (130 nucleotides [nt]), radiolabeled RNA substrates harboring three MS2 sites into oocytes expressing each of the fusion proteins (Fig. 2A). The 32P-labeled RNAs were extracted after a 16-h incubation and visualized on polyacrylamide gels. As expected, CeGLD-2, MmGLD2, HsGLD2, and ScTRF5 lengthened the substrate RNAs (Fig. 2B, lanes 2, 4–6).1 Five other proteins (ScTRF4, Hs4, Hs5, Ce2, and Ce6) were inactive in both the luciferase assay and on the 32P-RNAs (Fig. 2B, lanes 14–18). Surprisingly, SpCID1 and six other proteins (At1, Hs2, Hs3, Ce3, Ce4, and Ce5) that were negative in the luciferase assay also substantially elongated the substrate RNAs (Fig. 2B, lanes 7–13). Tethered SpCID1 carrying an active site mutation was inactive (not shown). Thus the tethered enzyme itself (rather than any proteins with which it might associate) was responsible for synthesis of the tails.
FIGURE 2.
RNAs are elongated by proteins that do not stimulate translation. (A) Protocol. A radiolabeled RNA containing three MS2 binding sites was injected into oocytes that were expressing a fusion protein. (B) Results. The radiolabeled RNA substrates were analyzed by electrophoresis through urea–polyacrylamide gels. The products are grouped by what proved to be their enzymatic activities (PAPs [lanes 2–6], PUPs [lanes 7–13], or inactive [lanes 14–18]). RNAs from two oocytes were loaded on each lane. (Lane 1) An uninjected input RNA. Markers on left indicate approximate sizes of tails.
The tails added in oocytes are poly(U)
To test whether the tails added to the substrate RNAs were poly(A), we first treated the labeled RNA products with RNaseH and oligo(dT). This treatment specifically removes poly(A) tails. As expected, the tails synthesized by CeGLD-2 and ScTRF5 were truncated (Fig. 3A, cf. lanes 1,2 and 5,6). In contrast, the tails added by SpCID1 and the other six proteins that did not stimulate translation were refractory to RNaseH and oligo(dT), suggesting that these tails were not poly(A) (Fig. 3A, lanes 7–20). Similarly, RNAs elongated by CeGLD-2 bound to biotinylated oligo(dT), while those that were generated by SpCID1, Ce3, Ce4, and Ce5 did not (Fig. 3B). The added tails were not DNA, since they were unaffected by treatment with RQ DNase (data not shown).
FIGURE 3.
Poly(A) analysis of the products. (A) RNaseH/oligo(dT) reactions. RNAs produced by each tethered protein, prepared as in Figure 2B, were treated with oligo(dT) and RNaseH and analyzed by electrophoresis and autoradiography. (−) No DNA oligo added; (+) reactions included oligo(dT)18. (B) Binding to oligo(dT). RNAs were fractionated using biotinylated oligo(dT). (−) RNA that did not bind to biotinylated oligo(dT); (+) RNA that did bind.
To determine the sequences of the added polynucleotides, we cloned and sequenced products synthesized by eight different tethered proteins. Clones representing RNA products were obtained using the protocol outline in Figure 4A (Charlesworth et al. 2004; Rouhana and Wickens 2007). In brief, RNAs from oocytes expressing a specific protein were ligated to a DNA anchor primer, P1, and the ligated products were reverse transcribed using an antisense anchor primer, P1′. The cDNAs were amplified using a gene-specific forward primer, P2, and P1′. The PCR products then were cloned and sequenced. Tethered HsGLD2 was used as a control.
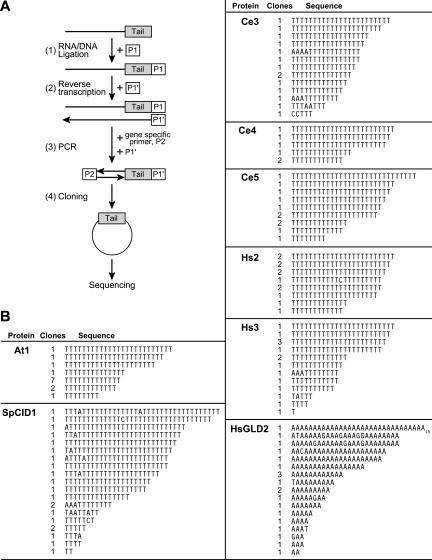
FIGURE 4.
