Abstract
Refractory to apoptosis induced by anticancer drugs is one of the major causes of drug resistance in human cancers. The involvement of noncoding RNA (ncRNA) in cancer cell drug resistance has not yet been reported. By using the technique of RT-PCR-based differential display, a novel gene, cancer up-regulated drug resistant (CUDR) gene, was found to be overexpressed in a doxorubicin-resistant subline of human squamous carcinoma A431 and A10A cells, which were also more resistant to drug-induced apoptosis. The full-length CUDR mRNA transcript is ∼2.2 kb as detected by Northern blot analysis and has no sequence homology with other genes identified so far. Interestingly, no distinct open reading frame was found throughout the CUDR cDNA sequence, and no recombinant protein was detected from in vitro translation or from a protein lysate of human cancer cells after CUDR transfection. Therefore, CUDR is likely to exert its function as a noncoding RNA. Stable transfection with the CUDR gene was found to induce resistance to doxorubicin and etoposide as well as drug-induced apoptosis in A431 cells. By Western blot analysis, down-regulations of caspase 3 were observed in CUDR transfectants. On the other hand, overexpression of CUDR promoted anchorage-independent growth in A431 cells. Results from the present study suggest that CUDR may likely regulate the drug sensitivity and promote cellular transformation at least through caspase 3-dependent apoptosis.
Keywords: ncRNA, CUDR, caspase 3, drug resistance, cellular transformation
INTRODUCTION
In the human genome ∼97%–98% of the transcriptional gene products do not code for proteins. These noncoding RNA (ncRNA) transcripts will function as riboregulators that likely exert their biological activities directly as RNAs (Eddy 2001; Erdmann et al. 2001). Many ncRNAs are processed (RNA splicing, polyadenylation, etc.) just like mRNAs, but they generally lack a substantial open reading frame (ORF) (Okazaki et al. 2002). In higher eukaryotes, ncRNAs participate in many complicated genetic phenomena such as cosuppression, transcriptional and post-transcriptional gene silencing, RNA interference, etc. (Lindbo et al. 1993; Fire et al. 1998; Montgomery et al. 1998). However, an involvement of ncRNAs in drug resistance of cancer cells has not yet been reported.
Cancer is one of the leading causes of death globally. One of the major cancer treatment methods is chemotherapy. Acquired multidrug resistance frequently occurs in cancer cells after exposure to chemotherapeutic agents. The development of drug resistance subsequently may lead to decreased response to cancer treatments and poor prognosis. In addition to overexpression of P-glycoprotein, MRP1, and down-regulation of topoisomerase II alpha, there are many additional ways for cancer cells to develop resistance to chemotherapeutic drugs, one of them being resistance to drug-induced apoptosis (Cole and Deeley 1995; Lage et al. 2000; Devarajan et al. 2002; Fojo and Bates 2003; Yang et al. 2005; Szakacs et al. 2006). Alteration of the expression of molecules involved in the apoptotic pathway may increase the resistance of cells to the drugs and thus affect the outcome of cancer therapy (Yang et al. 2001, 2005; Devarajan et al. 2002). In addition to therapeutic response, the process of apoptosis is also believed to have an important role in cancer development. The escape of cells from apoptotic cell death is one of the factors that favor the growth of the cancer cells. Correction between the expressions of genes in the apoptotic pathway with the incidence of cancer has also been suggested (Winter et al. 2001; Fong et al. 2006). There are two major pathways in apoptosis, intrinsic and extrinsic. Although the two pathways can be activated by different stimuli, both will activate caspase 3, the executor caspase, which subsequently leads to DNA fragmentation. Because of its role in apoptosis, the importance of caspase 3 in therapeutic resistance and cancer development has been suggested in many studies (Winter et al. 2001; Yang et al. 2001, 2005; Devarajan et al. 2002; Fong et al. 2006).
The technique of RT-PCR-based differential display was employed to explore the expressions of genes that may regulate the drug sensitivity of cells. By this technique, a novel gene was identified as overexpressed in the doxorubicin-resistant subline of human squamous carcinoma A431 cells. The gene was found to have no sequence homology with genes of identified biological functions, and it is also likely to function as ncRNA. This gene is named cancer up-regulated drug resistant (CUDR) gene because in the subsequent characterization the gene was confirmed to induce drug resistance and also to be up-regulated in human cancer tissues. In the present study, the function of CUDR in drug resistance and cellular transformation is suggested at least to be related to its effect on suppression of apoptosis through down-regulation of caspase 3. CUDR is believed not only to provide new insight concerning the function of ncRNA, but also to serve as a potential target of anticancer drug development.
