Abstract
Translation initiation driven by internal ribosome entry site (IRES) elements is dependent on the structural organization of the IRES region. Picornavirus IRES are organized in structural domains, in which the terminal stem–loops participate in functional RNA–protein interactions. However, the mechanistic role performed by the central domain during internal initiation has not been elucidated yet. Here we show that the foot-and-mouth-disease virus IRES contains a structural motif that serves in vitro as substrate for the Synechocystis sp. RNase P ribozyme, a structure-dependent endonuclease that participates in tRNA precursor processing. Recognition of the IRES substrate was dose dependent, required high magnesium concentration, and resulted in the formation of cleavage products with 5′ phosphate and 3′ hydroxyl ends. Mapping of the core recognition motif indicated that it overlapped with the apical region of the central domain. Two IRES constructs containing nucleotide substitutions in the apical region of the central domain that reorganized RNA structure displayed an altered pattern of cleavage by the cyanobacterial ribozyme generating new cleavage events in nearby residues. From these data it is inferred that the central domain of the IRES region has evolved a tRNA structural mimicry that renders it a substrate for RNase P ribozyme reaction. Recognition of this motif was affected in defective IRES mutants with a local RNA structure reorganization, suggesting that its structural preservation is required for IRES activity.
Keywords: translation control, IRES elements, RNA structure, RNase P ribozyme, tRNA, structural mimicry
INTRODUCTION
Translation initiation of picornavirus genomes is a cap-independent process, mediated by the internal ribosome entry site (IRES) that allows the recognition of an internal AUG codon (Hellen and Sarnow 2001). Picornavirus IRES consists of a cis-acting element composed of a sequence ∼450 nucleotides (nt) located immediately upstream of the functional start codon (Martinez-Salas et al. 2001). IRES elements also direct translation initiation in other viral RNAs, such as hepatitis C virus (HCV) and picornavirus-like insect dicistronic viruses (Sarnow 2003). IRES elements are organized as highly ordered RNA structures that contain several stable stem–loops (Martinez-Salas and Fernandez-Miragall 2004; Jan 2006). However, a unified model of IRES organization has not been found yet; on the contrary, IRES appear to be distributed into different classes that are well differentiated in terms of RNA structure and transacting factor requirements (Stoneley and Willis 2004; Baird et al. 2006).
Over the last years the RNA structural organization of the foot-and-mouth disease virus (FMDV) IRES has been studied using RNA probing analysis in conjunction with RNA–protein interactions and functional analysis. The FMDV IRES is composed of three differentiated regions. The first 85 nt on the 5′ end encompasses domain 2 (also termed H); it consists of a stem–loop that contains a binding site for the polypyrimidine tract-binding protein (PTB) (Luz and Beck 1991). Second, the central domain (termed 3 or I, encompassing nucleotides 86–299) is a self-folding region that adopts a cruciform RNA structure (Fernandez-Miragall and Martinez-Salas 2003) including internal RNA–RNA interactions (Fernandez-Miragall et al. 2006). Third, on the 3′ end of the IRES, domains 4 and 5 (or J–K and L, encompassing nucleotides 300–462) determine RNA–protein interactions involving translation initiation factors, eIF4G, eIF4B, eIF3, and auxiliary proteins as PTB (Lopez de Quinto and Martinez-Salas 2000; Pilipenko et al. 2000; Lopez de Quinto et al. 2001).
The role of the central domain in picornavirus IRES has remained elusive from the mechanistic point of view. This region contains two conserved motifs, GNRA and RAAA (where G stands for guanine; N, any nucleotide; R, purine; and A, adenine); mutations disrupting these motifs impaired IRES activity (Lopez de Quinto and Martinez-Salas 1997; Robertson et al. 1999), indicating its requirement for IRES activity. It has been shown that defective IRES elements bearing nucleotide substitutions in these motifs induced a reorganization of the RNA structure of the central domain (Fernandez-Miragall and Martinez-Salas 2003).
Recently, the HCV IRES was found to contain RNA structural motifs that served as substrates for the human RNase P (Nadal et al. 2002), and thus it was inferred that these IRES might contain structural elements that mimic the tRNA-like structure. This result was later expanded to pestivirus RNAs (Lyons and Robertson 2003). Similarly plant RNA viruses containing tRNA-like structures at the 3′ end were recognized in vitro by RNase P (Guerrier-Takada et al. 1988; Mans et al. 1990). In an independent approach, it has been described that IGR elements belonging to the Dicistroviridae family mimic the initiator tRNA during internal initiation (Spahn et al. 2004; Schuler et al. 2006) in a process independent of initiation factors and tRNAi (Wilson et al. 2000; Jan and Sarnow 2002; Jan et al. 2003; Pestova and Hellen 2003).
RNase P is an endonuclease present in all organisms involved in the processing of the tRNA precursor (Gopalan et al. 2002; Evans et al. 2006; Kazantsev and Pace 2006), resulting in the production of RNA fragments that specifically contain 5′P and 3′OH ends (Robertson et al. 1972; Gopalan et al. 2002). RNase P subunits consist of RNA and protein, and in bacteria RNA subunits are catalytically active in vitro (Guerrier-Takada et al. 1983). The RNase P ribozyme from Synechocystis sp., which does not require the CCA sequence at the 3′ end of the tRNA precursor (Pascual and Vioque 1999), recognizes a structural motif in the HCV IRES (Sabariegos et al. 2004).
Here we have used the RNase P ribozyme from Synechocystis sp. as a tool to investigate the presence of structural elements in the FMDV IRES. Of interest, we have found a structural region in the central domain that is recognized in vitro by this ribozyme in a dosage-dependent manner, either in the form of the full-length IRES or as short transcripts encompassing the apical region of the central domain. The reaction was dependent on high Mg2+ concentration and yielded cleavage products with 5′-P and 3′-OH ends. Under the same conditions, the 3′ region of the IRES did not serve as a substrate for this ribozyme. The use of IRES transcripts carrying nucleotide substitutions in critical residues of this region led to a modified pattern of ribozyme cleavage concomitantly with the presence of altered RNA organization in the defective IRES.
