Abstract
The discovery and characterization of microRNAs (miRNAs) and other families of short RNAs has led to a rapid expansion of research directed at elucidating their expression patterns and regulatory functions. Here, we describe a convenient, sensitive, and straightforward method to detect and quantitate specific miRNA levels in unfractionated total RNA samples. The method, based on splinted ligation, does not require specialized equipment or any amplification step, and is significantly faster and more sensitive than Northern blotting. We demonstrate that the method can be used to detect various classes of small regulatory RNAs from different organisms.
Keywords: miRNA, microRNA, small RNA, splinted ligation, Northern blot
INTRODUCTION
MicroRNAs (miRNAs) are short (∼21–23 nucleotides [nt]) sequence-specific regulatory molecules found in all multicellular organisms. Over the last few years, several hundred conserved and nonconserved miRNAs have been identified and characterized in animals and plants using cloning and prediction strategies. Recent evidence indicates that they play important roles in a wide range of biological processes including developmental timing, differentiation, and growth control.
Once identified, the temporal and spatial regulation of miRNA expression is generally investigated to better understand their biological function in vivo. To this end, numerous techniques for detecting miRNAs and other small RNAs have been developed including a variety of microarray-based (e.g., Krichevsky et al. 2003; Barad et al. 2004; Calin et al. 2004; Liu et al. 2004; Nelson et al. 2004; Shingara et al. 2005) and PCR-based approaches (Schmittgen et al. 2004; Chen et al. 2005; Jiang et al. 2005), rolling circle amplification (Jonstrup et al. 2006), an Invader assay (Allawi et al. 2004), an ELISA-based assay (Mora and Getta 2006), bead-based assays (Lu et al. 2005), single molecule detection (Neely et al. 2006), and the use of signal amplifying ribozymes (Hartig et al. 2004). Many of these techniques require specialized and expensive equipment, and others involve multiple steps and an amplification reaction. To date, the most straightforward and widely used assay for small RNA detection has been traditional Northern blotting. Nevertheless, even with improvements in sensitivity provided by LNA substituted probes (Valoczi et al. 2004), Northern blotting requires relatively large amounts of starting material and involves multiple, time-consuming handling steps. Therefore, we sought to develop an assay for small RNA detection that retained the simplicity of Northern blotting but eliminated its disadvantages. We show that a splinted ligation strategy fulfills these criteria. The described assay takes advantage of liquid hybridization kinetics and circumvents the transfer, prehybridization, and washing steps required for Northern blotting. Comparison of the two techniques reveals that the splinted ligation assay is approximately 50 times more sensitive than Northern blotting using DNA probes.
RESULTS AND DISCUSSION
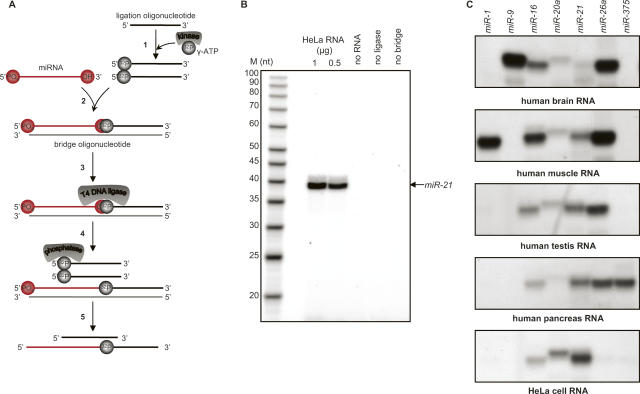
A schematic representation of the design of the splinted ligation strategy for miRNA detection is shown in Figure 1A. Briefly, an oligodeoxynucleotide (the bridge oligonucleotide) with perfect Watson–Crick complementarity to the target miRNA is designed. In addition to the region of complementarity to the target RNA, the oligonucleotide contains a 5′ extension of 14 nt of unrelated sequence; we arbitrarily chose an unstructured sequence derived from the coding sequence of GFP (see Materials and Methods). A second oligodeoxynucleotide complementary to the extension sequence (the ligation oligonucleotide) is synthesized and labeled at its 5′ end with 32P using polynucleotide kinase and [γ32P]-ATP. Concurrent annealing of the miRNA and ligation oligonucleotide to the bridge oligonucleotide yields a double-stranded structure with a nick on one strand. Because T4 DNA ligase can efficiently seal such nicks when the 3′ hydroxyl is presented as either RNA or DNA (Moore and Query 2000; Bullard and Bowater 2006), treatment with this enzyme creates a covalent phosphodiester linkage between the miRNA and the ligation oligonucleotide, thus labeling the target miRNA. Furthermore, because the labeled phosphate provided by the ligation oligonucleotide is rendered insensitive to phosphatase action, the label present on the unligated oligonucleotide can be removed by incubation with phosphatase after the ligation step. Following the splinted ligation reaction, labeled miRNA carrying a 14 nt extension and any residual labeled ligation oligonucleotide can then be fractionated by denaturing gel electrophoresis and visualized.
