Abstract
In trypanosomatids, uridylate residues are post-transcriptionally added to or deleted from pre-mRNAs during the complex process of RNA editing. Editing is carried out exclusively in the mitochondrion of these parasites and involves numerous proteins assembled into protein and ribonucleoprotein complexes. Previously we identified RNA-editing-associated protein -1 (REAP-1), an RNA binding protein found in the mitochondrion of Trypanosoma brucei. REAP-1 was shown to specifically recognize and bind to pre-mRNAs that require editing and was proposed to act as a recruitment factor to deliver pre-mRNAs to editing complexes. To help define the role of REAP-1, we have now constructed REAP-1 null mutants. We show that the null mutants, although viable, have a significant growth defect. RNA levels within the mitochondrion were evaluated using reverse transcriptase real-time PCR. Surprisingly, the results show that mitochondrial RNA levels are increased, regardless of the editing status of the RNA. All RNA tested, whether unedited, edited, or never edited were increased in the mutant cell line relative to wild-type levels. This study provides the first evidence for a role of REAP-1 in RNA metabolism.
Keywords: RNA editing, kinetoplastid, Trypanosoma brucei, REAP-1, RNA turnover, RNA chaperone
INTRODUCTION
Trypanosoma brucei is a blood-borne pathogenic parasite transmitted by tsetse flies. The parasite causes African trypanosomiasis, or sleeping sickness, an infection that is lethal if left untreated and for which there is no prophylactic chemotherapy (Stich et al. 2002). Additionally, drugs currently used for treatment are hampered by both toxicity and resistant parasites. Yet many aspects of the biology of T. brucei and related pathogenic organisms are unique, suggesting that suitable targets for safe and effective treatment should be identifiable. One such unique aspect of trypanosome biology is RNA editing, carried out within the mitochondrion of the parasite (Benne et al. 1986). Twelve of the genes encoded in the mitochondrion require some degree of RNA editing in order to generate transcripts that can be translated into functional proteins (for review, see Estevez et al. 1999; Lukes et al. 2005; Stuart et al. 2005).
In T. brucei and related kinetoplastids, editing consists primarily of insertion (∼90%) and, to a lesser degree, deletion of uridine nucleotides. Editing is achieved by a series of enzymatic steps: endonucleolytic cleavage of the pre-mRNA, uridine addition by terminal uridyl transferase or removal by uridine-specific exonuclease, and ligation to rejoin the two halves of the pre-mRNA. The location and number of the uridines to be added or removed by the editing reaction is directed by small guide RNAs (gRNAs), which recognize their cognate pre-mRNA through base-pairing interactions (Blum et al. 1990). While RNA editing in general is not restricted to kinetoplastids, uridine insertion and deletion editing is exclusive to these organisms. Furthermore, it has been shown that a number of proteins required to carry out the editing process are encoded by essential genes. Functional knockdown experiments using RNA interference (RNAi) revealed severe growth inhibition (Aphasizhev et al. 2002; Carnes et al. 2005; Trotter et al. 2005) or lethal effects (Rusche et al. 2001; Brecht et al. 2005) to the life-cycle stage found in the insect vector. Loss of gene expression for most of these genes, as well as others, is also lethal in the stage of the parasite that multiplies in the bloodstream (Schnaufer et al. 2001; Wang et al. 2003). Taken together, these results suggest that genes involved in RNA editing are attractive candidates for possible therapeutic targets.
Previously we described an RNA binding protein, RNA-editing-associated protein -1 (REAP-1), which specifically recognizes RNA substrates prior to editing (Madison-Antenucci et al. 1998; Madison-Antenucci and Hajduk 2001). We showed that REAP-1 is able to recognize and bind to unedited RNA substrates, thus distinguishing them from RNAs that do not require editing (never edited), or those that have already been edited. Based on this activity we proposed that REAP-1 could act as a recruitment factor to facilitate association between unedited RNAs and the complex of proteins that carry out the editing reaction. The core complex required to carry out the enzymatic steps of the editing reaction is ∼20S in size, contains at least 20 proteins (for review, see Lukes et al. 2005; Stuart et al. 2005) and is capable of carrying out an editing reaction in vitro when supplied with exogenous gRNA and pre-mRNA (Seiwert and Stuart 1994; Kable et al. 1996; Sollner-Webb 1996). Purification of the ∼20S complex by a number of laboratories, as well as glycerol gradient sedimentation analysis, indicated that REAP-1 is not a part of this core complex (Madison-Antenucci et al. 1998; Panigrahi et al. 2003). However, the inability of the core complex to carry out multiple rounds of editing as well as the association of endogenous pre-mRNA with a larger complex (Pollard et al. 1992) suggests that, although the enzymatic activities are contained within the core complex, the editing complex likely functions in vivo as a 35–40S complex. Undoubtedly, proteins outside the ∼20S core associate with editing complexes, either stably or transiently. In fact, accessory proteins that are not part of the 20S complex have been identified. These include stimulating factor RBP16 (Miller et al. 2006), regulatory factor MRP1/2 (Vondruskova et al. 2005), TBRGG1 (Vanhamme et al. 1998), and the pre-mRNA binding protein REAP-1.
Since REAP-1 is not a component of the 20S complex yet has a proposed role as a recruitment factor, it was unclear whether REAP-1 would be required for editing and thus be essential for parasite viability. To investigate this issue, we first attempted to achieve functional knockdown of REAP-1 using RNAi. Failing this, null mutants were generated using homologous recombination to replace the REAP-1 gene with selectable drug resistance markers. After confirming integration at the correct locus we verified loss of expression by Northern and Western blot analyses. Experiments showed that REAP-1 null mutants have a significant growth defect. To examine the effect of the loss of REAP-1 on mitochondrial RNAs, we performed reverse transcriptase real-time PCR to evaluate the levels of unedited and edited RNAs in wild-type and null mutant cells. The results show that RNA metabolism is perturbed in the absence of REAP-1 and, surprisingly, that the steady-state level of mitochondrial RNAs is increased, regardless of whether the RNA is in the unedited or edited form or is an RNA that does not undergo editing. The results suggest that REAP-1 is involved in mitochondrial RNA stability.