Tail sequencing. (A) Protocol. Luciferase reporter mRNA was injected and RNAs prepared after a 16-h incubation. Step (1): RNAs were ligated to the 3′-amino modified DNA primer P1. Step (2): Reverse transcriptase was used to synthesize DNAs complementary to the substrate RNAs. Oligonucloeotide P1′, which is complementary to primer P1, was used as a primer for this step. Step (3): cDNAs were amplified by PCR reactions using a substrate specific primer, P2, and P1′. PCR products contain tails of the ligated products. Step (4): PCR reaction products were cloned into vectors using blunt-end ligation and transformed in E. coli. DNA was prepared from independent E. coli transformants, and the sequence of the insert in the plasmid determined. (B) Results. The tethered enzymes add poly(U). Sequences obtained from the RNAs products of reactions using eight different tethered enzymes, as indicated in the figures. The sequences in the figure correspond to that present in the RNA. The number of clones obtained is given in the second column: thus, with At1, seven clones contained 13 T residues.
Each of the tethered enzymes added poly(U) (Fig. 4B). The cloned products of each enzyme possessed oligothymidine tracts, corresponding to poly(U) added to the 3′ end of the injected RNA. For example, we obtained 14 clones corresponding to the products of At1: All possessed oligo(U) tails, ranging from 8 to 25 nt. Similarly, 11 clones were derived from Ce5 products, and all possessed only oligo(U). The five other enzymes tested—SpCID1, Hs2, Hs3, Ce3, Ce4—behaved similarly. With several, a few cloned tails had other nucleotides rarely inserted within oligo(U) tracts (Fig. 4B). The average lengths of the cloned tails were shorter than those on the starting RNAs, consistent with the known loss of homopolymeric tracts during cloning in bacteria (S. Kushner, pers. comm.) As controls, tails added by HsGLD2 were poly(A), not poly(U) (Fig. 4B), and catalytically inactive, mutant forms of CeGLD-2 yielded only clones with no tails (not shown). We conclude that the new family of enzymes add poly(U) tails, and so refer to them as poly(U) polymerases (PUPs).
Activity in vitro
To investigate the novel polymerase activity in vitro, we used purified recombinant SpCID1 protein; we focused on SpCID1 because the PUPs of other species were expressed poorly in bacteria. His6-tagged SpCID1 was produced in Escherichia coli and purified using a Ni2+ column. A single major band was detected in the final protein solution by Coomassie Blue staining (data not shown). To examine its nucleotide specificity, SpCID1 protein was incubated with radiolabeled RNA substrates plus one of the four ribonucleoside triphosphates. Recombinant S. cerevisiae PAP1 (ScPAP1), a canonical nuclear PAP, was used as a positive control.
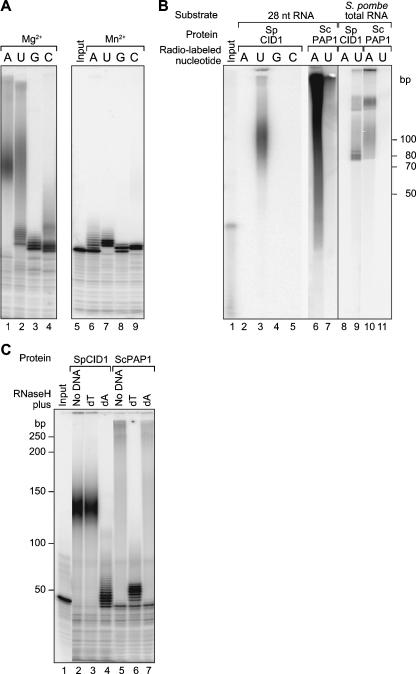
SpCID1 displayed more robust polymerase activity with Mg2+ than with Mn2+ (Fig. 5A, cf. lanes 1–4 and 6–9); ScPAP1 showed the opposite behavior (data not shown; Lingner et al. 1991). With Mg2+, SpCID1 was most active with ATP or UTP (Fig. 5A, lanes 1–4); ScPAP1 was highly specific for ATP (not shown). With Mn2+, SpCID1 was much less active, but UTP was preferred (Fig. 5A, lanes 6–9).
FIGURE 5.