FIGURE 1.
Detection of miRNAs using splinted ligation. (A) Schematic depiction of the assay process. As described in the text, the assay involves: (1) Labeling of the ligation oligonucleotide; (2) concurrent annealing of the ligation oligonucleotide and miRNA to a bridge oligonucleotide; (3) linking of the ligation oligonucleotide to the miRNA by DNA ligase; (4) removal of labeled phosphate from unligated oligonucleotide; and (5) fractionation on a denaturing gel. (B) Assay reactions were performed with the indicated amounts of HeLa cell total RNA as described in Materials and Methods. Samples were separated on 12% urea polyacrylamide gels prior to autoradiography. Lanes designated “no ligase,” “no bridge,” and “no RNA” were complete reactions in which the specified components were replaced by water; 1 μg of total RNA was used in the no ligase, and no bridge reactions. Lane M, 5′-end-labeled oligodeoxynucleotides of the indicated sizes (USB). The arrow indicates the position of a synthetic miR21 RNA ligated and analyzed in parallel. (C) Detection of selected human miRNAs in different tissues by splinted ligation. Assays were carried out as described in Materials and Methods with the indicated miRNA-specific bridge oligodeoxynucleotides: a common labeled ligation oligonucleotide, and 500 ng of total RNA from the indicated tissues.
Preliminary experiments employing synthetic RNAs indicated that the labeling scheme outlined above was efficient and specific (data not shown; see Figs. 1, 2). As expected from previous studies (Moore and Query 2000; Bullard and Bowater 2006), the ligase only sealed nicks when the 3′ and 5′ ends were perfectly paired and juxtaposed on the bridge oligonucleotide. Occasionally, we observed anomalous products that apparently resulted from aberrant ligase activities since they did not depend upon added RNA. These products were almost completely eliminated by blocking the 5′ and 3′ ends of the bridge oligonucleotide and the 3′ end of the ligation oligonucleotide with unligateable groups such as three carbon spacers (see Materials and Methods). Accordingly, all assays used blocked oligonucleotides. We also found that the efficiency of detection was insensitive to a wide range of concentrations of bridge and ligation oligonucleotides as long as they were equimolar. In addition, detection was unchanged when amounts of DNA ligase or time of ligation were varied. Therefore, we routinely use a fixed amount of bridge and ligation oligonucleotides and a fixed time and amount of DNA ligase; “standard” conditions are given in Materials and Methods.
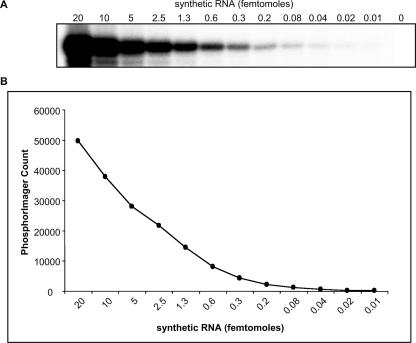
FIGURE 2.