RESULTS
REAP-1 is a single-copy gene
To evaluate whether or not REAP-1 is essential, we first attempted RNA interference to generate cell lines with reduced REAP-1 expression. Several transfections failed to generate viable cells whereas mock-transfected cells grew normally. After prolonged culturing, cells that survived transfection and selection were eventually obtained. These cells did not exhibit an altered growth phenotype upon treatment with tetracycline; however, further characterization showed that in the presence of tetracycline the levels of REAP-1 RNA were not reduced (data not shown), indicating that REAP-1 had not yielded to RNAi.
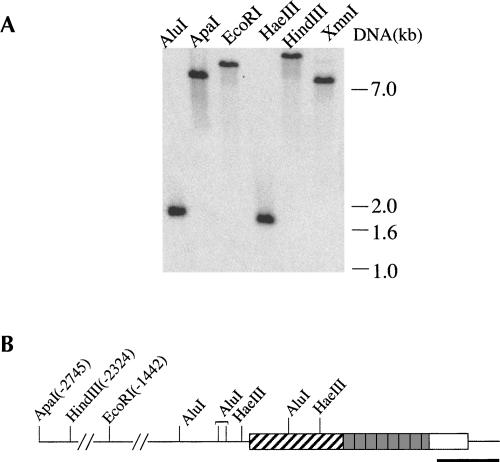
Difficulties encountered while trying to obtain stable transformants via RNAi could have been due to repression of REAP-1 target alone or could have been exacerbated by expression of T7 RNA polymerase and tetracycline repressor in the cell line used for RNAi experiments. Therefore the feasibility of constructing a REAP-1 null mutant starting from a wild-type cell line was investigated. Genomic DNA was isolated from wild-type cells to determine whether REAP-1 is a single-copy gene. The results of the Southern blot analysis are shown in Figure 1A. The DNA was digested with six different restriction enzymes and probed with a PCR-generated gene fragment that extends from the 3′ end of the coding region into the 3′ UTR. For five of the six restriction enzymes, at least one DNA restriction site is included in the genomic clone originally used to identify the REAP-1 gene. The positions of these sites are indicated in Figure 1B. For all six digests the REAP-1 probe hybridized to only one band, indicating that REAP-1 is, in fact, a single-copy gene. These data indicated that it is possible to replace REAP-1 by use of only two selectable markers.
FIGURE 1.
REAP-1 is a single copy gene. (A) Genomic DNA was isolated and digested with restriction enzymes as indicated at the top of each lane. The positions of DNA markers visualized by ethidium bromide staining of the gel prior to capillary transfer are indicated on the right. The probe used to detect REAP-1 containing fragments is indicated in the diagram below. (B) A diagram of the REAP-1 gene shows the RNA binding domain (diagonal stripes) and the 7 2/3 repeats of a well-conserved 21 amino acid sequence (shaded boxes). The heavy black line indicates the region of the gene used as a probe. Locations of restriction sites found within the genomic clone (Madison-Antenucci et al. 1998) are indicated. Positions for ApaI, HindIII, and EcoRI sites are given relative to the REAP-1 start codon.
REAP-1 gene replacement in double-knockout cell lines
In order to generate null mutants, the two alleles of REAP-1 were replaced with genes encoding resistance to neomycin and hygromycin. Antibiotic resistance genes were flanked upstream by an intergenic region from the procyclic acidic repetitive protein (PARP) locus to supply a splice acceptor site and downstream by an intergenic region from the tubulin array to provide a polyadenylation signal. These antibiotic cassettes were targeted to the REAP-1 locus by a 326 base-pair (bp) DNA fragment lying 82 bp upstream of the 45-kDa open reading frame and by a downstream DNA fragment that begins 234 bp upstream of the stop codon and ends 145 bp downstream. Single-knockout (SKO) mutants with either the neomycin or hygromycin cassette were easily obtained. The resulting cell lines were cloned and screened by PCR for correct integration (data not shown). Starting from SKO replacement cell lines using either neomycin or hygromycin, several attempts were made to generate the double-knockout (DKO) mutant, but they failed to generate stable transformants. Eventually, however, doubly resistant cells were obtained.
