Abstract
During translation, some +1 frameshift mRNA sites are decoded by frameshift suppressor tRNAs that contain an extra base in their anticodon loops. Similarly engineered tRNAs have been used to insert nonnatural amino acids into proteins. Here, we report crystal structures of two anticodon stem–loops (ASLs) from tRNAs known to facilitate +1 frameshifting bound to the 30S ribosomal subunit with their cognate mRNAs. ASLCCCG and ASLACCC (5′–3′ nomenclature) form unpredicted anticodon–codon interactions where the anticodon base 34 at the wobble position contacts either the fourth codon base or the third and fourth codon bases. In addition, we report the structure of ASLACGA bound to the 30S ribosomal subunit with its cognate mRNA. The tRNA containing this ASL was previously shown to be unable to facilitate +1 frameshifting in competition with normal tRNAs (Hohsaka et al. 2001), and interestingly, it displays a normal anticodon–codon interaction. These structures show that the expanded anticodon loop of +1 frameshift promoting tRNAs are flexible enough to adopt conformations that allow three bases of the anticodon to span four bases of the mRNA. Therefore it appears that normal triplet pairing is not an absolute constraint of the decoding center.
Keywords: frameshift suppressor tRNAs, ribosome, +1 frameshift, processivity errors, anticodon stem–loop
INTRODUCTION
The ribosome is a complex macromolecular machine that synthesizes protein with high fidelity using an RNA template consisting of a triplet code (for review, see Ogle and Ramakrishnan 2005). The bacterial ribosome consists of two subunits, each having distinct roles in protein synthesis: the 30S subunit directly monitors triplet codon–anticodon base-pairing while the 50S subunit catalyzes the chemical reaction of peptidyl transfer.
The ribosome maintains the triplet reading frame as the tRNAs, along with the mRNA, move from the aminoacyl (A) to the peptidyl (P) site. A shift in the reading frame would usually result in a prematurely terminated protein product ending in a stretch of incorrect residues (Manley and Gesteland 1978). Such frameshift errors would be expected to be far more deleterious than substitutions of single amino acids and, not surprisingly, such errors occur at a lower rate (Kurland 1992). However, there are also examples of “programmed frameshifting” that are required for biological function such as feedback regulation or production of a specific ratio of two different proteins (Farabaugh 1996; Gesteland and Atkins 1996). Frameshifting could involve several processes: The incoming aminoacyl-tRNA could recognize a codon larger or smaller than the canonical three bases, or bind to an out-of-frame codon. Likewise, the translocational step size could differ from the normal 3 nucleotides (nt). Finally, a translocated peptidyl-tRNA could slip on the mRNA and base pair out of frame (Stahl et al. 2002).
So-called frameshift suppressor tRNAs decode sites requiring a frameshift to restore the reading frame for the remainder of the coding sequence (Riyasaty and Atkins 1968; Riddle and Carbon 1973). Some +1 frameshift suppressor tRNAs that decode a quadruplet codon contain an additional nucleotide 5′ of the anticodon between positions 33 and 34 (Fig. 1; hereafter the additional base is referred to as 33.5). Many of these suppressor tRNAs isolated in both bacteria (Riyasaty and Atkins 1968; Yourno 1972; Riddle and Carbon 1973; Prather et al. 1981; Bossi and Smith 1984; Magliery et al. 2001) and yeast (Cummins et al. 1982; Gaber and Culbertson 1982; Sroga et al. 1992) display Watson–Crick complementarity at all four anticodon positions to their quadruplet codon, suggesting that formation of 4 base pairs (bp) might cause +1 frameshifting (Roth 1981). However, +1 frameshifting still occurs, albeit at lower efficiencies, when either anticodon base 33.5 or the fourth base of the codon is singly mutated to form noncanonical base pairs, indicating that Watson–Crick pairing between these bases is not crucial (Gaber and Culbertson 1984; Curran and Yarus 1987; Weiss et al. 1990).
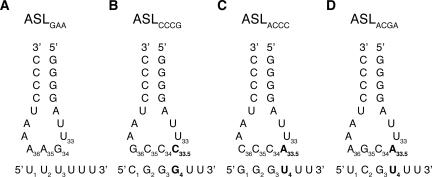
FIGURE 1.