SpCID1 activity in vitro. (A) Divalent metals. Purified recombinant SpCID1 was incubated with a 32P-labeled RNA substrate and one of four nucleoside triphosphates. Reactions contain either MgCl2 or MnCl2, as indicated above the lanes. (B) Preference for UTP and poly(U) synthesis in vitro. (Lane 1) End-labeled RNA corresponding to the substrate of the reactions in lanes 2–7. (Lanes 2–5) SpCID1 was incubated with an unlabeled RNA substrate and all four nucleoside triphosphates. Reactions contained either 32P-labeled ATP, UTP, GTP, or CTP, as indicated above each lane. Only UTP was significantly incorporated. (Lanes 6,7) Recombinant ScPAP1 was incubated with an unlabeled RNA substrate and the four nucleoside triphosphates plus either 32P-labeled ATP or UTP, as indicated. (Lanes 8–11) Similar reactions using total RNA from S. pombe as a substrate. (C) Oligo(dA)/RNaseH treatment confirms that the products of SpCID1 are poly(U). In vitro reactions were performed using radiolabeled RNA substrates and four NTPs. The products were treated with RNase H and either oligo(dA) or oligo(dT), as indicated. (Lane 1) substrate RNA (no A or U added); (lanes 2,5) no oligonucleotides added; (lanes 3,6) oligo(dT) added; (lanes 4,7) oligo (dA) added.
We next examined SpCID1 activity in the simultaneous presence of four nucleoside triphosphates. We incubated recombinant SpCID1 and unlabeled RNA substrates with all four unlabeled nucleoside triphosphates at 0.1 mM, in the presence of a 2.5 nM concentration of either 32P-ATP, 32P-UTP, 32P-CTP, or 32P-GTP. 32P-UTP was incorporated, while 32P-ATP was not (Fig. 5B, lanes 2–5). ScPAP1 behaved in the opposite fashion and added 32P-ATP much more efficiently than it did 32P-UTP (Fig. 5B, lanes 6,7).
We performed the same assay using S. pombe total RNA as substrates. SpCID1 incorporated 32P-UTP and ScPAP1 incorporated 32P-ATP. SpCID1 and ScPAP1 products differed in electrophoretic mobility (Fig. 5B, lanes 8–11), suggesting differences in their intrinsic RNA binding properties.
Polymers added in vitro are poly(U)
The data in Figure 5B strongly suggested that SpCID1 adds UMP monomers to RNA in vitro. To test this idea, we analyzed the products of SpCID1, obtained in the presence of all four nucleoside triphosphates, in two ways. First, we treated the labeled products with RNAseH plus either oligo(dT) or oligo(dA). The RNA products derived from SpCID1 were dramatically shortened in the presence of oligo(dA), suggesting that the tails were predominantly poly(U); consistent with this, treatment with oligo(dT) and RNaseH had no effect (Fig. 5C). The products of ScPAP1 behaved in the opposite manner, as they were trimmed in the presence of oligo(dT), but not oligo(dA). These data revealed that SpCID1 preferentially incorporated UTP into homopolymeric tracts in vitro.
Second, we cloned and sequenced products of in vitro reactions, derived from incubations in the presence of all 4 nt. SpCID1 products contained 1–6 uridine residues, while ScPAP1 products possessed poly(A) (not shown). The length of cloned tails was short for both SpCID1 and ScPAP1 products, presumably because homopolymeric regions are lost in E. coli. We conclude that SpCID1 has poly(U) polymerase activity in vitro.
DISCUSSION
In this report, we identify seven poly(U) polymerases from Arabidopsis, S. pombe, C. elegans, and Homo sapiens. These proteins add uridine monophosphate residues to the 3′ end of an RNA to form a 3′ terminal poly(U) tail. The poly(U) polymerase activity is observed both in oocytes and in vitro.