RESULTS
The FMDV IRES serves as a substrate for the Synechocystis RNase P ribozyme
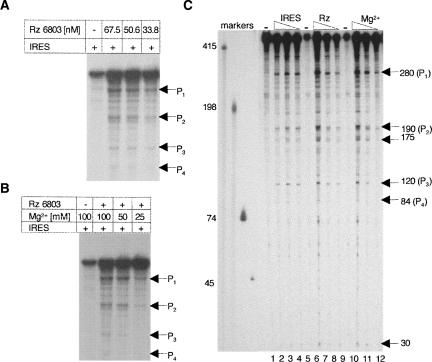
The IRES region of FMDV was transcribed using radiolabeled CTP and digested with Synechocystis RNase P ribozyme (hereafter, Rz 6803) varying the ribozyme concentration. As a control, a substrate aliquot was incubated in buffer without ribozyme. The reaction products were separated in 6% denaturing acrylamide gels (Fig. 1A). Four specific cleavage products absent in the sample incubated in buffer were identified, whose intensity decreased according to their size. The results indicated that the optimal ribozyme concentration was 67.5 nM. A similar assay using a decreasing concentration of substrate showed that 6.75 nM was enough to detect all digestion products.
FIGURE 1.
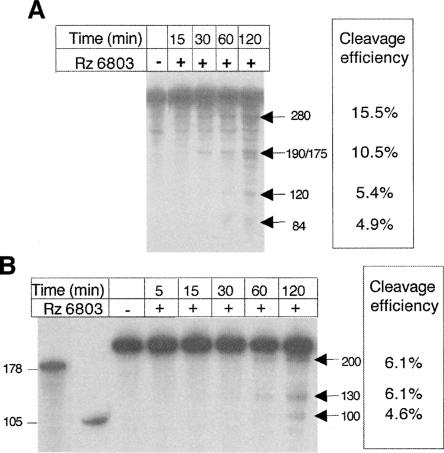
Cyanobacterial RNase P recognition of the FMDV IRES. (A) Ribozyme concentration-dependent cleavage of the FMDV IRES by the Synechocystis RNase P. A constant concentration of uniformly labeled FMDV IRES (6.75 nM) was incubated with increasing concentrations of Rz 6803; the digestion products were fractionated on 6% denaturing acrylamide gels and visualized by autoradiography. P1–P4 depict the most intense specific digestion products, absent in the control RNA lane incubated in the absence of ribozyme. (B) Influence of the Mg2+ concentration in the cleavage efficiency of the FMDV IRES by the Synechocystis RNase P. Uniformly labeled FMDV IRES (6.75 nM) was incubated with Rz 6803 in a buffer containing the indicated concentration of Mg2+; the digestion products were fractionated on 6% denaturing acrylamide gels and visualized on X-ray films. (C) Determination of the average products length. The digestion products obtained in reactions similar to those described in A and B were fractionated in long denaturing gels in parallel to known labeled transcripts indicated to the left of the autoradiogram, used as markers. The substrate concentration in lanes 2, 3, and 4 was 67.5–6.75 nM with a constant concentration of ribozyme (67.5 nM).
To define the optimal conditions of cleavage, we tested a range of Mg2+ concentrations in the incubation buffer (Fig. 1B). Ribozyme hydrolysis was more efficient at 100 mM Mg2+, with a significant decrease in cleavage efficiency when the concentration was 25 mM, as expected (Guerrier-Takada et al. 1986). Subsequent fractionation of the reaction in parallel to known RNA markers allowed an estimation of the size of the cleavage products (Fig. 1C). The size of the most intense cleavage products corresponded to ∼280 nt and a doublet of 190/175 nt. In addition, weak products of 120, 84, and 30 nt were observed. Of these products, the most intense added up to the entire IRES length (∼470 nt), suggesting that the remaining products could be generated in second site cleavage events.
Cleavage assays performed in parallel with the HCV IRES substrate as a positive control indicated that the efficiency of FMDV IRES cleavage was similar to that found for the entire HCV IRES (around 10% of the input substrate; data not shown).
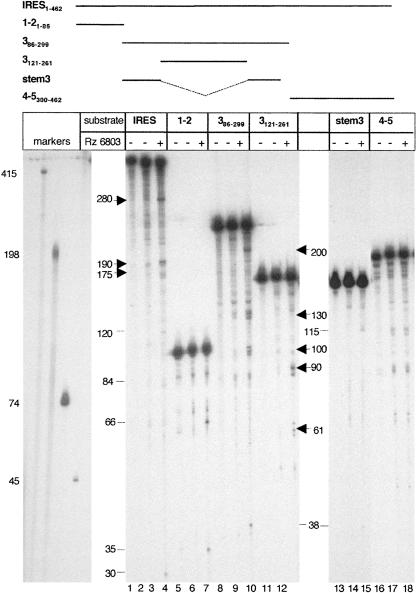
Identification of the core region recognized by RNase P in the FMDV IRES
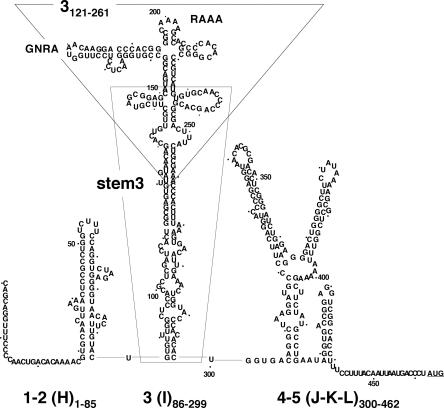
In order to map the motif recognized by the Rz 6803 in the FMDV RNA, we made use of transcripts that correspond to three distinct regions of the IRES, encompassing domains 1–2, 3, and 4–5 (Fig. 2). These RNAs were previously identified as stable stem–loops in RNA probing analysis as well as in RNA–protein binding assays (Martinez-Salas and Fernandez-Miragall 2004). The cleavage products were fractionated in denaturing gels, parallel to an RNA aliquot kept on ice for the entire duration of the assay. Another aliquot was incubated in reaction buffer with a high magnesium concentration, but in the absence of Rz 6803. The results obtained for the different transcripts together with the entire IRES are shown in Figure 3. Comparison of the treated with the untreated RNA identified several products in the entire IRES of ∼280 nt and a doublet of 190/175 nt (Fig. 3, lane 3), in agreement with Figure 1C. Two cleavage products of ∼66 and 35 nt were seen in substrate 1–2 (Fig. 3, lane 6). Digestion of transcript 3 resulted in fragments of ∼200, 130, 100, and 38 nt (Fig. 3, lane 9). Paired up, the size of these fragments was compatible with two internal cuts that add up the whole transcript (228 nt), which also occurred in the entire IRES RNA.
FIGURE 2.