Secondary structure of anticodon loops with corresponding cognate mRNAs that were complexed to the 30S A site. (A) Cognate anticodon–codon interaction between a normal ASL, ASLGAA, and mRNA UUU. (B–D) ASLs and mRNAs used in this study. These ASLs all contain eight rather than seven bases in their anticodon loop. The additional base is shown in bold.
Another explanation for how suppressor tRNAs with an extra base in the anticodon loop induce frameshifting is that they make a normal three-base codon–anticodon interaction in the A site but slip on the mRNA after translocation to the P site (Qian et al. 1998). The efficiency of suppression would then depend on the ability of the tRNA to slip during the translational pause while waiting for the next in-frame tRNA as well as on competition of tRNAs for the subsequent zero and +1 frame codons (Qian et al. 1998).
To expand the chemical group functionality of the genetic code, rare tRNAs modified to contain an extra nucleotide at base 33.5 can be charged with nonnatural amino acids. These designed tRNAs perform quadruplet decoding in vitro (Fig. 1B,C; Hohsaka et al. 1996; Murakami et al. 1998; Hohsaka et al. 2001) and multiple nonstandard amino acids can be incorporated into one protein. CGGG and GGGU were the most efficient codons (5′–3′ nomenclature for both anticodon and codon bases) with 76%–86% yield of nonnatural amino acid containing protein compared to wild-type protein (Hohsaka et al. 2001).
Recently, using a primer extension toeprint assay, one of these expanded tRNAs, tRNACCCG, as well as the corresponding anticodon stem–loop (ASL), was shown to cause four-base translocation of the mRNA when moving from the A to the P site (Phelps et al. 2006). Additionally, the tRNA, upon direct binding to the P site, positioned the mRNA in the +1 frame. These experiments demonstrated for the first time that this particular tRNA as well as the ASL contain all the essential elements for +1 frameshifting.
In order to understand the mechanism of four-base decoding, we solved crystal structures of the 30S subunit in complex with ASLCCCG, ASLACCC, (efficient at +1 frameshifting) (Hohsaka et al. 2001), and ASLACGA (no observable +1 frameshifting) (Fig. 1B–D; Hohsaka et al. 2001). These studies elucidate how such expanded ASLs are accommodated in the decoding center of the ribosome.
RESULTS AND DISCUSSION
The structures of the Thermus thermophilus 30S ribosomal subunit (30S) in complex with cognate and near-cognate ASL–mRNA pairs have elucidated molecular details of decoding (Ogle et al. 2001, 2002). In order to study the initial step of +1 frameshifting, we have solved the structures of the T. thermophilus 30S in complex with three different ASLs, each containing an extra base inserted at position 33.5 and their cognate mRNAs (Fig. 1B–D) in the presence of the antibiotic paromomycin. The corresponding tRNAs have been used in nonnatural amino acid incorporation studies; ASLCCCG and ASLACCC were found to promote +1 frameshifting in an in vitro translation system (Hohsaka et al. 2001) and four-base translocation as shown by toeprinting (Phelps et al. 2006), whereas ASLACGA does not promote +1 frameshifting in in vitro translation (Hohsaka et al. 2001). Crystallographic data are shown in Table 1. All three cognate ASL–mRNA structures give good difference Fourier density for the anticodon, the 3′ side of the anticodon stem, and the mRNA, while the density on the 5′ of the ASL is poor compared to canonical ASLs. Paromomycin was added because it has previously been shown to facilitate 30S domain closure, give better diffracting crystals, and a more well-defined ASL density (Ogle et al. 2002). The binding of paromomycin to the ribosome causes the bases of rRNA A1492 and A1493 to flip out to interact with the first two bases of the anticodon–codon helix in a position that is normally induced when a cognate tRNA–mRNA pair is recognized (Ogle et al. 2001). The ASLCCCG soak was also performed in the absence of paromomycin, but these crystals only diffracted to 3.8 Å resolution and the resulting difference density was too poor for interpretation (data not shown).
TABLE 1.