Detection sensitivity of the splinted ligation assay. (A) Serial twofold dilutions as indicated of a synthetic 22 nt RNA corresponding to the mature hsa-miR-124a were assayed by splinted ligation in the presence of a miR-124a-specific bridge oligonucleotide as described in Materials and Methods. (B) The miR-124a signals were quantified with a PharosFX Plus PhosphorImager (BioRad).
Figure 1B shows a representative assay in which expression of hsa-miR-21 was monitored in unfractionated RNA extracted from HeLa cells. A labeled band of the expected size was observed only if all of the assay components were present. The absence of background bands suggested that the assay was specific. To explore the specificity of the method further, we designed bridge oligonucleotides complementary to a variety of miRNAs whose expression pattern has been established. We then monitored their expression in different human tissues (Fig. 1C). The results were in agreement with data obtained by other means (Lagos-Quintana et al. 2002; Poy et al. 2004; Sempere et al. 2004; Baskerville and Bartel 2005). Specifically, hsa-miR-1 is muscle specific, and it was found to be highly expressed in muscle and undetectable in brain, testis, pancreas, and HeLa cell RNA. We also detected pancreatic islet-specific hsa-miR-375 miRNA only in the pancreas. Similarly, hsa-miR-9 is known to be brain-specific and the ligation assay detected it only in brain RNA, while hsa-miR-16, hsa-miR-20a, and hsa-miR-21, known to be ubiquitously expressed, were detected in all samples analyzed. We conclude from these results that the splinted ligation assay has high specificity.
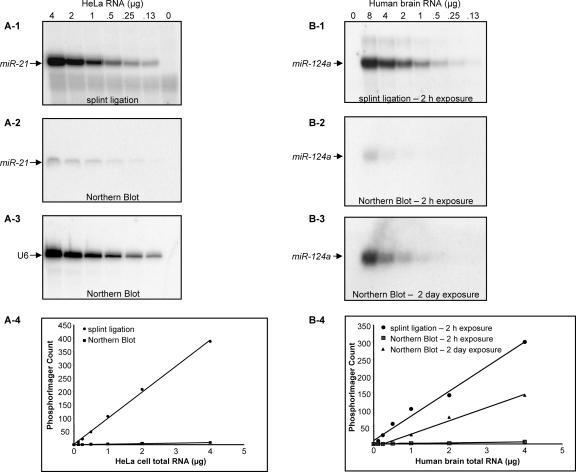
To examine the sensitivity of detection using the splinted ligation method, a synthetic RNA corresponding to mature hsa-miR-124a was synthesized and serial dilutions were analyzed (Fig. 2). As little as 0.3 fmol of the synthetic RNA could readily be detected under these idealized conditions and the signal increased linearly up to 20 fmol, the highest amount analyzed. These results indicate that the splinted ligation assay is both highly sensitive and quantitative. The detection limits of the splinted ligation approach suggested that it might be more sensitive than Northern blotting; therefore, it was of interest to compare the two procedures directly. As shown in Figure 3A, both techniques gave a clear signal for hsa-miR-21 in unfractionated HeLa cell RNA. However, splinted ligation was significantly more sensitive; quantitation of the signal by PhosphorImager revealed a consistent ∼50-fold enhancement in detection sensitivity over a 40-fold range in RNA concentration. A comparable increase in sensitivity was also observed when assessing hsa-miR124a expression in total brain RNA (Fig. 3B).
FIGURE 3.
Comparison of the detection sensitivity of Northern blots and splinted ligation. The indicated amounts of total HeLa cell RNA were assayed for the presence of hsa-miR-21 either by splinted ligation (A-1) or Northern blot (A-2) as described in Materials and Methods. In A-3, the Northern blot was overprobed with an oligonucleotide complementary to U6 snRNA. In A-4, signals from A-1 and A-2 were quantified with a Storm PhosphorImager (Amersham). In B, the indicated amounts of total RNA from human brain were assayed for the presence of hsa-miR-124a either by splinted ligation (B-1) or Northern blot (B-2,B-3) as described in Materials and Methods. (B-4) signals were quantitated as in A.