Secondary structure of FMDV IRES. RNA structure is depicted according to RNA probing analysis (Fernandez-Miragall et al. 2006). IRES domains are numbered 1–5 (or H–L); the nucleotides corresponding to the IRES transcripts used in this study are indicated as subscripts; a dot is used to mark positions every 10 nt. The conserved GNRA and RAAA motifs in domain 3 are highlighted. A broken line surrounds transcript 3121–261. A thin line surrounds transcript stem3, encompassing residues 85–299 with 151–227 deleted.
FIGURE 3.
The apical region of the central domain contains the main cleavage site in the FMDV IRES. The uniformly labeled transcripts, schematically represented in the top panel, were subjected to RNase P digestion parallel to the entire IRES. The reaction products were fractionated in 6% denaturing gels parallel to known markers. The nucleotide composition and RNA structure organization of the indicated transcripts are depicted in Figure 2. Products detected in RNA kept on ice (lanes 1,4,7,10,13,16), or incubated in buffer for the entire duration of the reaction (lanes 2,5,8,11,14,17) were used as negative reaction controls, and not considered as Rz products. Thick arrows in the left and right of each panel depict the reaction products characterized in this study, while thin lines depict reaction products not characterized.
Subsequently, shorter internal transcripts, 3121–261 and stem 3, corresponding to the apical and proximal region of the central domain (see Fig. 2), were used to more precisely allocate the recognition motif in this region. Of interest, the most intense cleavages were seen in the transcript 3121–261 corresponding exclusively to the apical region, giving rise to two products of 90 and 61 nt (Fig. 3, lane 12). In contrast, digestion of stem 3 resulted in only faint fragments (Fig. 3, lane 15). Transcripts shorter than 3121–261 were not studied because their RNA structure is not stable (O. Fernandez-Miragall, unpubl. data). Thus, with the exception of weak cleavages on the 5′ end of transcripts 1–2, 3, and stem 3 resulting in short fragments of ∼30–38 nt, no other ribozyme recognition motifs other than that located in the apical region were detected. In our assay conditions, transcript 4–5 corresponding to the 3′ region of the IRES was not a substrate for this enzyme since the products detected in lane 18 were also seen in the RNA aliquot incubated in buffer (Fig. 3, lane 17).
Therefore, taken together with the results shown in Figure 3, we conclude that the Rz 6803 recognition motif was located within the central domain of the FMDV IRES, in a region encompassing residues 121–261. In our previous studies it was shown that this region adopts a specific RNA folding that depends on the GNRA tetraloop (Fernandez-Miragall and Martinez-Salas 2003), an essential motif in the picornavirus IRES elements (Lopez de Quinto and Martinez-Salas 1997).
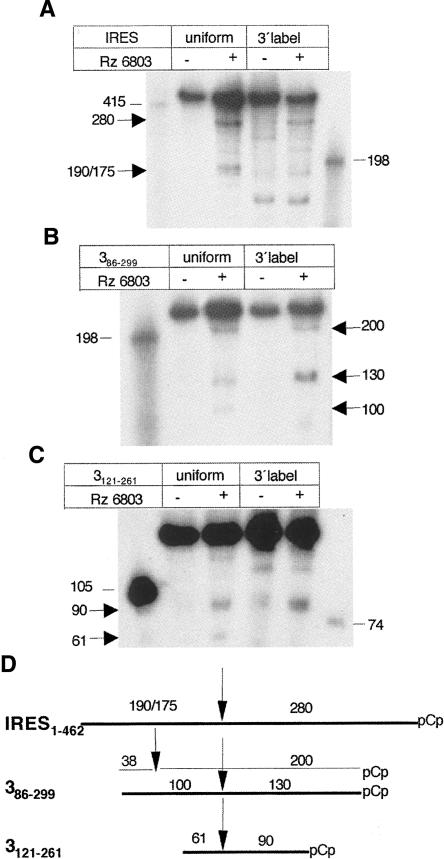
Mapping of the 5′ and 3′ ends of the ribozyme cleavage products in the entire IRES and the central domain transcripts reveals a shared cleavage site
Next, to map the relative position of 5′ and 3′ products on the Rz 6803 substrates, the complete IRES as well as two internal transcripts encompassing the apical region of the central domain were analyzed by taking advantage of the comparison between the uniform- and the 3′-end-labeled substrate. Whereas digestion of the uniformly labeled IRES transcript resulted in the production of two main products, 280 and 190, the 3′-end-labeled substrate led to the production of the 280 nt with a significant decrease in the 190 nt (Fig. 4A). This result indicated that the 280 nt fragment corresponded to the 3′ end of the IRES substrate.
FIGURE 4.
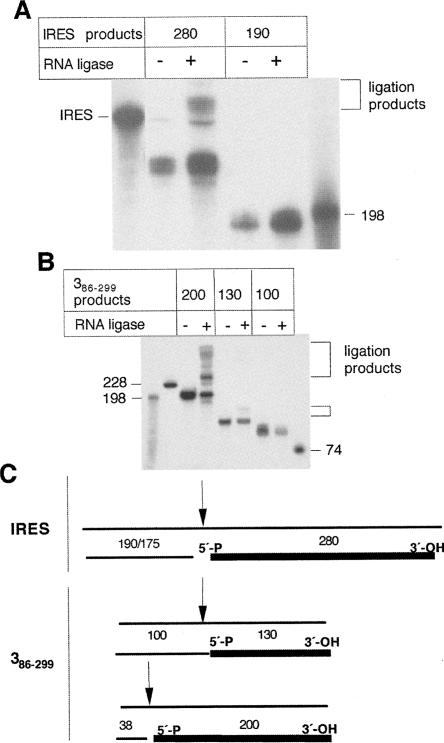
Mapping of the ribozyme cleavage products. Transcripts IRES (A), 386–299 (B), and 3121–261 (C) were labeled either uniformly or at the 3′ end using pCp and T4 RNA ligase prior to its incubation with the Rz. The reaction products were fractionated in denaturing gels. Arrows depict digestion products, while thin lines denote the RNA size markers position. (D) Alignment of the digestion products according to their relative orientation. Absence of a specific product detected in each pCp-labeled RNA relative to the uniformly labeled counterpart was taken as evidence of its 5′ position within the transcript under study. The size of each fragment is indicated in a number of nucleotides.