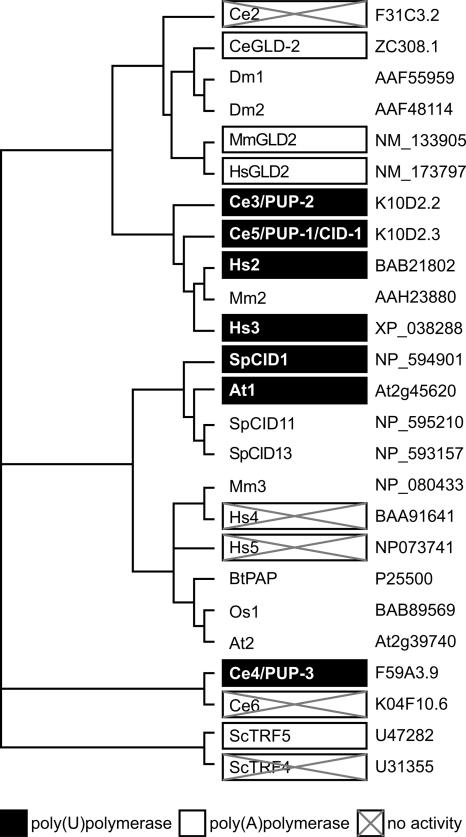
Our results expand the activities of the nucleotidyl transferase superfamily. The tethered assays identified a subfamily of PUPs that are clustered in sequence, with the exception of the more divergent Ce4 (Fig. 6). The PUPs include SpCID1, At1, Hs2, Hs3, Ce3, Ce4, and Ce5. The C. elegans PUPs have been formally named PUP-1 (K10D2.3, also called CID-1) (Olsen et al. 2006), PUP-2 (K10D2.2), and PUP-3 (F59A3.9) (see Fig. 6).
FIGURE 6.
Rooted phylogenetic tree of PUPs and PAPs and related proteins. (Black boxes) Proteins that add poly(U); (white boxes) proteins that add poly(A); (boxes with crosses) no activity detected in the tethered protein assay.
Certain GLD-2-related proteins (the PUPs) preferentially incorporate UTP, while others (the PAPs) incorporate ATP. Sequence comparisons do not reveal hallmarks that distinguish PUPs from PAPs, which might have identified residues that define nucleotide specificity. The PUPs do contain several residues that bind UTP in RNA editing TUTases (Deng et al. 2005), but these same residues are also found in PAPs. The biochemical basis of UTP specificity is understood in the case of one TUTase, RET2, from Trypansoma brucei (Deng et al. 2005). Its catalytic domain structure most closely resembles that of yeast nuclear PAP, although its overall shape is distinct. Purines are excluded sterically from their nucleotide binding sites, while a bound water molecule is positioned to coordinate with UTP and exclude CTP. Analysis of PUP structures should reveal whether they achieve specificity through a similar mechanism.
The tethered protein assays identified both PUPs and PAPs, but can yield false negatives. For example, ScTRF4 and Hs4, which were inactive in our tethered assays (Figs. 1, 2), are PAPs in vitro and in vivo (Kadaba et al. 2004, 2006; LaCava et al. 2005; Nagaike et al. 2005; Vanacova et al. 2005). Similarly, Hs5 was inactive as a tethered protein but is a nucleolar TUTase (Trippe et al. 2006). The tethered forms may be inactive because they require specific RNA structures for full activity: for example, purified ScTRF4 displays much higher activity on unmodified tRNAiMet than on native tRNAiMet (Vanacova et al. 2005), and Hs5 acts on the highly structured U6 snRNA (Trippe et al. 2006). Alternatively, these enzymes may require protein partners or simply misfold as chimeras.
Purified recombinant SpCID1 preferentially adds UTP to substrate RNA (Fig. 5B). While the purified protein can synthesize either poly(A) and poly(U) when provided with just one nucleoside triphosphate species (Read et al. 2002; this work), it preferentially synthesizes poly(U) in the presence of all four nucleoside triphosphates. Importantly, in oocytes, under physiological conditions, the PUPs synthesize poly(U), not poly(A).
The biological roles of certain PUPs may be conserved, since C. elegans and S. pombe PUPs have related functions in checkpoint control. C. elegans PUP-1/CID-1 (Ce5) is required for arrest in the S-phase in the presence of hydroxyurea (HU) (Olsen et al. 2006). C. elegans pup-1 also interacts genetically with chk-1, a known S/M checkpoint control gene. In S. pombe, cid1 mutants are sensitized to a combination of HU and caffeine that overrides the S/M checkpoint (Wang et al. 2000); similarly, SpCID1 overexpression partially suppresses the HU sensitivity of mutations in a protein kinase that is required for S/M and DNA damage checkpoint control (RAD3, RAD9, and RAD17) (for review, see Stevenson and Norbury 2006). The S. pombe cytoplasmic PAP, CID13, is also involved in checkpoint control (Read et al. 2002), hinting that the PAP and PUP pathways may interact. The related checkpoint phenotypes of PUPs likely reflect common RNA targets.