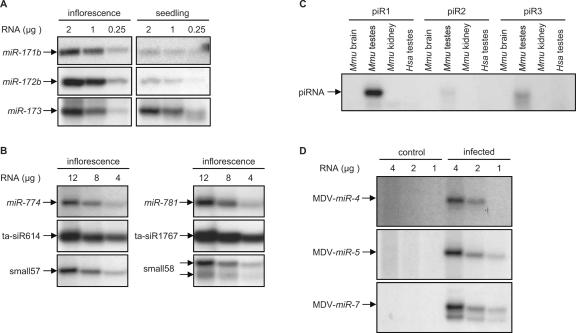
To this point, we have described the use of splinted ligation to detect animal miRNAs that are known to possess 5′-phosphate and 3′-hydroxyl termini. It was of interest to determine if we could monitor the expression of other small RNAs including plant miRNAs, which are known to be modified at their 3′ ends by 2′-O-methylation (Yu et al. 2005). Preliminary experiments using synthetic oligonucleotides with modified 3′ termini indicated that 2′-O-methylation did not interfere with splinted ligation (data not shown). Therefore, we used the technique to assay for expression of Arabidopsis miRNAs known to be methylated on the ribose of the last nucleotide (Yu et al. 2005). As shown in Figure 4A, AtmiR-171a, AtmiR-172b, and AtmiR-173 were readily detectable in total unfractionated Arabidopsis RNA derived from inflorescence and seedling tissues, and their expression patterns were consistent with previous observations (Reinhart et al. 2002; Aukerman and Sakai 2003). Additionally, Arabidopsis miRNAs, plant-specific trans-acting siRNAs (ta-siRNAs), and other small RNAs that are of relatively low abundance (Allen et al. 2005; Lu et al. 2006), were easily detected from a few micrograms of total RNA (Fig. 4B). Since detection of these RNAs by Northern analysis requires prior fractionation and enrichment for small RNAs from total RNA (Allen et al. 2005; Lu et al. 2006), we infer that splinted ligation is also more sensitive for the detection of small RNAs with modified 3′ ends compared to blotting.
FIGURE 4.
Detection of various classes of endogenous mammalian, plant, and viral small RNAs using splinted ligation. (A) The indicated amounts of total RNA prepared from the indicated Arabidopsis tissues were assayed for the presence of the indicated miRNAs by splinted ligation as described in Materials and Methods. (B) The indicated amounts of total Arabidopsis RNA were assayed for the presence of the indicated Arabidopsis small RNAs as described in Materials and Methods. (C) Four micrograms of total RNA from the indicated tissues were assayed for the presence of the indicated mouse piRNAs as described in Materials and Methods. (D) The indicated amounts of total RNA prepared from uninfected (control) or MDV-infected chicken embryo fibroblasts were assayed for the presence of the indicated MDV-miRNAs.
Finally, we used the ligation method to detect PIWI-associated small RNAs (piRNAs), a recently described class of germline-specific small RNAs (Giraud et al. 2006), and Marek's disease virus (MDV)-encoded miRNAs (Burnside et al. 2006). Although the exact status of the 3′ ends of piRNAs has not been established, analogous RNAs (rasiRNAs) in Drosophila are apparently modified (Vagin et al. 2006). Figure 4C shows that piRNAs are easily detected using the splinted-ligation strategy with as little as 4 μg of total RNA. Furthermore, this assay also validated the specific expression of MDV-miRNAs in infected chicken embryo fibroblasts (Fig. 4D). Together, these results further establish the general utility of the assay.
SUMMARY AND CONCLUSIONS
We have described a simple and straightforward assay for the detection of miRNAs and other small RNAs based on splinted ligation. The data support the conclusions that the assay is specific and quantitative. It does not require any specialized equipment and makes use of readily available enzymes. We have found it to be very useful for the analysis of multiple samples including gradient fractions (Maroney et al. 2006). Moreover, splinted ligation is considerably faster and more sensitive than Northern blotting while having the same applications such as expression analysis and validation of microarray studies.