RESULTS
Overexpression of CUDR in doxorubicin-resistant A10A cells
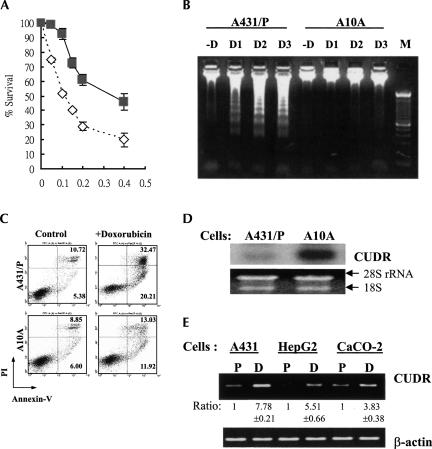
As shown in Figure 1A, A10A cells derived from human squamous carcinoma A431 cells are more resistant to doxorubicin than the parent cells. The IC50 for A431 parent cells and A10A cells are 0.1 μg/mL and 0.3 μg/mL, respectively. Moreover, A10A cells are also more resistant to doxorubicin-induced apoptotic cell death than the A431 parent cells. The extent of DNA fragmentation was more obvious in the A431 parent cells than in the A10A cells upon 48 h of doxorubicin treatment (Fig. 1B). In the case of phosphatidylserine externalization, another hallmark of apoptosis, the resistance to doxorubicin-induced apoptosis in A10A cells was also demonstrated by staining cells with annexin V/PI followed by flow cytometric analysis. As shown in Figure 1C, the percentage of apoptotic cells was significantly greater in A431 parent cells than in A10A cells after doxorubicin treatment.
FIGURE 1.
The expression of CUDR, doxorubicin sensitivity, and doxorubicin-induced cell death in A431 cells. (A) The sensitivity to doxorubicin of A431 parent cells and A10A cells as detected by MTT assay. Cells were exposed to different concentrations of doxorubicin for 48 h. A431 parent cells (open diamond); A10A cells (gray square). Results were the mean value from four independent experiments, and error bars show 95% confidence intervals. (B) Doxorubicin-induced DNA fragmentation in A431 cells. The cells were exposed to 0.025 μg/mL (D1), 0.05 μg/mL (D2), or 0.1 μg/mL (D3) doxorubicin for 48 h. Forty micrograms of genomic DNA were resolved in a 1.5% agarose gel. (M) 100-bp DNA marker. (C) Flow cytometric analysis of cells staining with annexin V/PI. The cells were treated with 0.05 μg/mL doxorubicin for 48 h and then analyzed for early apoptotic cells (bottom right quadrant) and late apoptotic or dead cells (top right quadrant); the percentages of cells in the two quadrants are shown. (D) Northern blot analysis of CUDR in A431 parent and A10A cells. The 18S and 28S rRNA demonstrated equal loading of total RNA in each lane. (E) CUDR expression in the parent (P) and the doxorubicin-selected subline (D) of A431, HepG2, and CaCO-2 cells by semiquantitative RT-PCR. The signal of each band was quantitated by Kodak Digital Science 1D image analysis software version 3.0 (Eastman Kodak). The relative expression of each band was calculated with respect to the expression of the parent cells (designated as 1). Data shown are the mean value of three independent experiments. The CUDR expression in the drug-resistant cells is significantly different from that of the respective parent cells (P < 0.05).
The observed drug resistance in A10A cells is unlikely related to the expression of the classical drug-resistance proteins, such as P-glycoprotein, MRP1, and topoisomerase II, as their expressions were similar between the two cell lines (data not shown). To find the underlying mechanism for the drug resistance in A10A cells, the technique of RT-PCR-based differential display was performed to compare the gene expression profile between the A431 parent cells and A10A cells. One of the clones, designated as CUDR, was found to be 12-fold (by densitometry analysis) overexpressed in A10A cells as confirmed by Northern blot analysis; a single mRNA transcript ∼2.2 kb was detected (Fig. 1D). By using semiquantitative RT-PCR, in addition to A431 cells, overexpression of CUDR was also observed in drug-resistant HepG2 cells and CaCO-2 cells, with respect to their parent counterpart (Fig. 1E).
