Abstract
snoRNAs are small protein-noncoding RNAs essential for pre-rRNA processing and ribosome biogenesis, and are encoded intronically in host genes (HGs) that are either protein coding or noncoding. mRNAs of protein-noncoding HGs differ in their nucleotide sequences among species. Although the reason for such sequential divergence has not been well explained, we present evidence here that such structurally different HGs have evolved from a common ancestral gene. We first identified two novel protein-noncoding HGs (mU50HG-a and mU50HG-b) that intronically encode a mouse ortholog of a human snoRNA, hU50. The sequences of mU50HG mRNA differed from that of hU50HG. However, a chromosome mapping study revealed that mU50HG is located at 9E3-1, the murine segment syntenic to human 6q15, where hU50HG is located. Synteny is a phenomenon whereby gene orthologs are arranged in the same order at equivalent chromosomal loci in different species; synteny between two species means it is highly likely that the genes have evolved from a common ancestral gene. We then extended this mapping study to other protein-noncoding snoRNA-HGs, and found again that they are syntenic, implying that they have evolved from genes of common ancestral species. Furthermore, on these syntenic segments, exons of adjacent protein-coding genes were found to be far better conserved than those of noncoding HGs, suggesting that the exons of protein-noncoding snoRNA-HGs have been much more fragile during evolution.
Keywords: intron, snoRNA, U50, snoRNA host gene, pseudogenes, synteny
INTRODUCTION
Small nucleolar RNAs (snoRNAs) are known to guide the post-transcriptional modifications of ribosomal and other RNAs. Such modifications are believed to play crucial roles in RNA folding as well as RNA–RNA and RNA–protein interactions. In addition, snoRNAs are thought to be involved in epigenetic modification of genes (Kiss et al. 2004). Hundreds of snoRNAs have been identified in a broad variety of organisms including yeast, plants, and vertebrates. They are well conserved phylogenetically among different species, suggesting their important physiological role. The evolution of snoRNA has recently been studied in detail. In nematodes, trans-duplication as well as cis-duplication of snoRNA host genes (HGs) with insertion of snoRNA genes close to the original loci are shown to have occurred during the course of evolution (Zemann et al. 2006). Luo and Li (2007) have suggested that a certain proportion of H/ACA-related snoRNA sequences in primates have arisen after rodent/primate divergence. Also, intronic snoRNA sequences are reportedly better conserved than exonic sequences in non-protein-coding HGs (Tycowsky et al. 1996; Weber 2006).
Most of the HGs of vertebrate snoRNAs are encoded intronically. A large proportion of snoRNA-HGs are known to encode proteins that operate in ribosomal biogenesis, translation, or other nuclear/nucleolar functions (for review, see Kiss 2002), while the remaining snoRNA-HGs are apparently protein noncoding (Tycowski et al. 1996; Pelczar and Filipowicz 1998; Smith and Steitz 1998). A few previous studies have compared the nucleotide sequences of mRNAs of human protein-noncoding snoRNA-HGs. Most of those studies demonstrated that the mRNA sequences of HGs are not homologous between human and mouse, while the intronically encoded snoRNAs are highly conserved (Tycowski et al. 1996; Smith and Steitz 1998). The only exception in this respect reported so far is the U87 HG, which is sequentially similar between human and mouse and is therefore speculated to have some functional activity apart from production of U87 snoRNA (Makarova and Kramerov 2005).
This marked diversity of HG mRNA among vertebrates has not been well explained. Recent studies have assumed that intronically encoded snoRNAs can be very mobile genetic elements, that they are readily insertable into introns of different genes, and that such mobility/insertability would explain the diversity of HG sequences among different species (Smith and Steitz 1997, 1998; Weber 2006; Zemann et al. 2006; Luo and Li 2007). However, none of those studies have presented direct evidence that noncoding HGs showing dissimilarity between mouse and human are orthologs, i.e., having originated from an identical ancestral gene and becoming separated during evolution. Considering the potentially high mobility of intronic snoRNA, it is still possible that such divergent HGs were originally different, and that intronic snoRNAs were inserted into the introns of these different HGs in a retroposon manner.