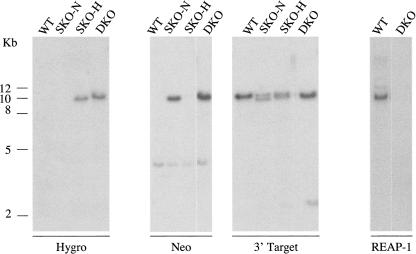
Due to the difficulty in obtaining null mutants, cells were analyzed by Southern blot analysis to insure that homologous recombination had occurred at the REAP-1 locus (Fig. 2). DNA was extracted from wild-type cells, SKO cells generated with neomycin (SKO-N) and with hygromycin (SKO-H), and from DKO cells. Genomic DNA was digested with HindIII and was analyzed using four separate probes. A probe specific for hygromycin hybridized to a band of ∼10 kb in SKO-H cells and DKO cells but, as expected, did not hybridize to DNA isolated from wild-type or SKO-N cells. The neomycin-specific probe cross-hybridized weakly to a 4 kb band in all cell lines, but it hybridized strongly to a band of 10 kb in only the SKO-N and DKO cells. All cell lines were also probed with the downstream DNA fragment used to target recombination to the REAP-1 locus. As expected, this probe hybridized to DNA from all cell lines. In wild-type cells the probe identified a single band while in the SKO-N cells it identified two bands. The second is slightly smaller than the band seen in wild-type cells. This band is of the size predicted to arise from the introduction of a new HindIII site at the junction between the PARP intergenic region and the REAP-1 targeting fragment. The resulting HindIII fragment is ∼1.1 kb smaller than the equivalent fragment in wild-type cells. Single-knockout cells generated via hygromycin resistance also showed two bands, although they were less well resolved. Introduction of the hygromycin cassette results in a new HindIII fragment ∼900 bp smaller than the wild-type fragment. In the DKO cells the two fragments, resulting from integration of one copy each of the hygromycin and neomycin cassettes, differ in size by only 230 bp and appear as a single band. To verify replacement of the REAP-1 gene, double-knockout cells were also examined with a probe that hybridizes to an internal region of REAP-1, which should not be present in DKO cells. As expected, when probed with this DNA fragment a band is detected in wild-type cells, but not in the DKO cells. Taken together the results confirm integration of the selectable markers at the expected locus, consistent with disruption of the REAP-1 gene.
FIGURE 2.
REAP-1 knockout mutants show loss of REAP-1 gene. Genomic DNA was isolated from wild-type cells (WT), REAP-1 single allele knockouts generated by replacement with a gene encoding resistance to neomycin (SKO-N) or to hygromycin (SKO-H), and REAP-1 double allele knockout (DKO) trypanosomes. Genomic DNA was digested with HindIII and probed as indicated at the bottom of each blot. Positions of DNA markers (kb) are indicated on the left. Probes were specific for the hygromycin gene (Hygro), neomycin gene (Neo), the DNA fragment used to direct homologous recombination to the 3′ end of the REAP-1 gene (3′ Target), or an internal region of the REAP-1 gene (REAP-1) lying between the two fragments used for homologous recombination.
REAP-1 RNA is undetectable in null mutants
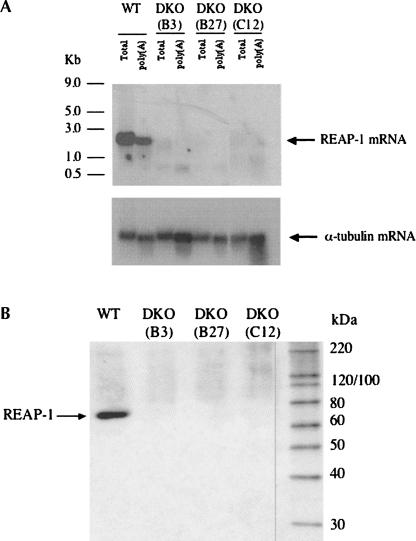
Using the DKO cell line verified by Southern blot analysis, several clonal populations were generated by limiting dilution. Three of these clonal cell lines were tested by Northern blot analysis to confirm loss of REAP-1 mRNA. As shown in Figure 3A, an RNA band of ∼2 kb is detected in both total RNA and polyadenylated RNA isolated from wild-type cells. None of the three clonal populations of cells showed hybridization to the REAP-1 specific probe either in total RNA or polyadenylated RNA. As a control to confirm equal loading and transfer, the blot was stripped and reprobed with a fragment of the gene encoding α-tubulin. The tubulin probe detected a band of approximately equal intensity in total and polyadenylated RNA isolated from all of the cell lines, showing that equivalent amounts of RNA were loaded and efficiently transferred. The results of the Northern blot analysis are consistent with those of the Southern blot and indicate loss of REAP-1 expression.
FIGURE 3.
REAP-1 is not detected in null mutants. (A)Total RNA and polyadenylated RNA (poly[A]) were isolated from wild-type and clonal populations of REAP-1 null mutant trypanosomes. Both total RNA and the poly(A) fraction from wild-type trypanosomes and three different null mutant clones were probed for the presence of REAP-1 RNA. The blot was stripped and then reprobed for α-tubulin RNA to verify consistent loading of RNA samples. (B) Western blot analysis shows whole-cell lysate from wild-type (WT) and REAP-1 clonal double-knockout cells probed with a polyclonal antibody to recombinant REAP-1. Total cell lysate is from 5 × 106 cells. The far right lane contains protein standards, and sizes are indicated (in kDa). An arrow indicates the position of REAP-1 detected in wild-type cells.
Monoclonal antibodies to REAP-1 recognize a second mitochondrial protein
The clonal DKO cell lines were also tested by Western blot analysis using two separate monoclonal antibodies that recognize the REAP-1 protein. Surprisingly, these antibodies did detect a protein in the null mutant, identical in size to that detected in wild-type cells (data not shown). Because both the Southern and Northern blot analyses had given consistent results showing that the REAP-1 gene had been lost, we speculated that the antibodies could be recognizing a second protein with a shared epitope. To address this issue the REAP-1 gene was cloned into an expression vector. Purified recombinant protein was used as an antigen to produce polyclonal antibodies (J. Hans, unpubl.). Both the original monoclonal antibodies and the polyclonal antibody raised against purified recombinant protein recognize recombinant REAP-1 protein. When wild-type cells and three separate DKO cell lines were probed with the polyclonal antibody, it was clear that the recognized protein is lost in the null cell lines (Fig. 3B). This is consistent with Southern and Northern blot results indicating loss of the REAP-1 gene. Additionally, results obtained with the two monoclonal antibodies indicate the presence of a second protein with a shared epitope (or epitopes). We are currently investigating the relationship between REAP-1 and this second protein. Nevertheless, Western blot analysis clearly showed that the polyclonal antibody against the purified recombinant protein specifically recognizes REAP-1 in a whole cell lysate, and that this protein is absent in the knockout cell lines.
