Abstract
Divalent ion sensitivity of hammerhead ribozymes is significantly reduced when the RNA structure includes appropriate tertiary stabilization. Therefore, we investigated the activity of the tertiary stabilized “RzB” hammerhead ribozyme in several nondivalent ions. Ribozyme RzB is active in spermidine and Na+ alone, although the cleavage rates are reduced by more than 1,000-fold relative to the rates observed in Mg2+ and in transition metal ions. The trivalent cobalt hexammine (CoHex) ion is often used as an exchange-inert analog of hydrated magnesium ion. Trans-cleavage rates exceeded 8 min−1 in 20 mM CoHex, which promoted cleavage through outersphere interactions. The stimulation of catalysis afforded by the tertiary structural interactions within RzB does not require Mg2+, unlike other extended hammerhead ribozymes. Site-specific interaction with at least one Mg2+ ion is suggested by CoHex competition experiments. In the presence of a constant, low concentration of Mg2+, low concentrations of CoHex decreased the rate by two to three orders of magnitude relative to the rate in Mg2+ alone. Cleavage rates increased as CoHex concentrations were raised further, but the final fraction cleaved was lower than what was observed in CoHex or Mg2+ alone. These observations suggest that Mg2+ and CoHex compete for binding and that they cause misfolded structures when they are together. The results of this study support the existence of an alternate catalytic mechanism used by nondivalent ions (especially CoHex) that is distinct from the one promoted by divalent metal ions, and they imply that divalent metals influence catalysis through a specific nonstructural role.
Keywords: catalytic mechanism, cobalt hexammine, divalent cations, hammerhead ribozymes
INTRODUCTION
Hammerhead ribozymes catalyze site-specific cleavage and ligation reactions in the replication cycles of some plant viroid satellite RNAs, and they have been found in several eukaryotic genomes. The minimal motif consists of three helices that intersect at a conserved catalytic core of 11 nucleotides (nt). In crystallographic and solution studies, these minimal hammerhead ribozymes assume a Y-shaped conformation, with stems I and II making a close approach (Pley et al. 1994; Tuschl et al. 1994; Scott et al. 1995; Penedo et al. 2004). Under standard in vitro assay conditions (10 mM Mg2+), a well-behaved, minimal hammerhead ribozyme can be expected to cleave its substrate at a rate of ∼1 min−1. Intracellular activity is typically modest or poor, largely due to low intracellular Mg2+ concentrations. Several studies during the last several years highlight the importance of peripheral sequences in natural and engineered hammerhead ribozymes for their in vitro activity under physiological Mg2+ conditions and for their activity inside cells (De la Pena et al. 2003; Khvorova et al. 2003; Saksmerprome et al. 2004; Yen et al. 2004). These peripheral elements pull the ribozyme into a compact structure that aligns the scissile phosphate for in-line displacement and that positions nucleobases G8 and G12 for acid–base chemistry (Martick and Scott 2006), as has been suggested by cross-linking studies and analysis of the pH dependence of cleavage kinetics (Han and Burke 2005; Heckman et al. 2005; Lambert et al. 2006a,b).
Site-specific Mg2+ binding within minimal hammerhead ribozymes has been suggested by crystallographic analysis, electron pair resonance studies, hydroxyl radical footprinting analysis, and thiophilic metal ion rescue studies (Scott et al. 1995; Wang et al. 1999; Hunsicker and DeRose 2000; Maderia et al. 2000; Hampel and Burke 2003; Vogt et al. 2003). A Mg2+-binding site adjacent to the tandem G•A mismatches is present in all the reported X-ray structures for minimal hammerhead ribozymes (Wedekind and McKay 1998), although an active site metal ion is not evident in the native (extended) Schistosoma structure (Martick and Scott 2006). Another specific metal binding site adjacent to the phosphodiester cleavage site has been implicated from thiophilic substitution studies of the nonbridging pro-Rp oxygen of the cleavage site phosphate group. Metal ions at both sites have been suggested to participate directly in the chemical step (Peracchi et al. 1997; Wang et al. 1999). By contrast, other biophysical techniques such as single molecule FRET and isothermal calorimetry support delocalized Mg2+ binding (Rueda et al. 2003; Mikulecky et al. 2004). By comparing the pH-rate profiles for Mg2+ and several divalent transition metal ions, we have proposed that divalent metal ions may support high rates of catalysis by perturbing the proton transfer character of guanosine nucleobases in the active site, perhaps through direct, inner-sphere coordination with the guanosines (Roychowdhury-Saha and Burke 2006).