We recently identified a new protein-noncoding HG, human U50 snoRNA host gene (hU50HG), which intronically encodes a human counterpart of U50 (Tanaka et al. 2000). The U50HG is a 5′-top gene encoding box C/D-type snoRNA U50 (Kiss-Laszlo et al. 1996) in its fifth intron. Using hU50 snoRNA as a probe, we isolated a mouse U50 ortholog (mU50) and its HGs (mU50HG-a and mU50HG-b). mU50 was shown to share high homology (86%) with hU50. Meanwhile, intensive homology researches showed that the exons of the mU50HGs did not share similarity with hU50HG. Subsequent chromosome mapping, however, showed that the mU50HGs were located on mouse chromosome 9E3.1, which is syntenic to human 6q15, where hU50HG is located. Synteny between species means that orthologous genes are located in the same order at a particular gene/chromosome site (MacAndrew 2002; Waterston et al. 2002; Sainz et al. 2006). Also, the syntenic segment is defined as a region in which a series of landmark genes shows a maximum degree of shared order on a given gene/chromosome in different species. Thus, it is highly likely that two genes showing synteny in two species are derived from a common ancestral gene (MacAndrew 2002; Waterston et al. 2002; Sainz et al. 2006). Thus, the above results for mU50HGs suggest that U50HGs of different species have evolved from common ancestral genes. Therefore, to clarify whether a similar situation has arisen for other snoRNA-HGs, we investigated four other noncoding snoRNA-HGs, together with four coding HGs. For this study, we used NCBI human and mouse genome databases, since it is now possible to conduct detailed analysis of segments showing synteny between humans and mice (NCBI Human–Mouse Homology Map, http://www.ncbi.nlm.nih.gov/Homology). The details are reported in this study.
For the purpose of comparison of oligonucleotide sequences as opposed to evolutionarily conserved sequences, the terms “similar” and “dissimilar” are used in this study instead of “homologous” or “homology.” This is because the terms homologous/homology have been used to describe both evolutionarily identical genes and similarity between sequences of the same species. Also, the term “ortholog” is used, instead of “homolog” to compare evolutionarily identical genes. When using the NCBI BLAST 2 software package for comparison of nucleotide sequences (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi), judgment of similarity/dissimilarity was based on the default level of the program. The identity of two sequences, when judged to be similar, was expressed in terms of the percentage of matched base pairs divided by total base pairs (bp/total bp). Comparison of two sequences was conducted on gene segments constituting both sequences rather than the whole sequences. In other words, judgment of similarity, as well as similarity in terms of percentage, can be made when one pair of exons, or even part of the exon pair, shares similarity for a certain length.
RESULTS AND DISCUSSION
Genomic structures and chromosomal localization of mU50HGs
Southern blot analysis of the mouse genome using the human U50 sequence as a probe demonstrated two bands, suggesting the existence of two HGs (data not shown). Successive cloning and sequencing of a mouse genomic library identified one mU50 (accession number AB116376) and two HGs (mU50HG-a, accession number AB116374; and mU50HG-b, accession number AB116375). Indeed, three identical mU50 sequences were detected in those two host genes (one in mU50HG-a and two in mU50HG-b) (Fig. 1). This mU50 sequence was found to share 86% similarity with hU50 when compared using NCBI BLAST 2 software. mU50 contained two sequences complementary to mouse 28S rRNA, along with the consensus sequences of the box C/D-type snoRNAs. No other sequences similar to mU50, as characterized by the presence of sequences complementary to mouse 28S rRNA and the box C/D structure, were detected on the NCBI and CERELA mouse genome databases, suggesting that mU50 is the only ortholog of hU50.