Data collection and refinement statistics
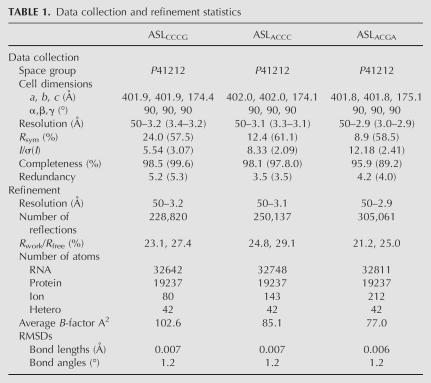
In the structures for ASLCCCG and ASLACCC, the 5′ ASL density for bases 28–32 is weak, but a path for the RNA backbone is seen in original, unbiased difference density maps. The model built for ASLCCCG contains ASL bases 33–40 and for ASLACCC contains ASL bases 33.5–40. Both contain mRNA bases 1–4 (Fig. 2C,E). As with a normal cognate ASL–mRNA complex, the rRNA bases A1492, A1493, and G530 interact with the codon–anticodon base pairs (Fig. 2D,F; Ogle et al. 2001).
FIGURE 2.
Comparison of a normal anticodon–codon interaction and extended anticodon–codon interactions in the 30S A site. Left panels show the mRNA on the left with the ASL anticodon located on the right. Right panels depict 180° vertical reorientations of the complexes, displaying the interactions of 16S rRNA bases A1492, A1493, G530, and C1054 (in gray) with the codon–anticodon helix. For C, E, and G, the final 3fo–2fc map is cut at 5 Å radius and displayed at 1.8 sigma. (A,B) Normal anticodon–codon interaction between ASLGAA (lime green) and its mRNA (pink) (Ogle et al. 2001). (C,D) ASLCCCG (blue) interaction with its cognate mRNA (red). (E,F) ASLACCC (cyan) interaction with its cognate mRNA (magenta). (G,H) ASLACGA (green) interaction with its cognate mRNA (orange).
However, unlike normal ASLs, only the first 2 bp of the codon–anticodon helix make standard Watson–Crick interactions: C1–G36 and G2–C35 in the ASLCCCG–mRNA structure (Fig. 2C) and G1–C36 and G2–C35 in the ASLACCC–mRNA structure (Fig. 2E). In both structures, the phosphate and ribose of the third anticodon base C34 are clearly visible in original difference density maps whereas the base is poorly ordered (Fig. 2C,E). In the ASLCCCG–mRNA structure, C34 does not form a base pair with the third codon base G3, but on the contrary, is within Watson–Crick base-pairing distance with the fourth base of the codon, G4 (Fig. 2C). A shift in C34 of ∼2 Å toward the codon makes this interaction possible, presumably due to the extension in the anticodon loop (Fig. 2C). In addition, G3 is in a syn conformation, thus precluding a standard Watson–Crick interaction with the anticodon bases. In contrast to ASLCCCG, the ASLACCC–mRNA structure shows neither G3 nor U4 of the codon forming canonical Watson–Crick base pairs with C34 (Fig. 2E). Instead, the base of C34 is equidistant from bases G3 and U4, suggesting a bifurcated interaction (Fig. 2E).
Another key difference relative to canonical ASLs is that ASLACCC and ASLCCCG do not display the characteristic U-turn in which the conserved base U33 stabilizes a sharp turn in the RNA backbone toward the 3′ end of the ASL by forming hydrogen bonds across the anticodon loop (Kim et al. 1974; Robertus et al. 1974). The U-turn has been suggested to be essential for translocation of normal tRNAs (Phelps et al. 2002; Phelps and Joseph 2005), but the present structures show that it is not essential for translocation in these particular tRNAs. In these two structures neither U33 nor the base at position 33.5 is in a position where it can interact with the other side of the anticodon loop (Fig. 2D,F). Instead, the base of 33.5 points in the opposite direction toward rRNA base 1052. In the absence of any cross-strand interaction in the anticodon loop, the backbones of ASLCCCG and ASLACCC adopt a wider conformation that allows the anticodon to span four bases of the codon (Fig. 2C,E).