Similar comparisons between uniform- and 3′-pCp-labeled substrates were carried out with transcripts 3 and 3121–261. The results obtained with substrate 3 showed that the products of 200 and 130 nt were present in both types of labeled substrates, while the 100 nt product disappeared in the 3′-end-labeled substrate (Fig. 4B). Subsequent analysis carried out in the short version of the apical region of RNA 3 showed that the 90 nt fragment was as detectable as in the uniformly labeled, while the 61 nt product was absent in the 3′-end-labeled RNA (Fig. 4C).
A summary of the RNase P recognition mapping deduced from the above data is shown in Figure 4D. The cleavage event around the internal cut in transcript 3121–261 can be aligned in the three substrates, demonstrating again the presence of a self-folding structural element present in the three overlapping RNAs. A second cleavage near the 5′ end that resulted in a fragment of 38 nt was detected in transcript 3. Small fragments of ∼30 nt were also observed in the uniformly labeled IRES transcript (not represented in Fig. 4D), as well as in substrates 1–2 and stem 3 (see Fig. 3). The study of the small fragments was not pursued because they were not found to result from cleavages in the same IRES residue when transcripts of different length were assayed.
A time course of the cleavage reaction carried out in the entire IRES (Fig. 5A) showed that the relative intensity of products of 280 and 190 nt normalized to their size was similar all along the incubation time, reaching values of ∼15.5% and 10.5%, respectively, at 120 min. At the same time point, the intensity of the products of 84 and 120 nt also normalized to their size was lower, ∼5%, suggesting that they may originate from a second cleavage event of the primary products. In the case of transcript 3, the intensity of all products was similar, 6.1, 6.1, and 4.6 (Fig. 5B), presumably arising from primary cleavage events. These results, together with those summarized in Figure 4D, further confirm a model in which the localization of the structural element recognized by Rz 6803 in all IRES transcripts resides within the apical region of the central domain.
FIGURE 5.
Time course of the ribozyme reaction with FMDV IRES substrates. Transcripts IRES (A) and 386–299 (B) were incubated with the Rz 6803 under optimal conditions; aliquots were withdrawn at the indicated times and fractionated on denaturing gels. The efficiency of cleavage was determined as the percent of label measured in a PhosphorImager in the product of interest relative to the total input normalized to the size of the transcript in each case.
Both the entire and the internal IRES substrates are specifically recognized by the cyanobacterial ribozyme
Previous work has characterized the specific formation of 5′-P and 3′-OH terminal ends during RNase P cleavage reaction (Robertson et al. 1972). To assess that the reaction products were specifically due to RNase P digestion, each of the Rz 6803 cleavage products was subjected to a self-ligation test. To this end, the 280 and 190 nt digestion products of the IRES substrate were excised and purified from a denaturing gel and subsequently tested for their capacity to circularize in the presence of RNA ligase. The results, shown in Figure 6A, indicated that the 280 nt fragment was able to form a product of retarded mobility in denaturing gels, compatible with a circular, covalently ligated RNA form. An aliquot of the same sample, incubated in the absence of ligase, did not contain this lower mobility form. In contrast, the 190 nt product was not able to self-ligate under the same conditions. We conclude that the 280 nt product contained 5′-P and 3′-OH terminal ends. The 5′-P end was fully consistent with the RNase P cleavage.
FIGURE 6.
The ribozyme cleavage products contain a 5′-phosphate end. (A) The indicated gel-purified uniformly labeled reaction products, corresponding to each end of the IRES transcript, were self-ligated in the presence of T4 RNA ligase. The ligation products were fractionated in denaturing gels, parallel to control samples incubated in the absence of ligase. (B) A similar study carried out with the three digestion products of the central domain, of 200, 130, and 100 nt. Brackets point to retarded ligation products; thin lines denote RNA size markers. (C) Summary of the ligation test assay. The gel-purified product resulting in the formation of ligation products, depicted by a thick line, contains 5′-P and 3′-OH residues. The size of each fragment (in a number of nucleotides) is indicated.
A similar test was performed with the reaction products of substrate 3. The three major products, 200, 130, and 100 nt, were excised from gels and self-ligated (Fig. 6B). Of interest, the fragments of 200 and 130 nt could be ligated, although with different efficiency. No sign of self-ligation was detected in the 90 nt product, even after long exposure times.
A summary of the 5′-P–3′-OH ligated fragments is represented in Figure 6C. These results are in good agreement with those shown in Figure 4D, and strongly favored a model in which a specific Rz 6803 cleavage event resided in the apical region of the central domain.
As a second way to verify the specificity of ribozyme recognition, we made use of a previously described RNase P substrate (Sabariegos et al. 2004). As indicated before, the efficiency of FMDV IRES cleavage by Rz 6803 was similar to that of the HVC IRES. Additionally, incubation of the uniformly labeled domain 3 against a 20-fold excess concentration of unlabeled HCV IRES resulted in about a twofold decrease in cleavage efficiency (data not shown). In contrast, no effects were detected with transcript 4–5, consistent with its lack of recognition by this ribozyme (Fig. 3).
Differential response to RNase P ribozyme cleavage of FMDV IRES mutants with an altered RNA organization of the central domain
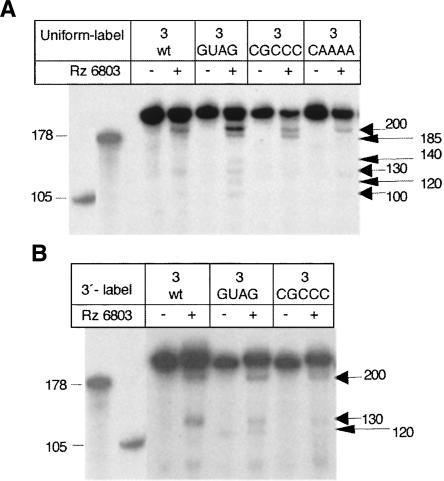
Based on results shown above, the RNase P ribozyme recognized a structural element in the apical region. To define the structural element recognized by the ribozyme more precisely, we took advantage of IRES mutants carrying base substitutions in the central domain. Two types of mutants were tested; the first type had the capacity to modify IRES structure, whereas the second one did not. Within the first group we tested a mutant affected in the conserved GNRA177–181 motif (see Fig. 2). Our previous studies have shown that the GUAG181 mutant RNA containing a single substitution in the GNRA motif impaired IRES activity and modified the RNA organization of the apical region (Fernandez-Miragall and Martinez-Salas 2003). Treatment of GUAG mutant RNA with Rz 6803 led to three new fragments of 185, 140, and 120 nt relative to the wild-type (wt) sequence (Fig. 7A). At the same time, the 200 nt product was more intense than that of the wt RNA. The use of a different mutant that contained a substitution in the nearby RAAA199–202 loop to CGCCC (see Fig. 2) accompanied by a higher accessibility of the apical region to nucleases and DMS (Fernandez-Miragall et al. 2006) also modified the Rz 6803 cleavage pattern, giving rise to one new fragment of 185 nt. Within the second group, mutations in the central domain that do not modify the RNA structure of the apical region, such as substitution in the ACCC234–237 loop to AAAA (Fig. 7A), did not modify the Rz 6803 cleavage pattern in a significant manner.