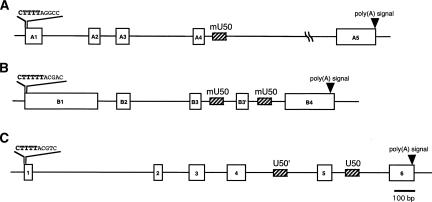
FIGURE 1.
Suggested genome structure of (A) mU50HG-a, (B) mU50HG-b, and (C) hU50HG. (Boxes) Exons; (oblique-line boxes) snoRNA sequences.
mU50HG-a is 14.2 kb, bearing five exons (A1, A2, A3, A4, A5; 96, 65, 75, 58, and 216 bp, respectively) and five introns (257, 89, 354, 51, and 12,969 bp, respectively) (Fig. 1). A TATA-like motif (TATAAA) and CAT-like box (CCAAT) are present in the 5′-upstream region of this gene. The GT-AG rule was well preserved at each exon–intron boundary. No obvious protein-coding frame was identified by the BLAST ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html; data not shown).
The other HG, mU50HG-b, is 1.8 kb, containing five exons, B1, B2, B3, B3′, and B4. Again, a TATA-like motif (TATAAA) and CAT-like box (CCAAT) are present in the 5′-upstream region. The GT-AG rule was well preserved at each exon–intron boundary. Again, no obvious protein-coding frame was identified by the BLAST ORF Finder. Significant sequence similarity was found between mU50HG-a and mU50HG-b at exons B3 and B3′ (88%), A4 and B3 (94%), and A4 and B3′ (95%), suggesting that these two HGs share a common ancestral gene.
To determine the chromosomal loci of the mU50HG-a and mU50HG-b genes, we conducted FISH analysis on normal mouse metaphase spreads. The specific hybridization signal for the mU50HG-a gene was detected on mouse chromosome 9 at band E3.1, while that for the mU50HG-b gene was detected on the same chromosome at band E1–E4. Further two-color analysis using FITC (mU50HG-a, green) and rhodamine (mU50HG-b, red) confirmed this result (data not shown). Interestingly, on the Human–Mouse Homology Map, these regions (mouse chromosome 9, band E3.1) are syntenic with human chromosome 6q, where hU50-HG is located (human 6q15). Here, synteny implies orthologs to locate in the same order on equivalent chromosomal loci of different species. In other words, genes in synteny between two species are suggested to have evolved from a common gene of their ancestor. Therefore, we expected that similarities must exist between the hU50HG and mU50HGs. However, the following result differed from that expected.
mRNA sequences of hU50HG and the two mU50HGs are dissimilar
The mRNA sequences of hU50HG and the two mU50HGs were compared using the NCBI BLAST 2 program, which revealed that there are no “similar” sequences between them. This is in sharp contrast to the high (86%) similarity between hU50 and mU50. To explain this paradox, we hypothesized that such protein-noncoding snoRNA HGs, such as U50HG, have evolved from an identical ancestral gene, and while its snoRNA-coding intron is important and thus well preserved, the exons are biologically useless and therefore labile to mutational pressure during genetic evolution. To confirm this idea, the following studies were conducted.
Protein-noncoding and protein-coding snoRNA host genes are syntenic between human and mouse
Their mRNA sequences show similarity between human and mouse for protein-coding genes but not for noncoding genes. Since hU50HG and mU50HGs were assigned to syntenic loci, we were interested in clarifying whether such synteny between human and mouse exists for other snoRNA-HGs. We studied eight snoRNA-HGs, four of which were protein coding and four noncoding. All eight genes were found to show perfect synteny (Table 1). Next, the sequences of these human and mouse HGs were determined. Interestingly, high similarities were noted for all four protein-coding snoRNA-HGs, whereas no similarities were found for the noncoding HGs (Table 1). This result supported the hypothesis that such protein-noncoding snoRNA HGs such as U50HG have evolved from an identical ancestral gene, and that its snoRNA-coding introns are well preserved, although their exons are very fragile.