Even though both ASLCCCG and ASLACCC have anticodon sequences complementary to a four-base codon, in the A site they do not form the four base pairs as proposed (Fig. 2 C,E; Roth 1981). The rRNA base C1054, a base known to interact with the third anticodon–codon base pair (Ogle et al. 2001), is stacked below anticodon base 34 and would need to move to create room for a fourth base pair (Stahl et al. 2002). Also, they do not display the standard three-base codon–anticodon interaction, as suggested later (Gaber and Culbertson 1984; Weiss et al. 1990; Stahl et al. 2002). Rather than a strict conservation of Watson–Crick base pairing, our structures suggest that a more important attribute is the ability of the ASL to span four bases of the codon. In the ASLCCCG–mRNA structure, a Watson–Crick base pair between the fourth base of the codon (G4) and the third base of the anticodon (C34) can form with three bases of the anticodon extending over four bases of the codon (Fig. 2C). However in the ASLACCC–mRNA structure, the fourth base of the codon (U4) is unable to form a Watson–Crick base pair with the third base of the anticodon (C34); this may explain the existence of a bifurcated interaction between both the third and fourth bases of the codon and C34 (Fig. 2E). These structures show that the wobble base of the anticodon has the ability to interact with the fourth base of the codon, exclusively or in combination with the third base. The additional base of the ASL prevents formation of a canonical U-turn, resulting in a more flexible anticodon loop that can extend over all four bases of the codon. Since both ASLCCCG and ASLACCC perform quadruplet decoding, these structures represent snapshots of the initial step of +1 frameshifting for two different tRNAs.
In contrast, the structure of ASLACGA, which has been shown not to perform +1 frameshifting (Hohsaka et al. 2001), forms a structure very similar to a canonical ASL (Ogle et al. 2001, 2002). rRNA bases A1492, A1493, and G530 make standard interactions with the codon–anticodon complex (Fig. 2G,H), and nucleotides 34–39 of the ASL and 1–3 of the mRNA superimpose almost perfectly with the codon–anticodon interaction of a canonical ASL (Fig. 2A,B). The ribose of A33.5 overlays with U33 of a normal ASL, resulting in a sharp turn of the backbone just as in the U-turn of normal tRNAs (Robertus et al. 1974). Thus, despite the base insertion, a canonical three-base anticodon is displayed by ASLACGA.
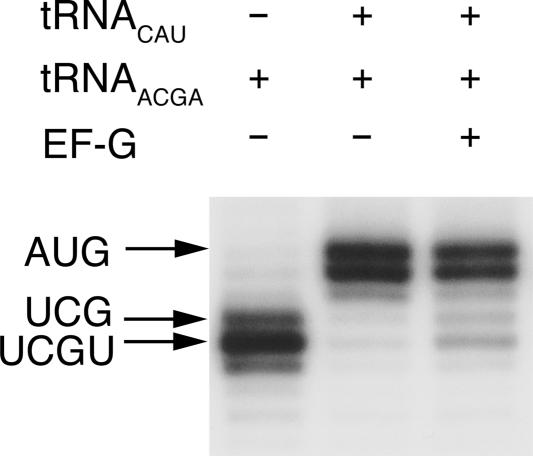
In addition, we determined the step size for translocation of tRNAACGA and ASLACGA using the primer-extension-based toeprinting technique (Hartz et al. 1988). This tRNA does not promote +1 frameshifting in vitro (Hohsaka et al. 2001). In toeprinting experiments translocation is very inefficient, but surprisingly the cognate mRNA is still translocated by four bases as tRNACCCG (Fig. 3; Phelps et al. 2006). However, ASLACGA does not undergo translocation at all (data not shown), possibly due to poor binding as shown with other extended tRNAs (Walker and Fredrick 2006). It is possible that, as suggested, in the in vitro translation assays tRNAACGA is out-competed for A-site binding by endogenous tRNAs present in the cell extract, explaining why tRNAACGA does not perform +1 frameshifting (Hohsaka et al. 2001). Under toeprinting conditions, when only one specific A-site tRNA is added, even a normal tRNA cognate for the +1 shifted A-site codon can promote four-base translocation (Phelps et al. 2006). The only known case of a similar +1 frameshift with an unpaired base between the A- and P-site codons occurs in the retrotransposon Ty3 in yeast, when the competing in-frame tRNA is rare (Farabaugh et al. 1993).
FIGURE 3.
Toeprint assay of translocation of tRNAACGA. Each band corresponds to a defined position of the ribosome on the mRNA, produced from a DNA oligonucleotide primer annealed to the mRNA upstream of the ribosome and extended by reverse transcriptase until it reaches the ribosome and falls off. The arrows indicate the position of the toeprint band corresponding to the mRNA codon in the P site. Positioning of the UCG codon in the P site would correspond to three-base translocation while UCGU in the P site corresponds to four-base translocation from the AUG codon. (Lane 1) mRNA positioning by direct P site binding of tRNAACGA. (Lane 2) mRNA positioning by P-site binding of tRNACAU and addition of tRNAACGA in the A site. (Lane 3) Same as lane 2 but after addition of EF-G + GTP.