MATERIALS AND METHODS
Oligonucleotide synthesis and preparation
Oligonucleotides were purchased from Integrated DNA Technologies. All oligonucleotides were purified by desalting or reverse-phase HPLC (Integrated DNA Technologies). The ligation oligonucleotide was 5′-end radiolabeled by incubating 20 pmol DNA, 2.5 μM [γ-32P]ATP (150 mCi/mL, Perkin-Elmer), 50 mM Tris-HCI (pH 7.5), 10 mM MgCl2, 5 mM dithiothreitol (DTT), 5% glycerol, and 20 units Optikinase (USB) in a reaction volume of 20 μL for 30 min at 37°C. The volume of labeled products was adjusted to 100 μL with RNase-free water and purified using a PrepEase Sequencing Dye Clean-Up Column (USB) according to the manufacturer's instructions. The volume of the eluate was adjusted to 200 μL with RNase-free water, extracted with phenol:chloroform:isoamyl alcohol, and ethanol precipitated with 20 μg glycogen. After precipitation, the pellet was resuspended to 100 nM with RNase-free water.
Bridge oligonucleotides were designed based on the miRNA sequences registered in the miRBase Sequence Database (http://microrna.sanger.ac.uk/sequences/). piRNA and ta-siRNA sequences used were published by Girard et al. (2006) and Allen et al. (2005), respectively. Oligonucleotides used in this study were as follows: hsa-miR-21 synthetic RNA, 5′-phos-uagcuuaucagacugauguuga-3′; hsa-miR-124 synthetic RNA, 5′-phos-uuaaggcacgcggugaaugcca-3′; hsa-miR-21 probe, 5′-GAATGTCATAAGCGTCAACATCAGTCTGATAAGCTA-3′; hsa-miR-124a probe, 5′-TGGCATTCACCGCGTGCCTTAA-3′; U6 probe (Maroney et al. 2000); and ligation oligonucleotide, 5′-CGCTTATGACATTC/dideoxyC/-3′. The following bridge oligonucleotides were synthesized with three carbon spacer groups (Integrated DNA technologies) at both the 5′ and 3′ ends:
hsa-miR-1, 5′-GAATGTCATAAGCGTACATACTTCTTTACATTCCA-3′;
hsa-miR-9, 5′-GAATGTCATAAGCGTCATACAGCTAGATAACCAAAGA-3′;
hsa-miR-16, 5′-GAATGTCATAAGCGCGCCAATATTTACGTGCTGCTA-3′;
hsa-miR-20a, 5′-GAATGTCATAAGCGCTACCTGCACTATAAGCACTTTA-3′;
hsa-miR-21, 5′-GAATGTCATAAGCGTCAACATCAGTCTGATAAGCTA-3′;
hsa-miR-26a 5′-GAATGTCATAAGCGGCCTATCCTGGATTACTTGAA-3′;
hsa-miR-124a, 5′-GAATGTCATAAGCGTGGCATTCACCGCGTGCCTTAA-3′;
hsa-miR-375, 5′-GAATGTCATAAGCGTCACGCGAGCCGAACGAACAAA-3′;
AtmiR-171b, 5′-GAATGTCATAAGCGCGTGATATTGGCACGGCTCAA-3′;
AtmiR-172b, 5′-GAATGTCATAAGCGATGCAGCATCATCAAGATTCT-3′;
AtmiR-173, 5′-GAATGTCATAAGCGGTGATTTCTCTCTGCAAGCGAA-3′;
AtmiR-774, 5′-GAATGTCATAAGCGGATGGCCATATGGGTAACCAA-3′;
AtmiR-781, 5′-GAATGTCATAAGCGTAAGTATCCAGAAAACTCTAA-3′;
ta-siR614, 5′-GAATGTCATAAGCGTGGAATACTTGAACTACCATC-3′;
ta-siR1767, 5′-GAATGTCATAAGCGTTCAAAATACACAAGTTCAAA-3′;
small 57, 5′-GAATGTCATAAGCGCATGTGCAAATGCTTCCCGCA-3′;
small 58, 5′-GAATGTCATAAGCGGTGAACTTTGATCTTGCGGTA-3′;
MDV-miR-4, 5′-GAATGTCATAAGCGGAAGGGTTCCGATACAGCATTAA-3′;
MDV-miR-5, 5′-GAATGTCATAAGCGACAGTAGACGACCACGATACACA-3′;
MDV-miR-7, 5′-GAATGTCATAAGCGCTGTAATCTCGTAGAGATCTCGA-3′;
piR1, 5′-GAATGTCATAAGCGAAACTATCTGAGCACCTGTGTTCATGTCA-3′;
piR2, 5′-GAATGTCATAAGCGACCAGCAGACACCGTCGTATGCATCACACA-3′;
piR3, 5′-GAATGTCATAAGCGACCACTAAACATTTAGATGCCACTCTCA-3′.