TABLE 1.
Chromosomal loci and mRNA sequence similarity of human and mouse snoRNA host genes
Protein-noncoding snoRNA-HGs are less well conserved than protein-coding genes located on the same chromosomal segment
Finally, to further substantiate our hypothesis, we studied the sequence similarity of human and mouse UHG, a protein-noncoding snoRNA-HG, and four coding genes located in the same syntenic segment. Here, UHG, rather than U50HG or other noncoding HGs, was selected because as many as four coding genes—Nxf1, Stx5a, Slc3a2, and Chrm1—are located in close proximity to UHG on the same chromosomal segment (Fig. 2; the sequence similarity of the corresponding human and mouse genes is shown in Table 2), while the number of coding genes is lower in H50HG. On the Human–Mouse Homology Map (http://www.ncbi.nlm.nih.gov/Homology), the four coding genes and UHG are located in the order cen–Nxf1–Stx5a–UHG–Slc3a2–Chrm1–ter on human chromosome 11q12–13, and in the reverse order on mouse chromosome 19A (Fig. 2). When the sequences were compared, high similarity between human and mouse was noted for all four coding genes, unlike the UHG exons. Therefore, this result provided further support for the speculation that protein-noncoding snoRNA-HGs are much more susceptible to oligonucleotide sequence changes than are protein-coding genes.
FIGURE 2.
A syntenic segment of human and mouse chromosomes bearing UHG, a protein-noncoding snoRNA-HG and four protein-coding genes. These five genes are arranged on this syntenic segment in the same order and in reverse positions. The sequence similarity of the corresponding human and mouse genes is shown in Table 2.
TABLE 2.
mRNA sequence similarity between human and mouse protein-coding orthologs that are in synteny with UHG
MATERIALS AND METHODS
Cloning of mouse genomic library
A library prepared from Sau3A partial digests of normal mouse genomic DNA using the lDASH II phage vector (Stratagene) was a gift from Y. Iwakura (Institute of Medical Science, University of Tokyo). A cDNA library was constructed from poly(A)+ RNA of MELgP3 cells using the lZIP-LOX phage vector (GIBCO BRL) in accordance with the manufacturer's instructions. Genomic DNAs and cDNAs of two murine genes, mU50HG-a and mU50HG-b, were isolated by plaque hybridization of 106 phage clones according to the standard procedure.
Sequence analysis
Subcloned genomic DNA, cDNA, and PCR products were sequenced with the use of a Thermo Sequence Fluorescent Labeled Primer Cycle Sequencing kit (Amersham). The M13 forward and M13 reverse primers were used for this sequencing. Sequence ladders were electro-resolved on 6% urea-denatured polyacrylamide gel and sequenced using an Autosequencer SQ5500 (Hitachi).
cDNA 5′-end amplification (5′-RACE)
In order to identify the 5′ terminals of the mU50HG-a and mU50HG-b transcripts, a 5′-RACE kit (Version 2.0; GIBCO BRL) was used. First-strand cDNA was synthesized from total RNA using a common primer to mU50HG-a and mU50HG-b (5′-ATCTCACTGGTCAGCATTCA-3′) in accordance with the manufacturer's instructions. A homopolymeric tail was added to the 3′ end of the cDNA using TdT and dCTP, and the dC-tailed cDNA was amplified using the bridged anchor primer supplied with the kit and the nested primers (5′-ATTCTGTCGGAAGCTTTGGG-3′ for mU50HG-a; 5′-CTCGTAGCTGCTTTGAAGAC-3′ for mU50HG-b). After reamplification of the PCR product using the AUAP primer supplied with the kit and specific nested primers (5′-AGAATGATGAGCCATGGTCC-3′ for U50HG-a; 5′-GTCTCTTTCGGTGACTCTAG-3′ for U50HG-b), the 5′-RACE product was cloned, using an original TA cloning kit (Invitrogen), and sequenced.