FIGURE 7.
RNase P recognition of IRES substrates containing an altered RNA structure in the central domain. (A) A uniformly labeled transcript carrying a single nucleotide substitution of GUAA to GUAG in the GNRA tetraloop (Fernandez-Miragall and Martinez-Salas 2003) was incubated with RZ 6803, parallel to wt RNA. A long arrow depicts three new digestion products of 185, 140, and 120 nt. The transcript 3 carrying a substitution of AAAAG to CGCCC in the RAAA motif was processed in the same way. A substitution mutant in the C-rich motif, C235–238 of this domain, was used as a control of a nonreorganized RNA structure. (B) Ribozyme 6803 digestion of 3′-pCp-labeled transcripts, processed as in A.
The relative position of products in GUAG and CGCCC RNAs was examined following Rz 6803 treatment of 3′-end-labeled fragments. The absence of three products with 185, 140, and 100 nt (Fig. 7B) demonstrated their 5′ terminal locations within the transcript.
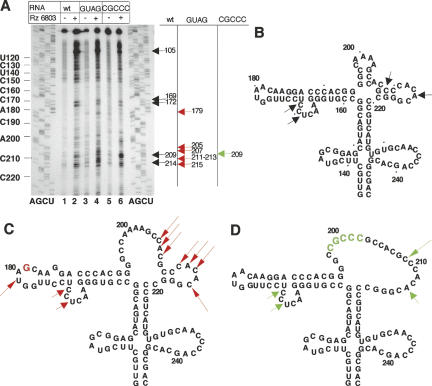
To identify more precisely the Rz 6803 cleavage sites, we performed primer extension analysis on the different substrates carrying the wild-type sequence, the GUAG or CGCCC substitutions (Fig. 8A). In comparison to the undigested RNA, an intense product was detected around residue C105 in the three transcripts. Similarly, two cuts appeared to affect residues C169 C172 in both wild-type and mutant sequences. In contrast, GUAG RNA showed new cuts at residue U179 as well as C207 to A214 (Fig. 8A). In the case of CGCCC RNA, an increase in the cleavage to C209 was detected.
FIGURE 8.
Primer extension analysis of the Rz cleavage sites. (A) Transcripts corresponding to the central domain bearing the wild-type sequence or two different nucleotide substitutions (GUAG in the GNRA tetraloop and CGCCC in the RAAA loop) were subjected to reverse transcriptase (RT) extension with a 5′-end-labeled primer. cDNA products were analyzed on denaturing 6% acrylamide gels parallel to a DNA sequence prepared with the same oligonucleotide. IRES residues are indicated on the left side of the gel every 10 nt. Arrows depict the position of cleavages specifically detected in the Rz digestion samples. Synthesis of the full-length cDNA product is detected at the top of the gel. (B) Position of the cleavage sites indicated on the RNA structure previously determined by RNA probing (Fernandez-Miragall and Martinez-Salas 2003; Fernandez-Miragall et al. 2006) obtained for wt sequence, GUAG mutant sequence (C), and CGCCC mutant sequence (D).
The main cleavage events found in the central domain of wild-type FMDV IRES are summarized in Figure 8B. There was good agreement between the primer extension analysis of the digestion products and the direct alignment of ribozyme hydrolysis fragments derived from labeled substrates as shown in Figures 3, 4, and 7. The strong stop near residue C105 at the top of the gel in Figure 8A is consistent with the appearance of fragments with 38 and 200 nt during Rz digestion of transcript 3 (Figs. 3, 4). The primer extension stop observed at residue C171 is compatible with the 130 and 100 nt fragments observed in Figures 3 and 4. Recognition of nt C209 and A214 will give rise to products of ∼140 nt, below the level of detection in Figures 3 and 4. Regarding the cleavage pattern of IRES mutants (summarized in Fig. 8C,D) the cleavage at residue U179 in the GUAG mutant was expected from the 120 nt fragment, and the cleavage at residues C207–A214 resulted in the 140 nt fragment. However, the new cleavage yielding the product of 185 nt in Figure 7 was not detected after primer extension mapping, presumably because of its close distance to the primer.
Taken together, we conclude that the core element recognized by Rz 6803 was located in the apical region of the central domain (see Fig. 2), and its recognition was affected in mutants with a local RNA structure reorganization (Fig. 8C,D), suggesting that its structural preservation is required for IRES activity.
DISCUSSION
In this study we have found a structural element in the FMDV IRES that is specifically recognized in vitro as a substrate for the RNA subunit of the cyanobacterial Synechocystis RNase P, a ribozyme that cleaves the tRNA precursor without a 3′-CCA sequence requirement (Pascual and Vioque 1999). The results obtained with the FMDV IRES demonstrated that the core element recognized in vitro by the Rz 6803 resided in the apical region of the central domain, within a transcript encompassing residues 121–261 of the IRES. The specificity of the reaction was evidenced by the self-ligation capacity of the 3′ end products arising in the Rz 6803 reaction and the competitive effect of HCV structural elements, previously demonstrated to be recognized as a substrate of this ribozyme (Sabariegos et al. 2004). The same structural element was recognized by the ribozyme irrespective of the substrate length; both the alignment of ribozyme hydrolysis products derived from different overlapping substrates and the primer extension analysis of the cleavage products pointed toward U169–C172 of the FMDV IRES as the cleavage site. These residues are located in a conserved bulge within the GNRA stem–loop (see Fig. 8B).
The central domain of the FMDV IRES, termed 3 or I, is a self-folding region that contains conserved motifs essential for IRES activity (Lopez de Quinto and Martinez-Salas 1997). According to RNA probing, the GNRA motif adopts a tetraloop conformation and mediates the local RNA structure (Fernandez-Miragall and Martinez-Salas 2003), presumably involving tertiary contacts (Fernandez-Miragall et al. 2006). As shown in this study, the core structural element recognized by the cyanobacterial Rz 6803 resides in this region. Ribozyme recognition remarks the unity of the structural entity in the apical region of domain 3. tRNA-like structures recognized by RNase P enzymes in various viral RNAs are located in the vicinity of RNA pseudoknots that create complex structural organizations (Mans et al. 1990). At present, it is not known if the apical domain threefolds in a pseudoknotted manner or if the GNRA–receptor interaction provides similar clues for Rz 6803 recognition.