DNA sequence analysis of CUDR
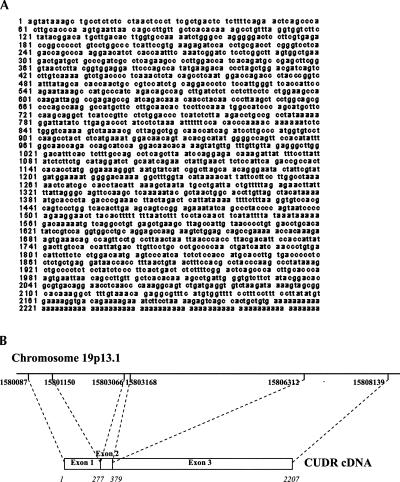
The full-length CUDR cDNA was isolated by screening with the human placenta arrayed cDNA library (Origene Technologies). Six clones were picked for DNA sequencing, and three of them had the longest DNA insert. The 5′ end was confirmed by RACE. The full-length CUDR gene sequence is shown in Figure 2A. The identity of the CUDR gene was revealed by mapping with the BLAST search program from NCBI. The gene structure of CUDR is shown in Figure 2B. The CUDR cDNA was found to have 99% identity with the human cDNA FLJ35802 fis clone (GenBank accession no. AK092401.1), and part of the DNA sequence of the chromosome 19 cosmid (GenBank accession no. AC004510), respectively. Moreover, the 5′ region of the CUDR cDNA (nucleotides 3–1294) had 99% identity with human urothelial cancer associated 1 (UCA1) mRNA (nucleotides 119–1409), which is 1.4 kb long (GenBank accession no. DQ3433132.1; Wang et al. 2006). Though the CUDR mRNA transcript was 2.2 kb, no distinct ORF was deduced throughout the cDNA sequence. Instead, a total of five putative small ORFs were predicted at the three reading frames. The full-length CUDR cDNA was subcloned into the pEXP2-DEST vector to perform in vitro protein translation. However, no protein was detected as compared with the protein detected from the β-galactosidase positive control (data not shown). On the other hand, to find out whether the predicted short ORFs were capable of translating into corresponding short peptides in human cells, all five putative ORF sequences as well as the full-length CUDR cDNA were amplified and subcloned separately into the mammalian expression vector pcDNA-DEST40 with a C-terminal 6×His V5 epitope. The constructs were then transfected into A431 cells and the total protein lysates were extracted. However, by Western blot analysis using anti-V5 antibody, the recombinant proteins from the CUDR cDNA and also the five ORF sequences were not detected in the cell lysates from any of these clones (data not shown). The results suggested that CUDR cDNA may not likely encode for any protein product.
FIGURE 2.
Sequence analysis of CUDR cDNA. The CUDR gene was mapped to human chromosome 19p13.1 and transcribed into mRNA transcript with three exons.
CUDR expression-induced resistance to apoptotic cell death
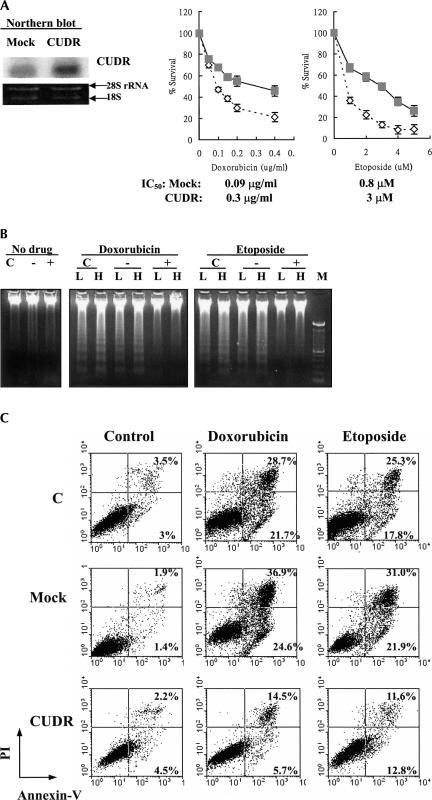
Overexpression of CUDR in the drug-resistant cells indicated that the gene may be important for drug resistance in cells. The underlying mechanism for CUDR-induced drug resistance was investigated by subcloning the full-length CUDR cDNA into the mammalian expression vector pcDNA3 for the establishment of pcDNA/CUDR stable transfectants. Compared with the mock transfectants (pcDNA empty vector transfection), stable CUDR transfectants were more resistant to cytotoxic drugs (doxorubicin and etoposide) as measured by an MTT assay (Fig. 3A). Cross-resistance of CUDR transfectants to other common anticancer drugs such as Taxol, vincristine, and cisplatin was also observed (data not shown). These results support the possibility that CUDR up-regulation plays an important role in inducing drug resistance in cancer cells.
FIGURE 3.
CUDR overexpression-induced resistance to apoptotic cell death in cells upon drug treatment. (A) Stable expression of CUDR in A431 cells. The CUDR expression in mock and CUDR/pcDNA transfectants was assessed by Northern blot analysis. Sensitivity of A431 stable transfectants to doxorubicin and etoposide as detected by MTT assay. Cells were exposed to various concentrations of drugs for 48 h. Mock (empty vector) transfection (open diamond); CUDR/pcDNA transfection (gray square). Results show the mean value from four independent experiments, and error bars show 95% confidence intervals. (B) Drug-induced apoptosis in A431 cells as detected by a DNA fragmentation assay. The parent (C), mock (−), and CUDR (+) transfectants were exposed to doxorubicin ([L] 0.025 μg/mL, [H] 0.05 μg/mL) or etoposide ([L] 1 μM, [H] 2.5 μM) for 48 h. (M) 100-bp DNA marker. (C) Doxorubicin- and etoposide-induced apoptotsis in A431 cells assessed by annexin V-GFP/propidium iodide staining followed by flow cytometric analysis. Cells were stained with annexin V-GFP/PI after drug treatments for 48 h, and then analyzed for early apoptotic cells (bottom right quadrant) and late apoptotic or dead cells (top right quadrant). The percentages of cells in the two quadrants are shown.