REAP-1 null mutants have a significant growth defect
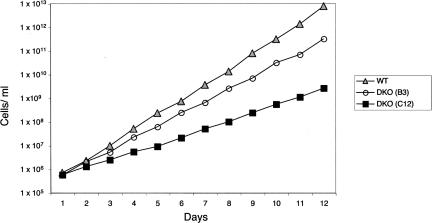
Although null mutants were obtained, the difficulty encountered in generating double-knockout cell lines suggested that a growth defect was likely to be associated with loss of REAP-1. Accordingly the growth rate of wild-type cells was compared with two of the clonal null mutant cell lines. As shown in Figure 4, wild-type cells grow at a significantly faster rate than either of the two clonal DKO cell lines. The wild-type cells doubled on average every 11.3 h, while the B3 cell line had a slightly longer doubling time, 13.5 h. The second null mutant cell line tested (C12) grew even more slowly, with a doubling time of 21.7 h, approximately twice as long as wild-type cells. It is unclear why growth of the DKO C12 cells is more severely retarded than that of the B3 cell line, although the difference was found to be reproducible. We also found that both null mutant cell lines showed enhanced sensitivity to cell density, and often did not survive if subcultured to a low cell density. In summary, although the null mutant cells are viable, the data indicate that loss of REAP-1 retards the growth rate of the parasite.
FIGURE 4.
Loss of REAP-1 inhibits growth of procyclic cells. Growth curves are shown for wild-type and the two REAP-1 null mutant clonal cell lines. Cultures were started at a concentration of 5 × 105 cells/mL and subcultured as necessary to maintain concentrations between 5 × 105 and 1 × 107. Wild-type cells were grown in SM media containing 10% FBS while DKO cell lines were maintained in SM containing 10% FBS, 15 μg/mL G418, and 25 μg/mL hygromycin. Numbers of wild-type cells (triangles), null mutant cell lines B3 (open circles), and C12 (filled squares) are plotted as the product of cell density and total cell dilution.
Mitochondrial RNA levels are increased
We have previously shown that REAP-1 protein specifically recognizes and binds to unedited pre-mRNA substrates preferentially over guide RNAs, RNAs that are already edited, or never-edited RNAs (Madison-Antenucci and Hajduk 2001). In order to evaluate what effect loss of REAP-1 might have, RNA was isolated from wild-type and two null mutant cell lines (B3, C12). Isolated RNA was further treated with DNase to remove contaminating genomic DNA, after which the purified RNA served as a template for reverse transcription reactions using random primers. The resulting cDNA was used as the template in real-time PCR reactions to determine relative RNA levels. Levels of unedited RNA, edited RNA, never-edited RNA, and REAP-1 mRNA were quantitated in wild-type cells and the two null mutant cell lines. RNA levels were calculated relative to 18S rRNA and to tubulin mRNA and ratios determined by the method of Pfaffl (2001) using PCR efficiencies calculated for each primer set (Ramakers et al. 2003).
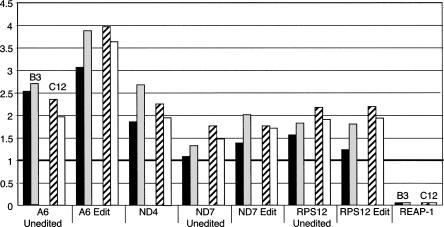
Results from real-time PCR confirmed loss of REAP-1 mRNA in the null mutants with an average ratio of <0.005 relative to wild-type cells (Fig. 5). For all RNAs tested, ratios calculated relative to 18S rRNA were consistent with those determined relative to tubulin mRNA. Similarly, results for the two mutant cell lines, DKO B3 and DKO C12, were typically in close agreement. Surprisingly, except for REAP-1, RNA levels in the DKO cell lines increased relative to wild type. Some increases were fairly modest, as in the case of unedited mRNA for NADH dehydrogenase subunit 7 (ND7), which showed a 1.5-fold increase over wild type. For other RNAs the increase was more dramatic. Edited RNA for ATPase subunit 6 (A6) was 3.5-fold higher in the null mutants. There was a greater increase in the level of edited A6 RNA than in the level of unedited RNA, which increased about 2.5-fold. Overall, there was a slight tendency for edited RNA levels to show a greater increase than unedited RNAs. However, this was not true in all cases, as the mRNA encoding ribosomal small subunit protein 12 increased about twofold for both unedited and edited transcripts. The results also show that never-edited RNAs were increased, as mRNA encoding NADH dehydrogenase subunit 4 (ND4), which is not edited, showed a twofold increase. The results indicate that RNA metabolism within the mitochondrion has been perturbed, and in a quite unexpected manner. Loss of REAP-1 leads to an increase in the steady-state levels of mitochondrial RNAs.
FIGURE 5.
Cells lacking REAP-1 show increased steady-state levels of mitochondrial RNA. Real-time PCR analysis of total RNA isolated from wild-type and REAP-1 double-knockout cell lines (B3 and C12). For each target RNA, the change in abundance in the B3 (left pair of bars) and the C12 (right pair of bars) null mutant cell lines is given relative to wild-type cells. Changes in abundance were normalized to 18S rRNA (black bars, B3, and striped bars, C12) and to tubulin (gray bars, B3, and white bars, C12). The target RNA is given below each set of bars. A thick black line at 1 indicates no change in RNA levels. RNA levels given are the average of at least four separate analyses.