Thus although the structural differences between ASLs from tRNAs that do +1 frameshifting (ASLCCCG and ASLACCC) and one that does not (ASLACGA) are suggestive, the fact that all three tRNAs show four-base translocation in toeprinting assays suggests that the situation with respect to +1 frameshifting is more complicated. The strength of the codon–anticodon interaction, the concentration of competing standard tRNAs, and the preferred structure of the expanded anticodon loop when bound to its mRNA in the context of the ribosomal A and P sites are all likely to contribute to frameshift efficiency.
Within the 70S ribosome, the A- and P-site tRNAs are positioned with an angle of 26° between their planes and the mRNA has a 45° kink between the two codons (Yusupov et al. 2001) that is stabilized by a magnesium ion (Selmer et al. 2006). In accordance with the roles of these two tRNA binding sites in decoding and reading-frame maintenance, they are quite different in character. In the A site, the geometry of the codon–anticodon base pair is strictly monitored in the two first positions (Ogle et al. 2001), while the ribosome only contacts the body of the tRNA from one side (Yusupov et al. 2001). Thus, correct codon–anticodon base pairing can give rise to a significant excess in binding energy that can be used to induce a transition of the ribosome from an open to a closed form (Ogle et al. 2002). By the time a tRNA has entered the P site, it has already been selected, and direct monitoring of the codon–anticodon pairing is no longer necessary. Accordingly, in the P site, the codon–anticodon helix rests on the 16S RNA bases 966 and 1400, defining the position of the wobble base pair next to the mRNA kink, without any contacts to the base pairs (Selmer et al. 2006). However, during the periods in the translation cycle when the A site is empty, it is important that the P site and mRNA not slip, so that the reading frame is maintained. As a result, the P-site tRNA is held rigidly in place, its anticodon stem is in contact with both 16S RNA and S13, and it is constrained by interactions on all three sides (Carter et al. 2000; Yusupov et al. 2001; Selmer et al. 2006). Together with the direct and magnesium mediated contacts between the P-site codon and 16S RNA, these interactions are important to stabilize the reading frame when the A site is empty (Selmer et al. 2006). In the A site, a normal three-base codon–anticodon helix fits between A1493 and C1054 (Ogle et al. 2001), and unless C1054 were to change conformation, there is no space for an extended anticodon stack beyond the wobble base.
The movement of the tRNA from A to P site is most likely defined by its interactions with the ribosome. Therefore a frameshift that occurs without slippage between codon and anticodon has to involve a nonstandard conformation of the anticodon loop and/or the mRNA at some stage. Superposition of the ASLCCCG of the present 30S structure into the P site of our recent high-resolution 70S structure (Selmer et al. 2006) suggests that with the current conformation and base pairing with the mRNA, the ASL would not form a base-stacking interaction with 16S bases G966 and C1400. To form such interactions, we can envision three scenarios. First, if a fourth base pair forms, leading to an extended base stack with G966 and C1400, this would prevent slippage of the mRNA back to the original reading frame. It appears likely that such an extended 4-bp codon–anticodon helix could be accommodated in the 70S structure. Second, if a normal 3-bp interaction forms, the codon–anticodon complex could move back to the normal reading frame and the standard interactions with 16S RNA could form. Perhaps this is how, for some expanded tRNAs, both three- and four-base decoding occurs (Curran and Yarus 1987). For the conformation we observe for the expanded ASL in the A site to be accommodated in the P site, the side chains of S9 Ser 126 and Arg 128 as well as the backbone of 16S RNA 1340–41 would have to rearrange slightly to prevent steric clashes. A third possibility is that the 16S P site changes its conformation to allow stabilizing interactions with the nonstandard codon–anticodon complex seen in the A site. However, since comparison of the empty 16S P site in the apo Escherichia coli 70S structure (Schuwirth et al. 2005) and the filled P site in the T. thermophilus 70S structure (Selmer et al. 2006) shows nearly identical conformations of G966 and C1400, it seems likely that these bases are part of a preformed site where the tRNA–mRNA complex is docked rather than accommodating different complexes by induced fit. Since there is a preference for complementarity between base 4 of the codon and base 33.5 for most +1 frameshift suppressor tRNAs, it is likely that a fourth base pair forms in the P site when possible. Such an interaction is also suggested by direct binding of tRNACCCG in the P site that positions the mRNA in the +1 frame (Phelps et al. 2006). In the 30S crystal structure, the spur of a symmetry related molecule mimics a P-site ASL (Carter et al. 2000). The fact that this noncognate ASL interacts similarly with the 30S P site (Selmer et al. 2006) suggests that the fourth base pair may not be essential for an extended stack to form, agreeing with the published substitution experiments (Gaber and Culbertson 1984; Curran and Yarus 1987; Weiss et al. 1990).