PCR and RT-PCR
PCR and RT-PCR were used to complement the library study and also to obtain probes for the two mU50HGs. PCR amplification was performed using the mF8 forward primer (5′-CTTTTTACGACGGAGCCTAA-3′) and the mR13 reverse primer (5′-CAATCACTGCCAGAATAAGG-3′) for confirmation of the mU50HG-b sequence; the p-014 forward primer (5′-ACTTAGATCAAACTGGCCTTACCA-3′) and the p-015 reverse primer (5′-AAAGGGCAATTGCACTCACACACAT-3′) for obtaining the U50HG-a probe AD4.6; and the p-004 forward primer (5′-TCCAAGGCGCGCGCTCCCACGCGTTGCTTCATGAA-3′) and the p-011 reverse primer (5′-AGGGGGCAGGTCCTCAGGTTTGGTAGCAAGTGCCT-3′) for obtaining the mU50HG-b probe BD0.9. In these reactions, 500 ng of mouse DNA was used as a template and amplified with 20 mM Tris-HCl (pH 9.0); 50 mM KCl; 1.5 mM MgCl2; 200 mM each dATP, dCTP, dGTP, and dTTP; and 0.05 U/μL Taq polymerase (TAKARA). PCR was performed using 1 cycle at 94°C for 60 sec, 30 cycles at 94°C for 30 sec, 56°C for 30 sec, 72°C for 60 sec, and 1 cycle at 72°C for 15 min.
RT-PCR was introduced to detect the mU50HG-b transcript. cDNA was synthesized from 5 mg of total RNA extracted from mouse spleen as the template, an oligo(dT) primer (5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTT-3′) (GIBCO BRL), and reverse transcriptase (SUPER SCRIPT II). The PCR reaction was performed with 2 mL of cDNA sample, mF4 forward primer (5′-GAGTAAATACAGATGCTCCGG-3′), and mR11 reverse primer (5′-ATGGCTAAGAGCACTGATGC-3′), under the conditions described above.
Database search
For in silico analysis, two databases, the NCBI genome database (http://www.ncbi.nlm.nih.gov/index.html) and CELERA DISCOVERY SYSTEM were used initially for sequence analysis of the human and mouse genomes and cDNAs. The data were reevaluated and reinforced partially by the recent NCBI database (http://www.ncbi.nlm.nih.gov/index.html). Open reading frames were searched with the use of the NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The NCBI BLAST 2 software package (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html) was used for comparison of human and mouse cDNAs. In this analysis, sequence similarity is expressed as a percentage, and when the percent value is lower than a given threshold, the outcome is indicated as “dissimilar.” Comparison of the two sequences was done on a gene segment base, exon sequence mostly, and a similarity by percent was shown when one segment/exon pair of two mRNAs shared similarity. For genes whose cDNA sequences have not been reported, the longest putative cDNA sequences were selected and submitted for study. For analysis of chromosome synteny, the NCBI Human–Mouse Homology Map (http://www.ncbi.nlm.nih.gov/Homology) was used.
ACKNOWLEDGMENTS
We thank Professor Yoichiro Iwakura for supplying the mouse genomic library, Dr. Masataka Asagiri and Dr. Takayuki Kannno for technical assistance, Professor Kiyoshi Takatsu for supplying the MELgP3 cells, Professor Haruo Saito and Dr. Tomoko Ohta for reviewing the manuscript, and Colin Crist for critical reading of the manuscript and valuable comments. This work was supported by grants from The Ministry of Education, Sports, Culture, Science and Technology of Japan (MEXT) to S.M. (12218207 and 14035103) and Y.N. (13051101 and 14035207); and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.209707.