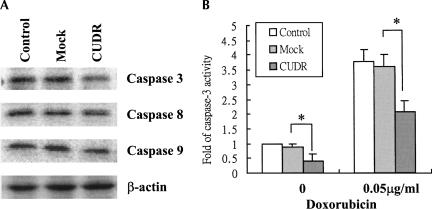
Refractory to apoptosis induced by anticancer drugs is one of the major causes for therapeutic resistance in cancers. To investigate if CUDR-induced drug resistance is mediated through the process of apoptosis, DNA fragmentation and phosphatidylserine externalization were assessed and compared between A431 mock transfectants and CUDR transfectants after drug treatment for 48 h. The CUDR-transfected A431 cells were found to be resistant to apoptotic cell death induced by doxorubicin and etoposide, as the DNA laddering was barely detectable (Fig. 3B). In addition, by flow cytometric analysis of cells staining with annexin V/PI, fewer apoptotic cells were detected in CUDR transfectants than in the mock and control cells after doxorubicin or etoposide treatment (Fig. 3C). The process of apoptosis may involve the activation of caspase 8 or 9, which subsequently will lead to direct proteolytic activation of the effector caspase, caspase 3, to induce apoptotic cell death. Compared with the mock transfectants, significant down-regulation of caspase 3 (∼70% by densitometry analysis) was observed in CUDR transfectants, while there were no or slight changes only in the levels of caspases 8 and 9 (Fig. 4A). In terms of caspase 3 enzymatic activity, the basal caspase 3 activity was about twofold higher in the mock transfectants than in the CUDR transfectants. After 48 h of doxorubicin treatment, an approximately fourfold increase in caspase 3 activity was observed in both the CUDR transfectants and the mock transfectants (Fig. 4B). By taking into account the differential in basal caspase 3 level, the specific activity of caspase 3 upon doxorubicin treatment was similar between the two groups of transfectants. Results support an idea that the protective effect of CUDR on doxorubicin-induced apoptosis in A431 cells may be related to its effect on the down-regulation of caspase 3 rather than on the process of drug-induced caspase 3 activation.
FIGURE 4.
(A) The basal protein levels of caspase 3, caspase 8, and caspase 9 in A431 cells. Twenty-five micrograms of total protein lysate were resolved in 15% SDS-PAGE and immunoblotted with the corresponding antibody by Western blot analysis. Experiments have been repeated at least three times, and the representative one is shown. (B) Effect of doxorubicin on caspase 3 activity in A431 cells. After treatment with 0.05 μg/mL doxorubicin for 48 h, the cells were trypsinized and lysed. Protein lysate was then subjected to a caspase 3 activity assay. Results show the mean value from four independent experiments, and error bars show 95% confidence intervals. (*) P < 0.05.
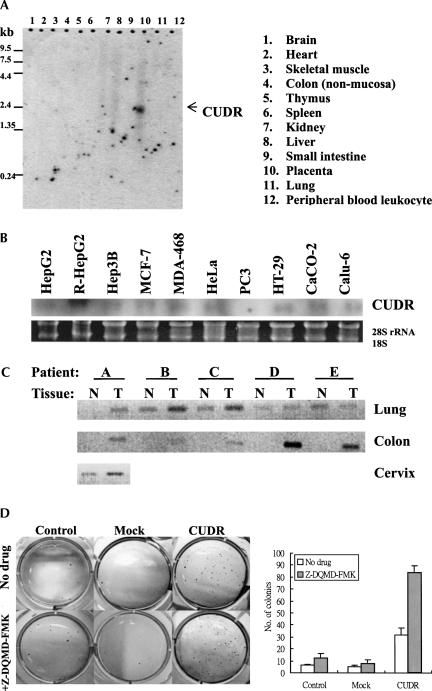
Differential expression of CUDR in human normal and cancer tissues
In the process of characterization of CUDR, its expression level in various human cancer cell lines and in normal tissues was examined. Results from Northern blot analysis indicated that CUDR was detectable only in human placenta but not in other normal tissues (Fig. 5A). For expression in human cancer cells, CUDR was detected in most cells of different tissue origins. CUDR mRNA was detected in human hepatocellular carcinoma HepG2 and Hep3B cells, human breast carcinoma MCF-7 and MDA-468 cells, human squamous carcinoma HeLa cells, human colon carcinoma HT-29 and CaCO-2 cells, as well as human lung carcinoma Calu-6 cells. However, CUDR was barely detectable in human prostate carcinoma PC3 cells (Fig. 5B). The differential expression of CUDR between human normal and cancer tissues is further demonstrated in tissues from cancer patients. Using PCR to compare the CUDR level between matched normal and tumor cDNA samples from the patient, it was indicated that CUDR was up-regulated in various tumor tissues: colon (five out of five), cervix (one out of one), and lung (four out of five), in comparison with respective normal tissue counterparts (Fig. 5C). The differential in CUDR expression between human normal tissues and human cancer tissues suggested that CUDR, in addition to its role in drug resistance, also may be important in carcinogenesis. To investigate whether CUDR may play some role in tumor formation, the anchorage-independent growth of A431 transfectants was examined by a soft agar colony formation assay. As shown in Figure 5D, the plating efficiency for CUDR transfectants was significantly higher than that for the mock transfectants and also for cells with no transfection. The results suggest that CUDR might likely promote anchorage-independent cell growth, a feature of malignant transformation, in A431 cells. Since refractory to apoptosis is one of the many factors for promoting tumorigenesis, down-regulation of caspase 3 by CUDR not only may lead to resistance to drug-induced cell death, but also may favor the process of tumor formation. The presence of 2 μM of caspase 3-specific inhibitor Z-DQMD-FMK in a soft agar colony formation assay resulted in an ∼1.5-fold to twofold increase in the number of colonies of A431 cells. These results support the idea that CUDR may favor the anchorage-independent growth of cells, possibly through down-regulation of caspase 3 expression.