Our structures show how expanded anticodon loops can be accommodated in the ribosomal A site. This demonstrates that although the decoding center is stringent with respect to the two first base pairs as with normal tRNAs, it allows wider ASLs to bind and form nonstandard pairing with the codon at the wobble position and the fourth base. Also, we demonstrate that tRNAs with expanded anticodon loops can adopt a variety of conformations in the context of the ribosomal A site. However, given the differences in structures among the three ASLs studied here, it is unclear if these structures are critical for frameshifting to occur in vivo or whether codon–anticodon interaction in the A site is related to translocational step size.
To test our structural hypotheses and clarify the role of the P site in +1 frameshifting, it is important to obtain high-resolution structures of these tRNAs after translocation. Such studies are best addressed in the context of well-diffracting crystals of the 70S ribosome bound to mRNA and tRNA (Selmer et al. 2006). Nevertheless, the work presented here yields molecular details of interactions between extended anticodon loop tRNAs involved in frameshift suppression with the decoding center of the 30S subunit.
MATERIALS AND METHODS
Materials and crystallization
T. thermophilus 30S ribosomal subunits were purified, crystallized, and cryoprotected as described (Clemons et al. 2001). ASL and mRNA oligonucleotides were chemically synthesized (Dharmacon). The ASL sequences were 5′-CCAGACUCCCGAAUCUGG-3′, 5′-CCAGACUACGAAAUCUGG-3′, and 5′-CAGACUACCCAAUCUGG-3′ (anticodon underlined) for ASLCCCG, ASLACGA, and ASLCCCA, respectively. The mRNA sequences were 5′-CGGGUU-3′, 5′-UCGUUU-3′, and 5′-GGGUAAA-3′ (codon underlined). The crystals were then gradually buffer exchanged to a final cryoprotectant solution containing 26% MPD, 100 mM MES-KOH (pH 6.5), 200 mM KCl, 75 mM NH4Cl, and 15 mM MgCl2. After cryoprotection, the crystals were soaked in a solution containing 80 μM paromomycin, 200 μM ASL, and 200 μM mRNA for at least 48 h as described (Ogle et al. 2001). Crystals were flash cooled in liquid nitrogen and stored for data collection.
Toeprint assay
70S ribosomes were purified from E. coli MRE600 cells (Powers and Noller 1990). mRNAUCGU and mRNACGGG were created by inserting the sequence for the specified 4-nt codon between the initiation codon (AUG) and the following tRNAPhe codon (UUU) in the gene32 mRNA. Toeprint assays were completed using previously detailed conditions (Hartz et al. 1988). Briefly, 10 pmol of activated 70S ribosomes were mixed with 20 pmol of mRNA, which had been annealed to the 32P radiolabeled AL2 primer. To position the P site on the mRNA, 20 pmol of tRNAfMet (Sigma) or 150 pmol ASL (Dharmacon) were added to the preprogrammed ribosome. For translocation reactions, 20 pmol of tRNAfMet were bound to the P site followed by the addition of 150 pmol of ASL or tRNA for binding to the A site. Translocation was initiated with the addition of 50 pmol EF-G•GTP followed by a 30-min incubation at 37°C. Reverse transcriptase (Seikagaku) was added and primer extension inhibition was evaluated on a 10% denaturing polyacrylamide gel.
Data collection and refinement
Crystals were initially screened at Daresbury 14.1 and data were collected at ESRF beamline ID14–4 and SLS beamline X10SA and processed with the XDS package (Kabsch 1993). For ASLACCC and ASLACGA data sets, we used lower exposures than in previous 30S data collection, which allowed us to merge fewer wedges for a complete data set. This is reflected in higher R sym values for the ASLCCCG data set where we used multiple crystals. CNS 1.1 was used for refinement (Brünger et al. 1998). O (Jones et al. 1991) and Coot (Emsley and Cowtan 2004) were used for visualization and building, and the CCP4 package (CCP4 1994) was used for various tasks. Figures were prepared using the Pymol package (DeLano 2002).