RNA sample preparation
HeLa cells were maintained in DMEM supplemented with 10% (v/v) FCS and harvested when ∼80% confluent. Cells were harvested and lysed with TRIzol (Invitrogen) according to the manufacturer's instructions. After precipitation with isopropanol, RNA was resuspended in buffer containing 1 mM EDTA, 0.3 M NaoAc, 0.2% SDS, and 20 mM Tris, pH 7.5 reextracted with phenol chloroform, and recovered by ethanol precipitation. Total RNA from human or mouse brain, kidney, muscle, pancreas, and testis was purchased from Ambion and OriGene. Total RNA from plant was prepared from 6-wk-old inflorescence tissue and 2-wk-old seedlings grown in vitro, using TRIzol reagent. Total RNA extracted from uninfected and MDV-infected chicken embryo fibroblast samples was kindly provided by Dr. Joan Burnside at the University of Delaware (Burnside et al. 2006).
RNA analysis
For ligation reactions, all reactions consist of 100 fmol bridge oligonucleotide, 100 fmol radiolabeled ligation oligonucleotide, total RNA sample, 75 mM KCl, 20 mM Tris (pH 8.0), 10 units T4 DNA ligase (USB), and 1× Ligate-IT buffer (USB) in a reaction volume of 15 μL. Before adding T4 DNA ligase and Ligate-IT buffer, the reaction mixture was denatured at 95°C for 1 min. After annealing at 65°C for 2 min and 37°C for 10 min, the ligase and the buffer were added to the reaction mixture and incubated at 30°C for 1 h. Reactions were terminated by heat inactivation at 75°C for 15 min and then treated with 1 unit of shrimp alkaline phosphatase (USB) at 37°C for 15 min. Reaction products were separated using denaturing 6% or 12% urea-polyacrylamide gels and quantified using a PhosphorImager.
For Northern blot analysis, total RNA samples were denatured, separated on denaturing 6% or 12% urea-polyacrylamide gels, transferred to either GeneScreen Plus membranes (Perkin-Elmer) or Zeta-Probe Blotting Membranes (BioRad), hybridized with a 5′-end radiolabeled deoxyoligonucleotide probe, prepared as described above in oligonucleotide synthesis and preparation, and quantified using a PhosphorImager.
ACKNOWLEDGMENTS
We thank Joan Burnside for the chicken embryo fibroblast samples, Jesse Fisher for technical assistance, and Orlando Gumbs and John Chase for helpful discussions. This work was supported by grants from NIH to T.W.N.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.518107.