FIGURE 5.
CUDR expression in human normal tissues and cancer tissues. (A) CUDR mRNA expression in different human normal tissues as detected by Northern blot analysis using the pre-made 12-lane human tissue Northern blot (Clontech). (B) CUDR expression in human cancer cell lines as detected by Northern blot analysis. (C) CUDR expression in cDNA pairs of matched normal (N) and tumor (T) tissues (Clontech) as detected by PCR. PCR was performed with a pair of CUDR-specific primers and then resolved in a 1.2% agarose gel. (A–E) Different patients designed by the manufacturer. The level of β-actin in each sample was also measured and did not demonstrate deviation among the samples (data not shown). (D) The effect of CUDR transfection on anchorage-independent cell growth of A431 cells with or without 2 μM caspase 3 inhibitor Z-DQMD-FMK as examined by soft agar colony formation assay. The number of colonies of each cell line is shown. Results show the mean value from three independent experiments, and error bars show 95% confidence intervals.
DISCUSSION
CUDR is believed to exert its function as an ncRNA in the regulation of drug resistance and cellular transformation. In addition to coding for protein products, much RNA may not be translatable and is considered ncRNA. Some of the ncRNA is found to have biological functions in cancer development, fetal development, cell growth, etc. However, the role of ncRNA in drug resistance has not yet been reported (Erdmann et al. 2001). At the time of annotation of any transcript in their database, the RIKEN standard in Tokyo classifies the transcript as ncRNA if the sequence of the putative ORF of the transcript is <300 base pairs (bp) (Okazaki et al. 2002). There are five putative ORFs in the 2.2-kb CUDR transcript, and their sequences are all <300 bp. The proteins supposedly encoded by these ORF sequences are not detected in cells after transfection with their sequences. Furthermore, no protein was detected by in vitro translation of CUDR cDNA. In Northern blot analysis, using any one of these five ORF sequences as probes, only a single transcript of 2.2-kb mRNA was detected. Transfection of cells with any one of these five ORF sequences did not alter the doxorubicin sensitivity of cells as demonstrated with full-length CUDR cDNA (data not shown). The absence of the distinct ORF and protein product prompts us to speculate that CUDR might function as ncRNA, and similar criteria also have been applied in the characterization of other known ncRNAs, e.g., H19 (Hurst and Smith 1999). By genomic analysis, CUDR has no sequence homology with any gene of known biological function in the Human Genome Database as well as miRNA currently deposited in the miRBase at http://microrna.sanger.ac.uk. The way that ncRNA regulates the expression of its effector genes is so far not clear. The RNA might interact with its binding proteins and then through some still unidentified pathways regulate the expression of effector genes, e.g., promoter methylation (Miyashiro et al. 2003; Vernucci et al. 2004). This kind of approach should be considered when examining the interaction between CUDR and caspase 3.
CUDR induces drug resistance in cells through suppression of drug-induced apoptosis. The functional role of CUDR in drug resistance is indicated by overexpression of the gene in several drug-resistant cells, e.g., A431 cells (Fig. 1C), HepG2 cells, and CaCO-2 cells (Fig. 1D). CUDR overexpression decreased both drug sensitivity and also drug-induced apoptosis of cells (Fig. 3). The regulatory role of CUDR in drug resistance is believed to be associated with the suppression effect of the gene on drug-induced apoptosis, probably through down-regulation of caspase 3. The role of caspase 3 in apoptosis is well documented, and its role in drug resistance is also confirmed in various cell systems (Yang et al. 2001, 2005; Devarajan et al. 2002). Preliminary results indicated that it is likely CUDR will not affect the activation of the enzyme, as the specific activity of caspase 3 appeared not to be altered by the level of CUDR (Fig. 4).