REFERENCES
- Bortolin, M.L., Kiss, T. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA. 1998;4:445–454. [PMC free article] [PubMed] [Google Scholar]
- Kenmochi, N., Kawaguchi, T., Rozen, S., Davis, E., Goodman, N., Hudson, T.J., Tanaka, T., Page, D.C. A map of 75 human ribosomal protein genes. Genome Res. 1998;8:509–523. doi: 10.1101/gr.8.5.509. [DOI] [PubMed] [Google Scholar]
- Kiss, T. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Kiss, A.M., Jaddy, B.E., Bertrand, E., Kiss, T. Human box H/ACA preudouridylation guide RAN machinery. Mol. Cell. Biol. 2004;24:5795–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., Bachellerie, J.P., Caizergues-Ferrer, M., Kiss, T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Luo, Y., Li, S. Genome-wide analysis of retrogenes derived from the human box H/ACA snoRNAs. Nucleic Acids Res. 2007;35:559–571. doi: 10.1093/nar/gkl1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAndrew, A. 2002. What does the mouse genome draft tell us about evolution? http://www.evolutionpages.com/Mouse%20genome%20home.htm.
- Makarova, J.A., Kramerov, D.A. Noncoding RNA of U87 host gene is associated with ribosomes and is relatively resistant to nonsense-mediated decay. Gene. 2005;363:51–60. doi: 10.1016/j.gene.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Pelczar, P., Filipowicz, W. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol. Cell. Biol. 1998;18:4509–4518. doi: 10.1128/mcb.18.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, L.H., Nicoloso, M., Michot, B., Azum, M.C., Caizergues-Ferrer, M., Renalier, M.H., Bachellerie, J.P. U21, a novel small nucleolar RNA with a 13 nt complementarity to 28S rRNA, is encoded in an intron of ribosomal protein L5 gene in chicken and mammals. Nucleic Acids Res. 1994;22:4073–4081. doi: 10.1093/nar/22.20.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz, J., Rovensky, P., Gudjonsson, S.A., Thorleifsson, G., Stefansson, K., Gulcher, J.R. Segmental duplication density decrease with distance to human–mouse breaks of synteny. Eur. J. Hum. Genet. 2006;14:216–221. doi: 10.1038/sj.ejhg.5201534. [DOI] [PubMed] [Google Scholar]
- Selvamurugan, N., Joost, O.H., Haas, E.E., Brown, J.W., Galvin, N.J., Eliceiri, G.L. Intracellular localization and unique conserved sequences of three small nucleolar RNAs. Nucleic Acids Res. 1997;25:1591–1596. doi: 10.1093/nar/25.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.M., Steitz, J.A. Sno Stoem in the nucleolus: New roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Smith, C.M., Steitz, J.A. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, R., Satoh, H., Moriyama, M., Satoh, K., Morishita, Y., Yoshida, S., Watanabe, T., Nakamura, Y., Mori, S. Intronic U50 small-nucleolar-RNA (snoRNA) host gene of no protein-coding potential is mapped at the chromosome breakpoint t(3;6)(q27;q15) of human B-cell lymphoma. Genes Cells. 2000;5:277–287. doi: 10.1046/j.1365-2443.2000.00325.x. [DOI] [PubMed] [Google Scholar]
- Tycowski, K.T., Shu, M.D., Steitz, J.A. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- Tycowski, K.T., Shu, M.D., Steitz, J.A. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weber, M.J. Small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006;2:e205. doi: 10.1371/journal.pgen.0020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yon, J., Jones, T., Garson, K., Sheer, D., Fried, M. The organization and conservation of the human Surfeit gene cluster and its localization telomeric to the c-abl and can proto-oncogenes at chromosome band 9q34.1. Hum. Mol. Genet. 1993;2:237–240. doi: 10.1093/hmg/2.3.237. [DOI] [PubMed] [Google Scholar]
- Zemann, A., Bekke, A., Kiermann, M., Brosius, J., Schmitz, J. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006;34:2676–2685. doi: 10.1093/nar/gkl359. [DOI] [PMC free article] [PubMed] [Google Scholar]