Identification of the natural RNA substrates of the PUPs is now critical. The targets of certain enzymes that add uridine residues to RNAs have been identified. For example, insertional editing in trypanosomes requires two specific TUTases, RET1 and RET2, to add uridine residues (Aphasizhev et al. 2002; Ernst et al. 2003), and oligouridylation of U6 snRNA is catalyzed by human U6-TUTase (Hs5) (Trippe et al. 2006). In contrast, the enzymes involved in oligouridylation of other RNAs in the microRNA and siRNA pathways are unknown. Arabidopsis miRNAs and siRNAs that are unmethylated receive one to five uridine residues through an unknown mechanism (Li et al. 2005). miRNA-directed cleavage products also get polyuridylated, an event that has been suggested to promote their decay (Shen and Goodman 2004). It will be of interest to determine whether the PUPs identified here participate in these processes, in mRNA control, or in other steps in RNA metabolism.
MATERIALS AND METHODS
DNA constructs
MS2 fusions
CeGLD-2, CeGLD-2 DA, HsGLD2, MmGLD2, Hs2–5, At1, and Ce2 constructs have been described by Kwak et al. (2004). ScTRF4 and ScTRF5 were PCR amplified from S. cerevisiae genomic DNA. ScTRF4 was inserted as a NheI–XhoI fragment, and ScTRF5 as a NheI–BamHI fragment, into p3HA-MSP. SpCID1 was PCR amplified from S. pombe cDNA and ligated into the NheI and XhoI sites of p3HA-MSP. Ce3, Ce4, partial Ce5 (361 aa–1421 aa) and Ce6 were PCR amplified from C. elegans cDNA. Ce3, partial Ce5, and Ce6 were inserted as NheI–XhoI fragments and Ce4 as an NheI–XbaI fragment, into the same sites of pCSMS2 (Rouhana et al. 2005).
mRNA reporter plasmids
pJK350 (β-galactosidase) and pLG-MS2 (luciferase), pLGMS2-LucHS (small RNA substrate) plasmids have been described previously by Dickson et al. (2001) and Kwak et al. (2004).
E. coli expression plasmids
To express SpCID1 in E. coli, the gene was PCR amplified from S. pombe cDNA and cloned into pET28a (Novagen), using the NheI and XhoI sites.
In vitro transcription
p3HA-MS2 based constructs were cleaved with an appropriate enzyme (HindIII for CeGLD-2 and CeGLD-2(D608A); ClaI for MmGLD2, HsGLD2, Hs2, Ce2, and SpCID1; BamHI for ScTRF5; SspI for ScTRF4, At1, and Hs5; PvuI for Hs3 and Hs4) and transcribed with Megascript T7 kit (Ambion). pCSMS2-based constructs were linearized with NotI and transcribed with Megascript SP6 kit (Ambion).
Reporter mRNAs were synthesized by transcription in vitro, as described by Kwak et al. (2004).
Western blotting
Western blotting was performed as described by Kwak et al. (2004). Proteins equivalent to three oocytes were loaded on each lane of SDS/PAGE gels. Anti-HA-tag antibody (α-HA11, Covance) was used to detect MS2 fusion proteins at 1:1000 dilution.
Tethered function assays and oocyte RNA analysis
Oocyte manipulations, injections, and enzyme assays were performed as described by Gray et al. (2000) and Dickson et al. (2001). RNA extraction, poly(A) selection, and electrophoresis were performed as described by Kwak et al. (2004).
Recombinant proteins
To purify recombinant SpCID1, pET28a-CID1 was transformed into BL21(DE3), and the protein expression was induced by addition of 0.5 mM IPTG for 6 h at 30°C. Cells were lysed by sonication in 20 mM Tris-HCl (pH 7.9), 5 mM immidazole, and 0.5 M NaCl. The lysate was loaded into a Ni2+ column (AP Biotech) and eluted with an immidazole gradient. Recombinant CID1 was eluted at ∼0.2 M immidazole range and the fraction was dialyzed in 20 mM Tris-HCl (pH 6.8), 100 mM KCl, 1 mM MgCl2, 1.5 mM DTT, and 50% glycerol at 4°C overnight.