The involvement of CUDR in carcinogenesis may be associated with its effect on caspase 3 expression. Defects in apoptosis are believed to be one of the possible causes of cancer formation. Therefore, this may explain why some of the genes in the apoptotic pathway have been shown to be associated with the development of cancer (Winter et al. 2001; Jaattela 2004; Fong et al. 2006). An increase in anchorage-independent growth by CUDR expression and the further enhancement by the caspase 3 inhibitor in A431 cells (Fig. 5D) indicated the possible role of CUDR in cancer development, and such an effect may be related to caspase 3 down-regulation. The exact role of CUDR in the process of cancer development is not clear, and whether or not it will regulate genes in addition to caspase 3 will require detailed investigation.
In the present study, the novel ncRNA, CUDR, is suggested to have important functions in drug resistance and cellular transformation, presumably through down-regulation of caspase 3. The gene might likely be a good potential target for drug development, as the drug identified can likely be applied as an anticancer drug per se and also as a modifier of drug resistance. Furthermore, as CUDR is rarely detectable in human normal tissues and will therefore likely have a low baseline level as compared with other current cancer biomarkers (e.g., CEA), the gene might therefore also be a promising biomarker for cancer progression and cancer therapeutic responses. In any case, extensive investigation is definitely needed to support the future use of CUDR in cancer therapy and cancer diagnosis.
MATERIALS AND METHODS
Cell lines and drugs
Human squamous carcinoma A431 cells were purchased from American Type Culture Collection. The doxorubicin-resistant subline, A10A, was established by sequential exposure of A431 cells to doxorubicin (Kwok and Sutherland 1991). The doxorubicin-resistant sublines of human hepatocellular carcinoma HepG2 cells and human colon carcinoma CaCO-2 cells were gifts from Professor K.P. Fung (Department of Biochemistry, The Chinese University of Hong Kong). All cells were cultured in a 37°C humidified incubator with 10% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% (HepG2 cells) or 10% (A431 and CaCO-2 cells) fetal bovine serum and 2 mM L-glutamine (Invitrogen). Doxorubicin and etoposide were purchased from Sigma. Caspase 3 inhibitor Z-DQMD-FMK was purchased from Calbiochem.
MTT cell viability assay
Eight thousand cells per well were seeded in a 96-well plate for 24 h and then exposed to various concentrations of drugs for 48 h. Cells were incubated in 50 μL of a 0.1 mg/mL solution of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) for 3 h at 37°C and lysed in 150 μL of dimethyl sulfoxide (DMSO) for 30 min at room temperature. The absorbance in each well was measured at 580 nm by a microplate reader.
RT-PCR-based differential display
The RNAimage Kit (GenHunter Corporation) was used. Briefly, 0.1 μg/μL DNA-free RNA was reverse transcribed with MMLV reverse transcriptase, 20 μM dNTPs, and one-base anchored oligo-dT primer (5′-AAGCTTTTTTTTTTTG-3′), followed by a PCR reaction with an arbitrary primer H-AP6 (5′-AAGCTTGCACCA-3′), 2 μM dNTPs, and [α-35S]dATP for 40 cycles of the following parameters: 30 sec at 94°C, 2 min at 40°C, 30 sec at 72°C, followed by 5 min at 72°C. RT-PCR products were resolved in a 6% denaturing polyacrylamide gel, blotted onto 3M paper, and dried under vacuum. The differentially expressed cDNA pattern was visualized by autoradiography. The bands that were identified to be differentially expressed were excised from the gel and extracted for PCR reamplification. The amplified PCR products were then subjected to TA cloning (Invitrogen). Positive clones were purified with a cDNA plasmid mini-preparation system (Qiagen). DNA sequences were deduced by automatic DNA sequencing.
Northern blot analysis
Cells in the exponential phase were lysed with TRI reagent (Molecular Research Center). Total RNA was extracted by 1-bromo-3-chloropropane (BCP) reagent (Molecular Research Center), precipitated with isopropanol, and solubilized in RNase-free water. Five micrograms of total RNA were run in a 1% agarose/7 M formaldehyde denaturing gel and blotted onto a Hybond N membrane (Amersham Pharmacia Biotech). The cDNA plasmid probes were labeled with the Redprime II random labeling system (Amersham Pharmacia Biotech) by [α-32P]dCTP. Hybridization was performed for 2 h at 65°C. After stringency washes, the blot was exposed to X-ray film with an intensifying screen at −80°C. The signal of each band was scanned and quantitated by Kodak Digital science 1D image analysis software version 3.0 (Eastman Kodak).