In favor of a structural element recognized by the ribozyme we also have found in this report that defective IRES mutants with a modified RNA structure in the apical region of the central domain responded differentially to ribozyme cleavage. Conversely, mutations that did not involve RNA reorganization did not alter the pattern of Rz 6803 cleavage either. It is of interest that IRES transcripts bearing nucleotide substitutions in the GNRA motif that resulted in a reorganization of RNA structure induced an increase in Rz 6803 recognition, accompanied by new cleavage sites located in nearby residues of the same region. In another FMDV IRES mutant bearing a CGCCC substitution at the most apical RAAA loop, a new product of the same length as in the GNRA substitution mutant was observed, indicating that this effect was not exclusively observed in the GUAG mutant. A related example was reported for HCV, in which a natural variant containing a single nucleotide substitution resulted in an enhanced second cut in a nearby residue by human RNase P (Piron et al. 2005).
It has been previously reported that RNase P recognizes transient RNA structures, present in riboswitches (Altman et al. 2005). Whether the secondary cleavage sites observed in the FMDV mutants studied here could be the result of recognition of transient RNA structures or a small percentage of the RNA molecules adopting a folding that serves as a substrate for the ribozyme more often that the wild type is not known at present. Since the efficiency of Rz 6803 cleavage of the FMDV IRES is low (about 10%), it is likely that that the ribozyme has a low affinity for this substrate or that the structural motif recognized by the Rz 6803 is present in only a small percentage of the molecules in solution. In this regard, cleavage of natural RNA substrates within bacterial polycistronic mRNAs has been described to occur less efficiently than tRNA precursors (Li and Altman 2003). On the other hand, the stem–loop located at the edge of the structural motif recognized by the ribozyme (see Fig. 2) may inhibit the RNase P reaction (Lee et al. 1997).
According to RNA probing analysis, both GUAG and CGCCC IRES transcripts adopted a folding pattern more accessible to nucleases and chemical reagents than the wild-type RNA. However, since not every single nucleotide is detected in the RNA probing procedure, we could not exclude the possibility that more residues than those depicted in Figure 8 are base paired, as suggested for CGCCC (Fernandez-Miragall et al. 2006), or conversely that the residues C209–210 are unpaired in GUAG RNA. The cleavage pattern of wt and CGCCC differed in the intensity of hydrolysis at C209, presumably due to the presence of several unpaired bases along this region. The cleavage pattern of GUAG mutant differed from the wt in two ways; on the one hand there was a moderate enhancement in the yield of the 200 nt fragment. On the other hand, three additional sites were recognized (Fig. 7) with the peculiarity that each hydrolysis site occurred at more than one contiguous nucleotide (see Fig. 8). This situation may be reminiscent of the modified enzyme–substrate interaction described in the 3′-CCA region of the tRNA precursor (Kikovska et al. 2005).
Domain 3 of the FMDV IRES is an integral part of the IRES element. Although nucleotide substitutions within this region abrogate IRES activity (Lopez de Quinto and Martinez-Salas 1997), it is important to note that domain 4–5, which contains the capacity to interact with key translation initiation factors (Lopez de Quinto and Martinez-Salas 2000; Pilipenko et al. 2000; Lopez de Quinto et al. 2001), does not show IRES activity (S. Lopez de Quinto and E. Martinez-Salas, unpubl.). However, the role performed by the central domain during internal initiation remains unknown.
Here we have shown that the FMDV IRES region contains a structural element within the FMDV RNA where the virus has evolved a tRNA structural mimicry that renders it a substrate for Rz 6803 reaction in vitro. This structural property of the FMDV IRES is shared with other IRES elements (Nadal et al. 2002; Lyons and Robertson 2003; Sabariegos et al. 2004). Indeed, the efficiency of Rz 6803 cleavage of FMDV and HCV IRES is similar and, according to our results of competition assays, a twofold decrease in cleavage efficiency was observed when the complete uniformly labeled FMDV IRES was incubated against 20-fold excess of unlabeled HCV IRES. However, in contrast to the HCV (Nadel et al. 2002) or the pestivirus IRES elements (Lyons and Robertson 2003), the Rz 6803 recognition motif in FMDV is not located at the very 3′ end of the IRES region. Although this result does not preclude the existence of other structural motifs near the 3′ end, undetected under our conditions, attempts to cleave the FMDV IRES with purified human RNase P were unsuccessful. These data point to a difference between distantly related IRES elements that may be relevant to understanding the different strategies used by viral RNAs to interact with the translational machinery.
FMDV, which is the prototypic member of the Aphthovirus genus in the Picornaviridae family, infects artiodactylae, mostly cattle, swine, sheep, and goats. The entire viral cycle occurs in the cytoplasm of infected cells. Therefore, viral RNA has no access to RNase P, which is localized in the cell nucleus. On the other hand, it is conceivable that the presence of a tRNA-like motif within the FMDV IRES will ensure an efficient recognition of the viral RNA by the translational machinery. Recent work performed with HCV and the IGR of Dicistroviridae has shown the capacity of these IRES elements to accommodate in the interface of the ribosomal subunits, but whereas the IGR occupies the A, P, and E sites, HCV only associates with the E site (Spahn et al. 2001; Boehringer et al. 2005; Costantino and Kieft 2005; Pfingsten et al. 2006; Schuler et al. 2006). These results led us to propose that specialized motifs of the IRES region mimic the tRNAi during protein synthesis. However, further work is needed to establish a functional connection between RNA organization detected by structural analysis and a presumed tRNA-like motif structure detected by ribozyme recognition.
MATERIALS AND METHODS
IRES constructs
The IRES of FMDV C-S8 and its domains, 1–2, 3, and 4–5, were cloned in the pGEM 3 vector (Promega) as described (Ramos and Martinez-Salas 1999). The central region, termed domain 3, was further deleted to generate the proximal region (termed 3121–261) and the apical region (stem 3, nucleotides 84–297Δ151–227) (Fernandez-Miragall et al. 2006). The GNRA and RAAA motif mutants are described by Lopez de Quinto and Martinez-Salas (1997). The IRES of hepatitis C virus was cloned in the same transcription vector (Lafuente et al. 2002).