RNA levels for a number of other mitochondrial transcripts were surveyed, including cytochrome oxidase II, cytochrome oxidase III, apocytochrome b (Cyb) NADH dehydrogenase subunit 3 (ND3), and mitochondrial unidentified reading frame 2 (MURF 2), and the results were consistent with those shown in Figure 5. In some cases the RNA levels remained the same or increased only slightly (CR4) while in other cases the increase was quite striking (MURF 2). Furthermore, none of the RNAs tested showed evidence of a decrease in RNA level. The results indicate that REAP-1 affects RNA stability and suggest the protein facilitates a decrease in steady-state RNA levels, most likely through increased turnover. Thus, REAP-1 may be involved in more that one aspect of RNA processing within the mitochondrion.
DISCUSSION
Previously we reported the identification of REAP-1, an RNA binding protein that specifically recognizes mitochondrial RNAs that require editing. In this study we examined the effects of loss of REAP-1 on cell viability and on mitochondrial RNAs. Possible deleterious effects due to loss of REAP-1 were suggested by the inability to generate stable RNAi knockdown cell lines. In addition, although single-knockout cell lines were easily obtained, numerous attempts were made before double-knockout cell lines were generated. We now show that loss of REAP-1 does, in fact, lead to a growth defect in procyclic cells. Two cloned null mutant cell lines display longer doubling times when compared to wild-type cells. Although REAP-1 does not appear to be essential, taken together the results indicate a significant growth defect when the protein is absent.
To further explore the effects of REAP-1 loss, we compared steady-state RNA levels of mitochondrial RNAs. RNA levels of pairs of unedited and edited RNAs were analyzed along with never-edited RNA (ND4). Surprisingly, all RNAs tested showed an increase in the relative abundance of RNA in null mutants compared to wild-type cells. There was no indication of a preferential increase in unedited RNAs nor of a decrease in edited RNAs. In fact an overall increase was observed among unedited, edited, and never-edited RNAs. This increase in steady-state RNA levels suggests that REAP-1 does not act strictly in RNA editing as previously thought. However, the results do not necessarily rule out a role in RNA editing either. It has previously been shown that RNAi-induced repression of core editing complex protein KREPB2 did not give rise to a growth effect and generated no change in the relative abundance of mitochondrial RNA (Trotter et al. 2005). Similarly, loss of KREL2, the 48 kDa RNA ligase, via RNAi-induced repression, produced no change in growth of procyclic or bloodstream form cultures and RNA levels remain unchanged (Trotter et al. 2005). However, the increase in RNA levels observed here, in the absence of REAP-1, does indicate a role in mitochondrial RNA metabolism and turnover.
There is evidence from other editing associated proteins, proteins that are not constituents of the 20S core editing complex, for dual roles in both editing and RNA metabolism. RBP16 is a mitochondrial RNA binding protein that has been shown to affect both RNA editing and stability. In the first report of an accessory editing factor with the capacity to stimulate in vitro editing (Miller et al. 2006), it was demonstrated that RBP16 significantly stimulates insertion editing of Cyb and to a somewhat lesser extent editing of A6 RNA. In addition to its role as a stimulatory editing factor, RBP16 influences the stability of both edited and never-edited RNAs. RNAi-induced knockdown of RBP16 expression leads to a decrease in the amount of Cyb-edited RNA and an increase in the level of unedited RNA (Pelletier and Read 2003). Inhibition of expression via RNAi also results in a decrease in levels of never-edited mRNAs COI and ND4, in a manner that parallels the decrease in RBP16 (Pelletier and Read 2003). Overexpression of RBP16 leads to a twofold increase in the levels of both unedited and edited CoIII and Cyb (Goulah et al. 2006), while other RNAs are unaffected either by knockdown or overexpression. Thus RBP16 stimulates insertion editing of at least two RNAs and influences the stability of several RNAs in a transcript-specific manner.
Mitochondrial RNA binding proteins MRP1 and MRP2 also affect both RNA editing and metabolism (Vondruskova et al. 2005). The level of edited Cyb RNA decreased during RNAi-induced repression of MRP1 and MRP2 together and singly, whereas the level of unedited RNA increased. The level of edited RPS12 RNA also decreased slightly in the same study. In contrast, the level of ND7-edited RNA increased only slightly when MRP2 was repressed but increased significantly (approximately fivefold) when expression of both MRP1 and MRP2 was repressed. Furthermore, the level of three never-edited RNAs was decreased after RNAi knockdown of the MRP proteins. For both RBP16 and MRP1/MRP2 the effect on RNA editing and stability is transcript specific, with different RNAs sometimes yielding opposite results.
Here we show that loss of REAP-1 leads to a global increase in unedited, edited, and never-edited RNAs alike. Increased steady-state levels of RNA in the absence of REAP-1 implicate the protein either in transcription or mRNA turnover. A role in transcription cannot formally be ruled out, as loss of a negative transcription factor could lead to an increase in RNA levels. However, mitochondrial polymerases are not typically multisubunit RNA polymerases, such as those found in the nucleus. Rather, mitochondrial polymerases thus far identified are single-subunit polymerases, and such a mitochondrial polymerase for T. brucei has been identified (Wang et al. 2000; Clement and Koslowsky 2001; Grams et al. 2002). It seems unlikely to us, based on what is known about mitochondrial transcription, that REAP-1 would be acting as a transcription factor. We therefore favor a model in which REAP-1 facilitates, in some manner, RNA processing and turnover.
Previous studies on RNA turnover in trypanosome mitochondria point out the role of the poly(A) tail, which may either stabilize or destabilize the RNA, depending upon that RNA's editing status (Kao and Read 2005). Although REAP-1 is an RNA binding protein that binds to the polypurine tracts of pre-mRNAs, the binding appears to be sensitive to the guanosine content of the RNA so that REAP-1 is unlikely to interact directly with the poly(A) tail. In binding experiments, which demonstrated the protein's high binding affinity for unedited RNA, there was no detectable binding to poly(A) (Madison-Antenucci and Hajduk 2001).