Determination of the full-length CUDR gene
The full-length CUDR gene was isolated by screening the human placenta arrayed cDNA library (Origene Technologies) and then confirmed by rapid amplification of cDNA ends (RACE) using CapFishing Full-length cDNA (Seegene). A pair of primers specific for CUDR cDNA and isolated from differential display was designed to identify positive clones from the cDNA library by PCR (forward: 5′-AATGGACAACAGTACACGC-3′; reverse: 5′-AAGGTGCCAGTTAGCGTAT-3′). Positive clones were then digested with NotI, and the longest desired cDNA clone was identified. The full-length DNA sequence of both strands was sequenced using library vector primers. Gene-specific primers 5′-GGAGATAGGAGAGGGGCAGCTTTATA-3′ and 5′-CCGTATAGAAGACCACCTAAACAGGCT-3′ were used in 5′-RACE PCR reactions. RACE-ready cDNA was prepared using 1 μg of total RNA extracted from human squamous carcinoma A431 cells according to the manufacturer's instructions. PCR products were resolved in agarose gel.
Stable transfection of CUDR cDNA
The full-length CUDR plasmid isolated from the human placenta library was digested with EcoRI and NotI restriction enzymes and subcloned into the pcDNA3 mammalian expression vector (Stratagene). Thereafter, the pcDNA3/CUDR construct was transfected into A431 cells with lipofectamine 2000 for 24 h and selected with 800 μg/mL G418 for 3 wk. Transfection with pcDNA3 empty vector (mock) acted as a control. Colonies were then trypsinized, and the positive clone was identified by RT-PCR for CUDR expression.
Semiquantitative reverse transcription–polymerase chain reaction (RT-PCR)
The first cDNA was synthesized by 1.5 μg of total RNA in the presence of an oligo-dT12–18 primer (Invitrogen) and ImProm II reverse transcriptase according to manufacturer's instructions (Promega Corporation). Primer sequences for 25 cycles of PCR amplification were as follows: CUDR (forward: 5′-GCACCCTAGACCCGAAA-3′; reverse: 5′-GCCACCTGGACGGATAT-3′), β-actin (forward 5′-GCGGGAAATCGTGCGTGACATT-3′, reverse 5′-GATGGAGTTGAAGGTAGTTTCGTG-3′). Annealing temperature was at 54°C, and PCR products were resolved in a 1.5% agarose gel.
DNA fragmentation assay
After drug treatment, cells were trypsinized, washed once with PBS, and then lysed in lysis buffer (5 mM Tris-HCl, 100 mM EDTA, 1% SDS, 0.4 μg/mL proteinase K) for 1 h at 45°C. Genomic DNA was isolated by phenol/chloroform extraction and then precipitated with absolute ethanol. RNA was removed by incubation of the DNA pellet in Tris-EDTA buffer (pH 8.0) with 0.4 μg/mL RNase A overnight. Thirty micrograms of DNA were run in a 1.5% agarose gel.
Flow cytometric analysis of apoptotic cell death
After drug treatment, cells were trypsinized, washed twice with PBS, and resuspended in binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2). The cells were then stained with Annexin V-GFP/PI for 15 min in the dark at room temperature. Cell populations were analyzed by a FASCort flow cytometer. Ten thousand cells were counted for each sample.
Caspase 3 activity assay
Cells were treated with doxorubicin for 48 h, harvested by trypsinization and then washed with ice-cold PBS. Thereafter, the cell pellets were lysed in lysis buffer (100 mM NaCl, 5 μg/mL aprotinin, 10 μg/mL leupeptin, 0.1 mM EDTA, 50 mM HEPES, 0.1% CHAPS, 0.1 mM DTT, 100 μM PMSF). Total cell lysates were first equilibrated with assay reaction buffer (100 mM NaCl, 50 mM EDTA, 50 mM HEPES, 0.1% CHAPS, 10 mM DTT, 10% glycerol) for 10 min at 37°C. The catalytic activity of caspase 3 in the protein lysate was measured by proteolytic cleavage of 10 μM of substrate Ac-DEVD-pNA for up to 3 h. The release of pNA, known as p-nitroaniline, was measured by a spectrophotometer at 405 nM, with no-lysate control as a blank.
Western blot analysis
Cells were lysed in Lamellae's lysis buffer containing 1% Triton X-100. Twenty-five micrograms of protein were resolved in a denaturing SDS-PAGE minigel and transferred to Immobilon-P nitrocellulose membrane (Millipore). Membranes were incubated with primary antibodies for caspases 3, 8, and 9 (Santa Cruz Biotechnology) for 2 h at room temperature, washed extensively with 0.1% Tween-20 in PBS, and then incubated with respective secondary antibody conjugated with horseradish peroxidase at 1:10,000 dilution. The signals were visualized with enhanced chemiluminescence (Amersham Life Science Inc.).
Soft agar colony formation assay
Soft agar plates were prepared in six-well plates with a bottom layer of 0.6% Noble agar in serum-free DMEM. One thousand cells in 0.3% Noble agar in DMEM supplemented with 10% fetal calf serum were seeded onto the bottom layer. Plates were then incubated for 3 wk in a 37°C incubator. The number of colonies was counted after staining with 0.05% crystal violet for 1 h.