In vitro transcription
Prior to RNA synthesis, plasmids were linearized to generate transcripts encoding the different domains or the full-length IRES. Transcription was performed for 1 h at 37°C using the Megashortscript kit (Ambion) as recommended by the manufacturer. When needed, RNA transcripts were uniformly labeled using (α32P)-CTP (5 μM, 400 Ci/mmol) and 50 U of T7 RNA polymerase (New England Biolabs) in the presence of 0.5–1 μg of linearized DNA template, 50 mM DTT, 0.5 mM rNTPS, and 20 U of RNasin (Promega). Reactions were incubated for 15 min with 1 U of RQ1 DNase (Promega) and unincorporated 32P-CTP eliminated by exclusion chromatography in TE-equilibrated sephadex columns (Amersham).
CTP-labeled RNAs of different lengths, 415, 198, 105, 74, or 45 nt, were prepared as marker size; they correspond to transcripts S, 3′ end, domain 1–2, SL2Δpoly(A), or 3160–196, respectively (Fernandez-Miragall et al. 2006; Serrano et al. 2006).
RNA 3′-end labeling
For 3′-end labeling, RNA transcripts (1 μM) were labeled with 20 U of T4 RNA ligase (New England Biolabs) and [32P]pCp 5′triphosphate (1 μM, 3000 Ci/mmol). The reaction was carried out in 10 μL of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM DTT, 1 mM ATP, 10% DMSO, and 20 U RNasin. The reaction mixture was incubated during 24 h at 16°C, and the unincorporated isotope eliminated by exclusion chromatography in TE-equilibrated Sephadex columns (Amersham).
Synthesis of the ribozyme
Plasmid pT76803, digested with DraI restriction enzyme, was used as a DNA template for the T7-dependent transcription of Synechocystis sp. PCC6803 RNase P RNA (Vioque 1992). Transcription was performed using the Megashortscript kit (Ambion). RNA concentration was determined by OD measurement and its activity titrated on the tRNA precursor obtained from pTyr and HCV transcripts as described (Nadal et al. 2002). Small aliquots were then kept at −70°C until further use.
RNase P ribozyme cleavage
Ribozyme cleavage assays were made essentially as described (Sabariegos et al. 2004) with the exception that the substrate was denatured 3 min at 95°C prior to its addition to the ribozyme reaction. Briefly, the cyanobacterial P RNA (67.5 nM) was incubated in assay buffer (50 mM Tris-HCl at pH 7.5, 100 mM MgCl2, 1 M KCl, 4% polyethylene glycol) for 15 min at 37°C before addition of the substrate. The reaction was incubated for 2 h at 37°C, and then stopped by addition of 2 vol of denaturing loading buffer (90% formamide, 1 mM EDTA at pH 8, 0.1% xylencianol, 0.1% bromophenol). Reaction products were separated on 6% acrylamide 7 M urea gels and visualized by autoradiography. Labeled transcripts of known length, 415, 198, 105, 74, and 45 nt, run in parallel wells, were used as markers. The efficiency of the ribozyme cleavage reaction was measured in a PhosphorImager in triplicate experiments when needed. The average cytosine content in the FMDV IRES is 27%. Cleavage efficiency was calculated as the percentage of intensity in the product of interest relative to the input sample, normalized to the RNA size.
RNA ligation
The uniformly labeled RNA products of interest resulting from ribozyme cleavage reactions were fractionated by gel electrophoresis, visualized in short exposure times, and excised from the gel. The RNA was allowed to elute from the gel slices overnight at 37°C in TE buffer containing 0.5% SDS. Following ethanol precipitation, each RNA was incubated with 20 U of T4 RNA ligase during 24 h at 16°C. As a control, an aliquot of the same RNA was incubated in the absence of T4 RNA ligase. The ligase reaction was stopped with denaturing loading buffer, heated at 92°C for 2 min, and then examined on denaturing 6% acrylamide, 7 M urea gels and visualized by autoradiography.
Primer extension analysis
The RNA cleavage reaction was made as described above using the desired unlabeled transcript (6.75 nM) as a substrate. Antisense oligonucleotides, complementary to FMDV IRES residues 296–287 (5′-CCCGGGTGTGGGTACC-3′) or 185–165 (5′-CTTGTCGCCAAGGAGGAGTTC-3′) were labeled at the 5′ end using (γ-32P)-ATP and T4 polynucleotide kinase (Boehringer Mannheim). Then, the ribozyme-treated RNA (about 5 ng) was denatured 3 min at 95°C and cooled down on ice. Annealing and extension of the labeled antisense primer was carried out in 15 μL of reverse transcriptase (RT) buffer (20 mM Tris-HCl at pH 7.5, 15 mM MgCl2, 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.01% [v/v] NP-40, 50% [v/v] glycerol) in the presence of 100 U of Superscript II RT (Life Technologies) and 1 mM each dNTP during 1 h at 45°C. The RNA template was then hydrolyzed, and RT-extension products were fractionated in denaturing 6% acrylamide 7 M urea gels as described (Fernandez-Miragall and Martinez-Salas 2003). To prepare the sequence ladder, the Thermosequenase cycle sequencing kit (USB) was used with the same 5′-labeled antisense oligonucleotide used for primer extension and plasmid pGEM-IRES DNA as a template.
ACKNOWLEDGMENTS
We are grateful to J. Ramajo for technical assistance, O. Fernandez-Miragall for early contributions to this work, and C. Gutierrez and A. Vioque for helpful suggestions on the manuscript. This work was supported by grant BFU-2005-00948 and by an Institutional grant from Fundación Ramón Areces.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.506607.