ACKNOWLEDGMENTS
We thank Frank V. Murphy IV, Sabine Petry, Albert Weixlbaumer, and John Weir for help in data collection. We also thank Raimond Ravelli at ESRF Beamline 14-4, Clemens Schulze-Briese at SLS Beamline X10SA, and Mike McDonald at Daresbury Laboratory Beamline 14-1. This work was supported by the Medical Research Council (United Kingdom) (V.R.), NIH grants GM67624 (V.R.) and GM65265 (S.J.), the Agouron Institute (V.R.), and Human Frontier Science Programs RGY23/2003 (T.S.) and RGY16 (S.J). C.M.D is supported by an American Cancer Society Postdoctoral Fellowship and M.S. is supported by a fellowship from the Wenner-Gren Foundations. Atomic coordinates for ASLCCCG (accession code 2uxd), ASLACCC (accession code 2uxb) and ASLACGA (accession code 2uxc) have been deposited in the Protein Data Bank.
Footnotes
Arti cle published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.367307.
REFERENCES
- Bossi, L., Smith, D.M. Suppressor sufJ: A novel type of tRNA mutant that induces translational frameshifting. Proc. Natl. Acad. Sci. 1984;81:6105–6109. doi: 10.1073/pnas.81.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clove, G.M., Delano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B.T., Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Clemons W.M., Jr, Brodersen, D.E., McCutcheon, J.P., May, J.L., Carter, A.P., Morgan-Warren, R.J., Wimberly, B.T., Ramakrishnan, V. Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: Purification, crystallization, and structure determination. J. Mol. Biol. 2001;310:827–843. doi: 10.1006/jmbi.2001.4778. [DOI] [PubMed] [Google Scholar]
- Cummins, C.M., Donahue, T.F., Culbertson, M.R. Nucleotide sequence of the SUF2 frameshift suppressor gene of Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 1982;79:3565–3569. doi: 10.1073/pnas.79.11.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, J.F., Yarus, M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987;238:1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. The PyMOL molecular graphics system. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Emsley, P., Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Farabaugh, P.J. Programmed translational frameshifting. Annu. Rev. Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- Farabaugh, P.J., Zhao, H., Vimaladithan, A. A novel programmed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: Frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber, R.F., Culbertson, M.R. The yeast frameshift suppressor gene SUF16-1 encodes an altered glycine tRNA containing the four-base anticodon 3′-CCCG-5′. Gene. 1982;19:163–172. doi: 10.1016/0378-1119(82)90002-6. [DOI] [PubMed] [Google Scholar]
- Gaber, R.F., Culbertson, M.R. Codon recognition during frameshift suppression in Saccharomyces cerevisiae . Mol. Cell. Biol. 1984;4:2052–2061. doi: 10.1128/mcb.4.10.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland, R.F., Atkins, J.F. Recoding: Dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- Hartz, D., McPheeters, D.S., Traut, R., Gold, L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Hohsaka, T., Ashizuka, Y., Murakami, H., Sisido, M. Incorporation of nonnatural amino acids into streptavidin through in vitro frameshift supression. J. Am. Chem. Soc. 1996;118:9778–9779. [Google Scholar]
- Hohsaka, T., Ashizuka, Y., Taira, H., Murakami, H., Sisido, M. Incorporation of nonnatural amino acids into proteins by using various four-base codons in an Escherichia coli in vitro translation system. Biochemistry. 2001;40:11060–11064. doi: 10.1021/bi0108204. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.-Y., Cowan, S.W., Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- Kim, S.H., Sussman, J.L., Suddath, F.L., Quigley, G.J., McPherson, A., Wang, A.H., Seeman, N.C., Rich, A. The general structure of transfer RNA molecules. Proc. Natl. Acad. Sci. 1974;71:4970–4974. doi: 10.1073/pnas.71.12.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland, C.G. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- Magliery, T.J., Anderson, J.C., Schultz, P.G. Expanding the genetic code: Selection of efficient suppressors of four-base codons and identification of “shifty” four-base codons with a library approach in Escherichia coli . J. Mol. Biol. 2001;307:755–769. doi: 10.1006/jmbi.2001.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley, J.L., Gesteland, R.F. Suppression of amber mutants in vitro induced by low temperature. J. Mol. Biol. 1978;125:433–447. doi: 10.1016/0022-2836(78)90309-1. [DOI] [PubMed] [Google Scholar]