ACKNOWLEDGMENTS
We thank Professor G.S. Guldan (Department of Biochemistry, The Chinese University of Hong Kong) for proofreading the manuscript. The present study was supported by an Earmarked Grant from the Hong Kong Research Grants Council (CUHK4024/01M) and a Direct Grant from the Chinese University of Hong Kong.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.359007.
REFERENCES
- Cole, S.P., Deeley, R.G. Multidrug resistance mediated by the ATP-binding cassette transporter protein MRP. Bioessays. 1995;20:931–940. doi: 10.1002/(SICI)1521-1878(199811)20:11<931::AID-BIES8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Devarajan, E., Sahin, A.A., Chen, J.S., Krishnamurthy, R.R., Aggarwal, N., Brun, A.M., Sapino, A., Zhang, F., Sharma, D., Yang, X.H., et al. Down-regulation of caspase 3 in breast cancer: A possible mechanism for chemoresistance. Oncogene. 2002;21:8843–8851. doi: 10.1038/sj.onc.1206044. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. Noncoding RNA genes and the modern RNA world. For. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Erdmann, V.A., Barciszewska, M.Z., Hochberg, A., de Groot, N., Barciszewski, J. Regulatory RNAs. Cell. Mol. Life Sci. 2001;58:960–977. doi: 10.1007/PL00000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montomery, M.K., Kostas, S.A., Driver, S.E., Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fojo, T., Bates, S. Strategies for reversing drug resistance. Oncogene. 2003;22:7512–7523. doi: 10.1038/sj.onc.1206951. [DOI] [PubMed] [Google Scholar]
- Fong, P.Y., Xue, W.C., Ngan, H.Y., Chiu, P.M., Chan, K.Y., Tsao, S.W., Cheung, A.N. Caspase activity is down-regulated in choriocarcinoma: A cDNA array differential expression study. J. Clin. Pathol. 2006;59:179–183. doi: 10.1136/jcp.2005.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, L.D., Smith, N.G.C. Molecular evolutionary evidence that H19 mRNA is functional. Trends Genet. 1999;15:134–135. doi: 10.1016/s0168-9525(99)01696-0. [DOI] [PubMed] [Google Scholar]
- Jaattela, M. Multiple cell death pathways as regulators of tumour initiation and progression. Oncogene. 2004;23:2746–2756. doi: 10.1038/sj.onc.1207513. [DOI] [PubMed] [Google Scholar]
- Kwok, T.T., Sutherland, R.M. Epidermal growth factor reduces resistance to doxorubicin. Int. J. Cancer. 1991;49:73–76. doi: 10.1002/ijc.2910490114. [DOI] [PubMed] [Google Scholar]
- Lage, H., Helmbach, H., Dietel, M., Schadendort, D. Modulation of DNA topoisomerase II activity and expression in melanoma cells with acquired drug resistance. Br. J. Cancer. 2000;82:488–491. doi: 10.1054/bjoc.1999.0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo, J.A., Silva-Rosales, L., Proebsting, W.M., Dougherty, W.G. Induction of a highly specific antiviral state in transgenic plants: Implications for regulation of gene expression and virus resistance. Plant Cell. 1993;5:1749–1759. doi: 10.1105/tpc.5.12.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro, K.Y., Beckel-Mitchener, A., Purk, T.P., Becker, K.G., Barret, T., Liu, L., Carbonetto, S., Weiler, I.J., Greenough, W.T., Eberwine, J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Montgomery, M.K., Xu, S., Fire, A. RNA as a target of double-stranded RNA-mediated genetic interference in caenorhabditis elegans. Proc. Natl. Acad. Sci. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Szakacs, G., Paterson, J.K., Ludwig, J.A., Booth-Genthe, C., Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- Vernucci, M., Cerrato, F., Pedone, P.V., Dandolo, L., Bruni, C.B., Riccio, A. The 5′ end of the KCNQ1OT1 gene is hypomethylated in the Beckwith–Wiedemann syndrome. Hum. Mol. Genet. 2004;13:353–361. [Google Scholar]
- Wang, X.S., Zhang, Z., Wang, H.C., Cai, J.L., Xu, Q.W., Li, M.Q., Chen, Y.C., Qian, X.P., Lu, T.J., Yu, L.Z., et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin. Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- Winter, R.N., Kramer, A., Borkowski, A., Kyprianou, N. Loss of caspase-1 and caspase-3 protein expression in human prostate cancer. Cancer Res. 2001;16:1227–1232. [PubMed] [Google Scholar]
- Yang, X.H., Sladek, T.L., Liu, X., Butler, B.R., Froelich, C.J., Thor, A.D. Reconstitution of caspase 3 sensitizes MCF-7 breast cancer cells to doxorubicin- and etoposide-induced apoptosis. Cancer Res. 2001;61:348–354. [PubMed] [Google Scholar]
- Yang, X.H., Edgerton, S., Thor, A.D. Reconstitution of caspase-3 sensitizes MCF-7 breast cancer cells to radiation therapy. Int. J. Oncol. 2005;26:1675–1680. [PubMed] [Google Scholar]