REFERENCES
- Altman, S., Wesolowski, D., Guerrier-Takada, C., Li, Y. RNase P cleaves transient structures in some riboswitches. Proc. Natl. Acad. Sci. 2005;102:11284–11289. doi: 10.1073/pnas.0505271102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird, S.D., Turcotte, M., Korneluk, R.G., Holcik, M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer, D., Thermann, R., Ostareck-Lederer, A., Lewis, J.D., Stark, H. Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: Remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Costantino, D., Kieft, J.S. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, D., Marquez, S.M., Pace, N.R. RNase P: Interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Fernandez-Miragall, O., Martinez-Salas, E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA. 2003;9:1333–1344. doi: 10.1261/rna.5950603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Miragall, O., Ramos, R., Ramajo, J., Martinez-Salas, E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA. 2006;12:223–234. doi: 10.1261/rna.2153206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan, V., Vioque, A., Altman, S. RNase P: Variations and uses. J. Biol. Chem. 2002;277:6759–6762. doi: 10.1074/jbc.R100067200. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N., Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Haydock, K., Allen, L., Altman, S. Metal ion requirements and other aspects of the reaction catalyzed by M1 RNA, the RNA subunit of ribonuclease P from Escherichia coli . Biochemistry. 1986;25:1509–1515. doi: 10.1021/bi00355a006. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., van Belkum, A., Pleij, C.W., Altman, S. Novel reactions of RNAase P with a tRNA-like structure in turnip yellow mosaic virus RNA. Cell. 1988;53:267–272. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- Hellen, C.U., Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Jan, E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Jan, E., Sarnow, P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- Jan, E., Kinzy, T.G., Sarnow, P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev, A.V., Pace, N.R. Bacterial RNase P: A new view of an ancient enzyme. Nat. Rev. Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- Kikovska, E., Brannvall, M., Kufel, J., Kirsebom, L.A. Substrate discrimination in RNase P RNA-mediated cleavage: Importance of the structural environment of the RNase P cleavage site. Nucleic Acids Res. 2005;33:2012–2021. doi: 10.1093/nar/gki344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente, E., Ramos, R., Martinez-Salas, E. Long-range RNA–RNA interactions between distant regions of the hepatitis C virus internal ribosome entry site element. J. Gen. Virol. 2002;83:1113–1121. doi: 10.1099/0022-1317-83-5-1113. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Kindelberger, D.W., Lee, J.Y., McClennen, S., Chamberlain, J., Engelke, D.R. Nuclear pre-tRNA terminal structure and RNase P recognition. RNA. 1997;3:175–185. [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Altman, S. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc. Natl. Acad. Sci. 2003;100:13213–13218. doi: 10.1073/pnas.2235589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto, S., Martinez-Salas, E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto, S., Martinez-Salas, E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA. 2000;6:1380–1392. doi: 10.1017/s1355838200000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto, S., Lafuente, E., Martinez-Salas, E. IRES interaction with translation initiation factors: Functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA. 2001;7:1213–1226. doi: 10.1017/s1355838201010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz, N., Beck, E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 1991;65:6486–6494. doi: 10.1128/jvi.65.12.6486-6494.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, A.J., Robertson, H.D. Detection of tRNA-like structure through RNase P cleavage of viral internal ribosome entry site RNAs near the AUG start triplet. J. Biol. Chem. 2003;278:26844–26850. doi: 10.1074/jbc.M304052200. [DOI] [PubMed] [Google Scholar]
- Mans, R.M., Guerrier-Takada, C., Altman, S., Pleij, C.W. Interaction of RNase P from Escherichia coli with pseudoknotted structures in viral RNAs. Nucleic Acids Res. 1990;18:3479–3487. doi: 10.1093/nar/18.12.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas, E., Fernandez-Miragall, O. Picornavirus IRES: Structure function relationship. Curr. Pharm. Des. 2004;10:3757–3767. doi: 10.2174/1381612043382657. [DOI] [PubMed] [Google Scholar]
- Martinez-Salas, E., Ramos, R., Lafuente, E., Lopez de Quinto, S. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 2001;82:973–984. doi: 10.1099/0022-1317-82-5-973. [DOI] [PubMed] [Google Scholar]
- Nadal, A., Martell, M., Lytle, J.R., Lyons, A.J., Robertson, H.D., Cabot, B., Esteban, J.I., Esteban, R., Guardia, J., Gomez, J. Specific cleavage of hepatitis C virus RNA genome by human RNase P. J. Biol. Chem. 2002;277:30606–30613. doi: 10.1074/jbc.M203595200. [DOI] [PubMed] [Google Scholar]
- Pascual, A., Vioque, A. Substrate binding and catalysis by ribonuclease P from cyanobacteria and Escherichia coli are affected differently by the 3′ terminal CCA in tRNA precursors. Proc. Natl. Acad. Sci. 1999;96:6672–6677. doi: 10.1073/pnas.96.12.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten, J.S., Costantino, D.A., Kieft, J.S. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Pestova, T.V., Kolupaeva, V.G., Khitrina, E.V., Poperechnaya, A.N., Agol, V.I., Hellen, C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes & Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- Piron, M., Beguiristain, N., Nadal, A., Martinez-Salas, E., Gomez, J. Characterizing the function and structural organization of the 5′ tRNA-like motif within the hepatitis C virus quasispecies. Nucleic Acids Res. 2005;33:1487–1502. doi: 10.1093/nar/gki290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, R., Martinez-Salas, E. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES) RNA. 1999;5:1374–1383. doi: 10.1017/s1355838299991240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, H.D., Altman, S., Smith, J.D. Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid presursor. J. Biol. Chem. 1972;247:5243–5251. [PubMed] [Google Scholar]
- Robertson, M.E., Seamons, R.A., Belsham, G.J. A selection system for functional internal ribosome entry site (IRES) elements: Analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabariegos, R., Nadal, A., Beguiristain, N., Piron, M., Gomez, J. Catalytic RNase P RNA from Synechocystis sp. cleaves the hepatitis C virus RNA near the AUG start codon. FEBS Lett. 2004;577:517–522. doi: 10.1016/j.febslet.2004.10.059. [DOI] [PubMed] [Google Scholar]
- Sarnow, P. Viral internal ribosome entry site elements: Novel ribosome–RNA complexes and roles in viral pathogenesis. J. Virol. 2003;77:2801–2806. doi: 10.1128/JVI.77.5.2801-2806.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, M., Connell, S.R., Lescoute, A., Giesebrecht, J., Dabrowski, M., Schroeer, B., Mielke, T., Penczek, P.A., Westhof, E., Spahn, C.M. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- Serrano, P., Pulido, M.R., Saiz, M., Martinez-Salas, E. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA–RNA interactions with the 5′ end region. J. Gen. Virol. 2006;87:3013–3022. doi: 10.1099/vir.0.82059-0. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Kieft, J.S., Grassucci, R.A., Penczek, P.A., Zhou, K., Doudna, J.A., Frank, J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Jan, E., Mulder, A., Grassucci, R.A., Sarnow, P., Frank, J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Stoneley, M., Willis, A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Vioque, A. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 1992;20:6331–6337. doi: 10.1093/nar/20.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., Pestova, T.V., Hellen, C.U., Sarnow, P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]