- Murakami, H., Hohsaka, T., Ashizuka, Y., Sisido, M. Site-directed incorporation of p-nitrophenylalanine into streptavidin and site-to-site photoinduced electron transfer from a pyrenyl group to a nitorphenyl group on the protein framework. J. Am. Chem. Soc. 1998;120:7520–7529. [Google Scholar]
- Ogle, J.M., Ramakrishnan, V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons W.M., Jr, Tarry, M.J., Carter, A.P., Ramakrishnan, V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Murphy, F.V., Tarry, M.J., Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Phelps, S.S., Joseph, S. Nonbridging phosphate oxygen atoms within the tRNA anticodon stem–loop are essential for ribosomal A-site binding and translocation. J. Mol. Biol. 2005;349:288–301. doi: 10.1016/j.jmb.2005.03.079. [DOI] [PubMed] [Google Scholar]
- Phelps, S.S., Jerinic, O., Joseph, S. Universally conserved interactions between the ribosome and the anticodon stem–loop of A-site tRNA important for translocation. Mol. Cell. 2002;10:799–807. doi: 10.1016/s1097-2765(02)00686-x. [DOI] [PubMed] [Google Scholar]
- Phelps, S.S., Gaudin, C., Yoshizawa, S., Benitez, C., Fourmy, D., Joseph, S. Translocation of a tRNA with an extended anticodon through the ribosome. J. Mol. Biol. 2006;360:610–622. doi: 10.1016/j.jmb.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Powers, T., Noller, H.F. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc. Natl. Acad. Sci. 1990;87:1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather, N.E., Murgola, E.J., Mims, B.H. Nucleotide insertion in the anticodon loop of a glycine transfer RNA causes missense suppression. Proc. Natl. Acad. Sci. 1981;78:7408–7411. doi: 10.1073/pnas.78.12.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, Q., Li, J.N., Zhao, H., Hagervall, T.G., Farabaugh, P.J., Bjork, G.R. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell. 1998;1:471–482. doi: 10.1016/s1097-2765(00)80048-9. [DOI] [PubMed] [Google Scholar]
- Riddle, D.L., Carbon, J. Frameshift suppression: A nucleotide addition in the anticodon of a glycine transfer RNA. Nat. New Biol. 1973;242:230–234. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- Riyasaty, S., Atkins, J.F. External suppression of a frameshift mutant in salmonella. J. Mol. Biol. 1968;34:541–557. doi: 10.1016/0022-2836(68)90179-4. [DOI] [PubMed] [Google Scholar]
- Robertus, J.D., Ladner, J.E., Finch, J.T., Rhodes, D., Brown, R.S., Clark, B.F., Klug, A. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Roth, J.R. Frameshift suppression. Cell. 1981;24:601–602. doi: 10.1016/0092-8674(81)90086-6. [DOI] [PubMed] [Google Scholar]
- Schuwirth, B.S., Borovinskaya, M.A., Hau, C.W., Zhang, W., Vila-Sanjurjo, A., Holton, J.M., Cate, J.H. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy, F.V.T., Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R., Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Sroga, G.E., Nemoto, F., Kuchino, Y., Bjork, G.R. Insertion (sufB) in the anticodon loop or base substitution (sufC) in the anticodon stem of tRNAPro 2 from Salmonella typhimurium induces suppression of frameshift mutations. Nucleic Acids Res. 1992;20:3463–3469. doi: 10.1093/nar/20.13.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, G., McCarty, G.P., Farabaugh, P.J. Ribosome structure: Revisiting the connection between translational accuracy and unconventional decoding. Trends Biochem. Sci. 2002;27:178–183. doi: 10.1016/S0968-0004(02)02064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, S.E., Fredrick, K. Recognition and positioning of mRNA in the ribosome by tRNAs with expanded anticodons. J. Mol. Biol. 2006;360:599–609. doi: 10.1016/j.jmb.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, R.B., Dunn, D.M., Atkins, J.F., Gesteland, R.F. Ribosomal frameshifting from −2 to +50 nucleotides. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]
- Yourno, J. Externally suppressible +1 “glycine” frameshift: Possible quadruplet isomers for glycine and proline. Nat. New Biol. 1972;239:219–221. doi: 10.1038/newbio239219a0. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]