REFERENCES
- Allawi, H.T., Dahlberg, J.E., Olson, S., Lund, E., Olson, M., Ma, W.P., Takova, T., Neri, B.P., Lyamichev, V.I. Quantitation of microRNAs using a modified Invader assay. RNA. 2004;10:1153–1161. doi: 10.1261/rna.5250604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J., Sakai, H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barad, O., Meiri, E., Avniel, A., Aharonov, R., Barzilai, A., Bentwich, I., Einav, U., Gilad, S., Hurban, P., Karov, Y., et al. MicroRNA expression detected by oligonucleotide microarrays. System establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville, S., Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard, D.R., Bowater, R.P. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem. J. 2006;398:135–144. doi: 10.1042/BJ20060313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside, J., Bernberg, E., Anderson, A., Lu, C., Meyers, B.C., Green, P.J., Jain, N., Isaacs, G., Morgan, R.W. Marek's disease virus encodes MicroRNAs that map to meq and the latency-associated transcript. J. Virol. 2006;80:8778–8786. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G.A., Liu, C.G., Sevignani, C., Ferracin, M., Felli, N., Dumitru, C.D., Shimizu, M., Cimmino, A., Zupo, S., Dono, M., et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Ridzon, D.A., Broomer, A.J., Zhou, Z., Lee, D.H., Nguyen, J.T., Barbisin, M., Xu, N.L., Mahuvakar, V.R., Andersen, M.R., et al. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, A., Sachidanandam, R., Hannon, G.J., Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Hartig, J.S., Grune, I., Najafi-Shoushtari, S.H., Famulok, M. Sequence-specific detection of MicroRNAs by signal-amplifying ribozymes. J. Am. Chem. Soc. 2004;126:722–723. doi: 10.1021/ja038822u. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Lee, E.J., Gusev, Y., Schmittgen, T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonstrup, S.P., Koch, J., Kjems, J. A microRNA detection system based on padlock probes and rolling circle amplification. RNA. 2006;12:1747–1752. doi: 10.1261/rna.110706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky, A.M., King, K.S., Donahue, C.P., Khrapko, K., Kosik, K.S. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Liu, C.G., Calin, G.A., Meloon, B., Gamliel, N., Sevignani, C., Ferracin, M., Dumitru, C.D., Shimizu, M., Zupo, S., Dono, M., et al. An oligonucleotide microchip for genome-wide miRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J., Getz, G., Miska, E.A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B.L., Mak, R.H., Ferrando, A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lu, C., Kulkarni, K., Souret, F.F., Muthu-Valliappan, R., Tej, S.S., Poethig, R.S., Henderson, I.R., Jacobsen, S.E., Wang, W., Green, P.J., et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney, P.A., Romfo, C.M., Nilsen, T.W. Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell. 2000;6:317–328. doi: 10.1016/s1097-2765(00)00032-0. [DOI] [PubMed] [Google Scholar]
- Maroney, P.A., Yu, Y., Fisher, J., Nilsen, T.W. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Moore, M.J., Query, C.C. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- Mora, J.R., Getta, R.C. Enzymatic microRNA detection in microtiter plates with DNA dendrimers. Biotechniques. 2006;41:420–424. doi: 10.2144/000112270. [DOI] [PubMed] [Google Scholar]
- Neely, L.A., Patel, S., Garver, J., Gallo, M., Hackett, M., McLaughlin, S., Nadel, M., Harris, J., Gullans, S., Rooke, J. A single-molecule method for the quantitation of microRNA gene expression. Nat. Methods. 2006;3:41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- Nelson, P.T., Baldwin, D.A., Scearce, L.M., Oberholtzer, J.C., Tobias, J.W., Mourelatos, Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- Poy, M.N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P.E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.R., Weinstein, E.G., Rhoades, M.W., Bartel, B., Bartel, D.P. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, T.D., Jiang, J., Liu, Q., Yang, L. A high-throughput method to monitor the expression of microRNA precursor. Nucleic Acids Res. 2004;32:43–53. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere, L.F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., Ambros, V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingara, J., Keiger, K., Shelton, J., Laosinchai-Wolf, W., Powers, P., Conrad, R., Brown, D., Labourier, E. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA. 2005;11:1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, V.V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Valoczi, A., Hornyik, C., Varga, N., Burgyan, J., Kauppinen, S., Havelda, Z. Sensitive and specific detection of microRNAs by Northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R.W., Steward, R., Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]