The simplest explanation for increased RNA levels in the null mutant would be if REAP-1 were a ribonuclease. However, we see no evidence of ribonuclease activity with recombinant REAP-1 either in RNA binding experiments or gel mobility shift assays. Further, the fact that the protein contains no recognizable motifs or homology with known proteins makes it difficult to speculate on other possible activities.
One possibility is that REAP-1 acts as an RNA chaperone. Chaperones are not known to share a particular conserved motif, although they do contain some type of RNA binding domain. The fact that regions of disordered secondary structure are also common has led to speculation that disorder permits RNA chaperones to bind with multiple RNA partners or with RNAs existing in multiple conformations. Predictions of disorder when applied to REAP-1 do indicate an extensive disordered region, although this is also a characteristic of many RNA binding domains including KHI, KH2, and RGG domains, which are not limited to RNA chaperones.
RNA chaperones act in numerous processes in the life of an RNA. Such a role for REAP-1 could potentially account for two seemingly opposing observations. REAP-1 shows a preference for binding to unedited RNAs, yet the level of all mitochondrial RNAs are increased in the protein's absence. To reconcile these observations, we propose that REAP-1 acts as a chaperone to bind with high affinity and deliver unedited RNAs to editing complexes and also bind with lower affinity to edited and never-edited RNAs, facilitating turnover. In order to test this model for the role of REAP-1 in mitochondrial RNA metabolism it will be necessary to identify both the in vivo RNA binding partners and the proteins with which REAP-1 interacts. Both of these areas are currently being pursued.
MATERIALS AND METHODS
Trypanosome cell growth and transfections
T. brucei (TREU 667) procyclic form cells were grown in SM medium (Cunningham 1977) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS). Mutant cell lines bearing the neomycin gene replacement cassette were maintained in medium also containing 15 μg/mL G418 while those bearing the hygromycin gene replacement cassette were grown in medium containing 25 μg/mL hygromycin. Except where indicated otherwise, null mutant cell lines were maintained in medium containing both antibiotics and 15% (v/v) FBS to improve viability.
T. brucei cells used for transfection (Beverley and Clayton 1993; Kelly et al. 1995) were collected by centrifugation, 10 min at 2000g, and washed twice with cold phosphate-buffered saline. Cells were resuspended in Opti-MEM (Gibco/Invitrogen) at a density of 4 × 107 cells/mL based on hemocytometer counts. Cells (0.5 mL) were placed in a chilled 0.4 cm gap electroporation cuvette with 1 μg of linear plasmid DNA in 100 μL Opti-MEM. Transfection was achieved by two pulses of 1.4 kV, 25 μF applied 10 sec apart. Cells were then transferred to culture flasks containing 10 mL of medium. Cells were grown for 24–48 h without selection and then transferred to media containing the appropriate antibiotic. Transfected cells were initially selected using 50 μg/mL G418 and/or 25 μg/mL hygromycin. Cells were collected by centrifugation and resuspended in fresh media as needed. Clonal populations were selected by limiting dilution of transfectants supplemented with wild-type cells to decrease cell death at low parasite concentration.
For growth experiments cells were grown in the appropriate media and maintained at a concentration between 5 × 105 and 1 × 107. Cell counts were performed using a Beckman Z1 Coulter Counter and determined in triplicate, with a cutoff value of 3.0–3.5 μM.
Plasmid constructs
Constructs used for targeted gene disruption were generated from plasmids carrying a gene conferring resistance to either neomycin or hygromycin, pKONeo, and pKOHygro, respectively. Targeting to the REAP-1 locus was achieved by insertion of a 326-bp DNA fragment upstream of the 45-kDa REAP ORF amplified by PCR using 5′-CCGGTTTTTTTG/TCGACCGCGCC-3′ plus 5′-CCACGACGAGAGGA/AGCTTGCACG-3′ and a 382 bp fragment spanning the stop codon, amplified with 5′-TCTTCGGT/CTAGAGAGGCGCATTAAGTC-3′ plus 5′-CCTTAGACTCACTGCGAGCT/CTCACCAA-3′. The upstream targeting fragment was digested with SalI and HindIII (at the italicized bases) and ligated into pKONeo and pKOHygro linearized with the same restriction enzymes. Restriction sites for the downstream targeting fragment were XbaI and SacI.
Genomic DNA isolation and Southern blot analysis
High-molecular-weight DNA was isolated using proteinase K and phenol extractions as described (Sambrook and Russsell 2001) with minor modifications. Cells were resuspended in 10 mM Tris-HCl, 1 mM EDTA (pH 8.0) at a concentration of 2 × 108/mL. Phenol extraction was followed by phenol:CHCl3 (1:1, v/v) extraction and DNA was precipitated from ethanol with 2 M ammonium acetate.
Genomic DNA (10 μg) was digested with the appropriate restriction enzyme, phenol extracted, and ethanol precipitated prior to electrophoresis on a 0.6% agarose gel. Following electrophoresis the DNA was transferred to GeneScreen Plus using the alkaline transfer protocol and the aqueous hybridization method recommended by the manufacturer.
RNA isolation and Northern blot analysis
RNA was isolated based on the method of Chomczyski and Sacchi (1987). Cells were resuspended in Tripure reagent (Roche) at a concentration of 5 × 107/mL. Following isopropanol precipitation RNA was resuspended in 10 mM Tris, 1 mM EDTA (pH 8.0) and extracted first with phenol and then with phenol:CHCl3 (1:1, v/v). RNA was precipitated from ethanol with 0.3 M sodium acetate. Polyadenylated RNA, poly(A)+, was purified from 50–100 μg of total RNA using MicroPoly(A) Pure (Ambion) as recommended.