S. cerevisiae nuclear PAP was purchased from USB Corporation.
In vitro assays and RNA analysis
RNA substrates
Chemically synthesized RNA corresponding to gld1-1 sequence (IDT DNA technologies) was end labeled with gamma 32P-ATP and T4 kinase. The sequence of this RNA was 5′-UUACCAUAGAAUCAUGUGCCAUACAUCA (Bernstein et al. 2005).
In vitro polymerization (polyuridylation)
Twenty-microliter reactions contained 32P-end-labeled gld1–1 RNA, 20 mM Tris-HCl (pH 7), 60 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 7.5 μg of BSA, 40 U RNasin, 1 mM rNTPs, and either 0.7 mM MnCl2 or 1.1 mM MgCl2. We added 170–340 ng of purified SpCID1 or 300 U of ScPAP1 (USB Corporation) to the reactions, which then were incubated at 30°C for 1 h. In some cases (Fig. 5), 1 μCi of α32P-NTP was added and unlabeled gld1-1 RNA or S. pombe total RNA was used as a substrate.
RNaseH analysis
In vitro polymerization reactions were stopped by boiling for 10 min. Fourteen microliters of the reactions were used for 40 μL of RNaseH reactions. RNaseH treatment was performed in 200 mM KCl, 1 mM EDTA, 20 mM Tris-HCl (pH 8.0), 30 mM MgCl2, 20 U RNasin, and 3 U of RNaseH. We added 0.5 μg of oligo(dT)18 or oligo(dA) 18 to the reactions. Reactions were incubated at 37°C for 75 min and stopped by addition of 2× gel loading dye.
RT-PCR-based tail sequencing
The tail sequencing was performed essentially as described previously, with some modifications (Charlesworth et al. 2004; Rouhana and Wickens 2007). For sequencing of oocyte products, 4 μg of total oocyte RNA was ligated to 0.4 μg of a 3′ amino modified P1 DNA anchor primer (5′-P-GGTCACCTTGATCTGAAGC-NH2). For in vitro product sequencing, 2 μL of boiled in vitro reaction were added to a ligation reaction containing 10 ng of P1 primer. The ligated products were reverse transcribed using P1′ primer (5′-GCTTCAGATCAAGGTGACC-3′). PCR reactions were performed using P1′ primer and either luciferase polylinker primer (5′-ACCTCTCTCTCTCTCTCAGGGCTGATTACTAG-3′) for oocyte products or gld1-1 primer (5′- TTACCATAGAATCATGTGCCATACATCA-3′) for in vitro products. PCR products were purified with PCR Clean Up kit (Qiagen) and ligated into pGEMT-easy vector (Promega). The ligation products were transformed to DH5α and E. coli colonies picked for sequence analysis. DNA plasmids were prepared using the FastPlasmid mini kit (Eppendorf) and sequenced using the SP6 promoter primer.
ACKNOWLEDGMENTS
We appreciate Labib Rouhana's advice and help with the cloning of tailed products and Judith Kimble's helpful comments on the work and manuscript. We also are grateful to Laura Vanderploeg of the UW Biochemistry Media Laboratory for help with the figures. This work was supported by an NIH Grant to M.W., and by a Mary Shine Peterson fellowship from the UW Biochemistry Department to J.E.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.514007.
For simplicity, we refer to proteins from different species using the abbreviated genus and species, then a number (e.g, Hs1); At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Sp, Schizzosaccharomyces pombe; Sc, Saccharomyces cerevisiae; Mm, Mus musculus; Hs, Homo sapiens. For proteins that already have a formal name, we use capital letters (e.g., TRF5), sometimes preceded by the species (e.g., ScTRF5), as needed for clarity. Gene names are in lowercase italics.