Total RNA (5 μg) or poly(A)+ RNA (2 μg) was mixed with an equal volume of methyl mercury loading dye (Maniatis et al. 1982) and separated by electrophoresis on a 1% agarose gel. Following electrophoresis the gel was rinsed in H2O and soaked in 3 M NaCl, 0.01 N NaOH for 20 min. Transfer to GeneScreen Plus was by capillary blot using 10× SSC. RNA was fixed to the membrane by UV cross-linking. Prehybridization was performed as recommended by the supplier of the membrane and included 100 μg/mL denatured sheared salmon sperm DNA.
Probes for Southern and Northern blots
Probes were amplified by PCR using 5′-CGGACCGCAAGGAATCGGTC-3′ plus 5′-CTTCTGCGGGCGATTTGTGTAC-3′ to generate a 531-bp probe for hygromycin, 5′-GATTGAACAAGATGGATTG-3′ plus 5′-GAAGAACTCGTCAAGAAGG-3′ to generate a 790-bp probe for neomycin, and 5′-GCAGCGGTAGGACTGGTGGGCT-3′ plus 5′-CTTGTTTGTCCTGCACGTTACC-3′ to generate a 478-bp probe for REAP-1. Probes were labeled with [α-32P]dCTP using random hexamer primers and Klenow DNA polymerase (Feinberg and Vogelstein 1983).
Western blot analysis
T. brucei cell counts were performed on a Beckman Z1 Coulter cell counter. Cells were collected by centrifugation and resuspended in phosphate-buffered saline at a concentration of 3 × 105. Cells were lysed in SDS gel-loading buffer at 100°C for 3 min and loaded directly on an SDS-PAGE gel. Following electrophoresis proteins were transferred to nitrocellulose, blocked for 1 h in 20 mM Tris, 500 mM NaCl, and 0.05% Tween-20 (v/v) (pH 7.5) containing 5% (w/v) powdered milk. For detection with monoclonal antibodies, incubation with the primary antibody was at a 1:1000 dilution at 4°C overnight; the secondary antibody was peroxidase-conjugated goat anti-mouse (1 mg/mL) diluted 1:50,000. Polyclonal antibodies were diluted 1:2000 and used with secondary goat anti-rabbit horseradish peroxidase conjugated antibody at a dilution of 1:20,000. Both were detected with SuperSignal West Pico (Pierce) chemiluminescent substrate.
Reverse transcriptase and real-time PCR
Total RNA, isolated as described above, was treated with DNA-free (Ambion) as recommended by the supplier to remove contaminating DNA. Treated RNA was converted to cDNA using reverse transcriptase as follows. The final reaction contained 0.5 μg RNA, 2.5 μM random hexamer oligonucleotides, 2.5 mM of each dNTP, 10 U RNase inhibitor, 30 U MultiScribe reverse transcriptase in 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 5.5 mM MgCl2 with a total volume of 25 μL. RNA was denatured at 65°C for 5 min and then chilled on ice prior to addition to the reaction, which was further incubated at 25°C for 10 min and then at 37°C for 60 min. The reaction was stopped by treatment at 95°C for 5 min. Control reactions lacked reverse transcriptase.
Real-time PCR reactions were performed using a 1:16 dilution of cDNA in H2O for all targets with the exception of the more abundant 18s rRNA and tubulin, for which cDNA was diluted 1:50. RT-PCR reactions were performed with 2.5 μL of diluted cDNA, and 300 nM forward and reverse primers in a 25 μL reaction with Power SYBR Green (Applied Biosystems). Samples were incubated at 50°C for 2 min, 95°C for 10 min, and followed by 40 cycles of 95°C 15 sec and 60°C 1 min. Fluorescence was detected during the 1 min, 60°C cycle. A dissociation step confirmed the presence of a unique amplicon. Control reactions were performed using cDNA reactions lacking reverse transcriptase. Primers were as previously described (Carnes et al. 2005) with the addition of REAP-1 specific primers, 5′-GCCATTGTTCAGTCGGTTC-3′ plus 5′-GGAGAGGGCTTCCGATGATC-3′. RNA ratios were determined by the method of Pfaffl (2001) using PCR efficiencies calculated for each primer set (Ramakers et al. 2003).
ACKNOWLEDGMENTS
This work was supported by Wadsworth Center New Investigator Funds (to S.M.-A.) and by National Institutes of Health Grant AI21401 (to S.L.H.). The authors thank J. Bangs (University of Wisconsin) for pKONeo and pKOHygro knockout vectors, M. Marchewka (Wadsworth Center) for technical assistance, and J. Keithly (Wadsworth Center) for helpful discussions and comments.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.486107.
REFERENCES
- Aphasizhev, R., Sbicego, S., Peris, M., Jang, S.H., Aphasizheva, I., Simpson, A.M., Rivlin, A., Simpson, L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- Benne, R., Van den Burg, J., Brakenhoff, J.P., Sloof, P., Van Boom, J.H., Tromp, M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Beverley, S.M., Clayton, C.E. Transfection of Leishmania and Trypanosoma brucei by electroporation. Methods Mol. Biol. 1993;21:333–348. doi: 10.1385/0-89603-239-6:333. [DOI] [PubMed] [Google Scholar]
- Blum, B., Bakalara, N., Simpson, L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Brecht, M., Niemann, M., Schluter, E., Muller, U.F., Stuart, K., Goringer, H.U. TbMP42, a protein component of the RNA editing complex in African trypanosomes, has endo-exoribonuclease activity. Mol. Cell. 2005;17:621–630. doi: 10.1016/j.molcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Carnes, J., Trotter, J.R., Ernst, N.L., Steinberg, A., Stuart, K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei . Proc. Natl. Acad. Sci. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Clement, S.L., Koslowsky, D.J. Unusual organization of a developmentally regulated mitochondrial RNA polymerase (TBMTRNAP) gene in Trypanosoma brucei . Gene. 2001;272:209–218. doi: 10.1016/s0378-1119(01)00538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, I. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J. Protozool. 1977;24:325–329. doi: 10.1111/j.1550-7408.1977.tb00987.x. [DOI] [PubMed] [Google Scholar]
- Estevez, A.M., Kierszenbaum, F., Wirtz, E., Bringaud, F., Grunstein, J., Simpson, L. Knockout of the glutamate dehydrogenase gene in bloodstream Trypanosoma brucei in culture has no effect on editing of mitochondrial mRNAs. Mol. Biochem. Parasitol. 1999;100:5–17. doi: 10.1016/s0166-6851(99)00024-9. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P., Vogelstein, B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Goulah, C.C., Pelletier, M., Read, L.K. Arginine methylation regulates mitochondrial gene expression in Trypanosoma brucei through multiple effector proteins. RNA. 2006;12:1545–1555. doi: 10.1261/rna.90106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grams, J., Morris, J.C., Drew, M.E., Wang, Z., Englund, P.T., Hajduk, S.L. A trypanosome mitochondrial RNA polymerase is required for transcription and replication. J. Biol. Chem. 2002;277:16952–16959. doi: 10.1074/jbc.M200662200. [DOI] [PubMed] [Google Scholar]
- Kable, M.L., Seiwert, S.D., Heidmann, S., Stuart, K. RNA editing: A mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- Kao, C.Y., Read, L.K. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei . Mol. Cell. Biol. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, J.M., Taylor, M.C., Rudenko, G., Blundell, P.A. Transfection of the African and American trypanosomes. Methods Mol. Biol. 1995;47:349–359. doi: 10.1385/0-89603-310-4:349. [DOI] [PubMed] [Google Scholar]
- Lukes, J., Hashimi, H., Zikova, A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci, S., Hajduk, S.L. RNA editing-associated protein 1 is an RNA binding protein with specificity for preedited mRNA. Mol. Cell. 2001;7:879–886. doi: 10.1016/s1097-2765(01)00231-3. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci, S., Sabatini, R.S., Pollard, V.W., Hajduk, S.L. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): A novel editing complex protein with repetitive domains. EMBO J. 1998;17:6368–6376. doi: 10.1093/emboj/17.21.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison-Antenucci, S., Grams, J., Hajduk, S.L. Editing machines: The complexities of trypanosome RNA editing. Cell. 2002;108:435–438. doi: 10.1016/s0092-8674(02)00653-0. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., Sambrook, J., Russell, D. Molecular cloning, a laboratoy manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1982. [Google Scholar]
- Miller, M.M., Halbig, K., Cruz-Reyes, J., Read, L.K. RBP16 stimulates trypanosome RNA editing in vitro at an early step in the editing reaction. RNA. 2006;12:1292–1303. doi: 10.1261/rna.2331506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A.K., Schnaufer, A., Ernst, N.L., Wang, B., Carmean, N., Salavati, R., Stuart, K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, M., Read, L.K. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei . RNA. 2003;9:457–468. doi: 10.1261/rna.2160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, V.W., Harris, M.E., Hajduk, S.L. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 1992;11:4429–4438. doi: 10.1002/j.1460-2075.1992.tb05543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers, C., Ruijter, J.M., Deprez, R.H., Moorman, A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Rusche, L.N., Huang, C.E., Piller, K.J., Hemann, M., Wirtz, E., Sollner-Webb, B. The two RNA ligases of the Trypanosoma brucei RNA editing complex: Cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Russsell, D. Molecular cloning, a laboratoy manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Schnaufer, A., Panigrahi, A.K., Panicucci, B., Igo R.P., Jr, Wirtz, E., Salavati, R., Stuart, K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei . Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Seiwert, S.D., Stuart, K. RNA editing: Transfer of genetic information from gRNA to precursor mRNA in vitro. Science. 1994;266:114–117. doi: 10.1126/science.7524149. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb, B. Trypanosome RNA editing: Resolved. Science. 1996;273:1182–1183. doi: 10.1126/science.273.5279.1182. [DOI] [PubMed] [Google Scholar]
- Stich, A., Abel, P.M., Krishna, S. Human African trypanosomiasis. BMJ. 2002;325:203–206. doi: 10.1136/bmj.325.7357.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, K.D., Schnaufer, A., Ernst, N.L., Panigrahi, A.K. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Trotter, J.R., Ernst, N.L., Carnes, J., Panicucci, B., Stuart, K. A deletion site editing endonuclease in Trypanosoma brucei . Mol. Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Vanhamme, L., Perez-Morga, D., Marchal, C., Speijer, D., Lambert, L., Geuskens, M., Alexandre, S., Ismaili, N., Goringer, U., Benne, R., et al. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem. 1998;273:21825–21833. doi: 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- Vondruskova, E., van den Burg, J., Zikova, A., Ernst, N.L., Stuart, K., Benne, R., Lukes, J. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei . J. Biol. Chem. 2005;280:2429–2438. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Morris, J.C., Drew, M.E., Englund, P.T. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wang, B., Ernst, N.L., Palazzo, S.S., Panigrahi, A.K., Salavati, R., Stuart, K. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei . Eukaryot. Cell. 2003;2:578–587. doi: 10.1128/EC.2.3.578-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]