REFERENCES
- Aphasizhev, R., Sbicego, S., Peris, M., Jang, S.H., Aphasizheva, I., Simpson, A.M., Rivlin, A., Simpson, L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- Aravind, L., Koonin, E.V. DNA polymerase β-like nucleotidyltransferase superfamily: Identification of three new families, classification, and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard, D.C., Ryan, K., Manley, J.L., Richter, J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Bernstein, D., Hook, B., Hajarnavis, A., Opperman, L., Wickens, M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, A., Cox, L.L., MacNicol, A.M. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J. Biol. Chem. 2004;279:17650–17659. doi: 10.1074/jbc.M313837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, J., Ernst, N.L., Turley, S., Stuart, K.D., Hol, W.G. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei . EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, K.S., Thompson, S.R., Gray, N.K., Wickens, M. Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J. Biol. Chem. 2001;276:41810–41816. doi: 10.1074/jbc.M103030200. [DOI] [PubMed] [Google Scholar]
- Egecioglu, D.E., Henras, A.K., Chanfreau, G.F. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, N.L., Panicucci, B., Igo R.P., Jr, Panigrahi, A.K., Salavati, R., Stuart, K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Gray, N.K., Coller, J.M., Dickson, K.S., Wickens, M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L., Sander, C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- Kadaba, S., Krueger, A., Trice, T., Krecic, A.M., Hinnebusch, A.G., Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae . Genes & Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba, S., Wang, X., Anderson, J.T. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk, L.C., Kimble, J. Genetic regulation of entry into meiosis in Caenorhabditis elegans . Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Keller, W., Martin, G. Gene regulation: Reviving the message. Nature. 2002;419:267–268. doi: 10.1038/419267a. [DOI] [PubMed] [Google Scholar]
- Kwak, J.E., Wang, L., Ballantyne, S., Kimble, J., Wickens, M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava, J., Houseley, J., Saveanu, C., Petfalski, E., Thompson, E., Jacquier, A., Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Li, J., Yang, Z., Yu, B., Liu, J., Chen, X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis . Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J., Radtke, I., Wahle, E., Keller, W. Purification and characterization of poly(A) polymerase from Saccharomyces cerevisiae . J. Biol. Chem. 1991;266:8741–8746. [PubMed] [Google Scholar]
- Martin, G., Keller, W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Nagaike, T., Suzuki, T., Katoh, T., Ueda, T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- Nakanishi, T., Kubota, H., Ishibashi, N., Kumagai, S., Watanabe, H., Yamashita, M., Kashiwabara, S., Miyado, K., Baba, T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev. Biol. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Olsen, A., Vantipalli, M.C., Lithgow, G.J. Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans . Science. 2006;312:1381–1385. doi: 10.1126/science.1124981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read, R.L., Martinho, R.G., Wang, S.W., Carr, A.M., Norbury, C.J. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, C.C., Campbell, J.L. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae . Genetics. 2006 doi: 10.1534/genetics.106.065987. doi: 10.1534/genetics.106.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana, L., Wickens, M. Autoregulation of GLD-2 cytoplasmic poly(A) polymerase. RNA. 2007;13:188–199. doi: 10.1261/rna.333507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana, L., Wang, L., Buter, N., Kwak, J.E., Schiltz, C.A., Gonzalez, T., Kelley, A.E., Landry, C.F., Wickens, M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005;11:1117–1130. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, S., Chabes, A., McDonald, W.H., Thelander, L., Yates, J.R., Russell, P. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell. 2002;109:563–573. doi: 10.1016/s0092-8674(02)00753-5. [DOI] [PubMed] [Google Scholar]
- Shen, B., Goodman, H.M. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Stevenson, A.L., Norbury, C.J. The Cid1 family of noncanonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- Suh, N., Jedamzik, B., Eckmann, C.R., Wickens, M., Kimble, J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe, R., Guschina, E., Hossbach, M., Urlaub, H., Luhrmann, R., Benecke, B.J. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova, S., Wolf, J., Martin, G., Blank, D., Dettwiler, S., Friedlein, A., Langen, H., Keith, G., Keller, W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.W., Norbury, C., Harris, A.L., Toda, T. Caffeine can override the S-M checkpoint in fission yeast. J. Cell Sci. 1999;112:927–937. doi: 10.1242/jcs.112.6.927. [DOI] [PubMed] [Google Scholar]
- Wang, S.W., Toda, T., MacCallum, R., Harris, A.L., Norbury, C. Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase δ or ε is inactivated. Mol. Cell. Biol. 2000;20:3234–3244. doi: 10.1128/mcb.20.9.3234-3244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Eckmann, C.R., Kadyk, L.C., Wickens, M., Kimble, J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans . Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]