Insight into the role(s) of divalent ions in ribozyme catalysis can be gained by studying catalysis in solutions with other cations used in place of the divalent metal ions. Modest catalytic activity has also been observed for minimal hammerhead ribozymes in monovalent ions, polyamines, and exchange-inert cations (Murray et al. 1998; Curtis and Bartel 2001; O'Rear et al. 2001). Activity in monovalent cations correlated in log-linear fashion with ionic radius and approached that observed in Mg2+, suggesting that hydrated divalent ions do not act directly as a general base in the reaction (Curtis and Bartel 2001). However, a comparison of activities among a series of ribozyme variants in 4 M Li+ and in 10 mM Mg2+ suggested that important interactions were absent in the monovalent reactions (O'Rear et al. 2001). Polyamines are abundant inside cells, where they influence gene expression, DNA condensation, and RNA folding (Koculi et al. 2004). In ribozyme catalysis, they are primarily used to augment self-cleavage or substrate cleavage in the presence of divalent cations (Dahm and Uhlenbeck 1991; Suh et al. 1993; Earnshaw and Gait 1998; Akashi et al. 2002). Very slow cleavage was reported for a minimal hammerhead ribozyme in 0.5 mM spermine (Dahm and Uhlenbeck 1991). Cobalt hexammine (CoHex, Co [NH3]6 3+) is a structural analog of fully hexahydrated Mg2+ (Mg [H2O]6 2+) and—to a lesser degree—of partially dehydrated Mg2+ (Mg[H2O]n 2+); thus, insight into the role of Mg2+ can be obtained from an analysis of the effects of substituting CoHex in place of Mg2+. The interactions of CoHex with RNA have been well documented in pseudoknots (Gonzalez and Tinoco 1999; Nixon et al. 1999), G–U tandem repeats (Kieft and Tinoco 1997), stem–loop structures (Rudisser and Tinoco 2000), and helices (Butcher et al. 2000). In these contexts, CoHex exhibits delocalized, outer-sphere contacts with RNA similar to those provided by Mg (H2O)n 2+. In contrast to water molecules bound to Mg2+, the NH3 ligands of CoHex exchange very slowly (k exchange ∼10−10 s−1) (Basolo and Pearson 1988), effectively preventing access to inner-sphere coordination. The consequence of cobalt hexammine binding on ribozyme activity has been investigated in several ribozymes. In the hairpin ribozyme (Hampel and Cowan 1997) and in an in vitro selected acyl-transferase ribozyme (Suga et al. 1998), CoHex promotes efficient cleavage in the absence of divalent metal ions. In the Neurospora crassa VS ribozyme, it enhances activity in the presence of Mg2+ but cannot support catalysis as the sole metal ion (Maguire and Collins 2001). In the HDV ribozyme (Nakano et al. 2000) and in a minimal hammerhead ribozyme (Horton and DeRose 2000), CoHex inhibits activity in the presence of other divalent ions, suggesting a competition for binding. Previous studies with minimal hammerhead ribozymes showed very slow cleavage activity (∼0.0071 min−1) at high (>100 mM) cobalt hexammine concentrations (Curtis and Bartel 2001).
Artificial tertiary stabilizing motifs that function in the context of trans-cleaving versions of two natural cis-cleaving molecules were recently identified using a selection-amplification approach (Saksmerprome et al. 2004). A hammerhead ribozyme, designated RzB (Fig. 1), carrying one of these artificial tertiary motifs exhibits exceptionally high rates in the range of 100–3000 min−1 in 1 mM divalent cations (Roychowdhury-Saha and Burke 2006). We observed an increase in thermal stability upon tertiary stabilization both for ribozyme RzB (Saksmerprome et al. 2004) and for an unrelated kinase ribozyme, Kin.46 (Cho and Burke 2006). Another tertiary-stabilized hammerhead ribozyme, derived from Schistosoma mansonii, has been analyzed extensively. This ribozyme exhibited a single-step folding process in the presence of low concentrations of Mg2+ ion ([Mg2+]1/2 ≈ 0.16 mM) (Penedo et al. 2004), in contrast to the two-stage folding process detected in minimal hammerhead ribozymes (Hammann and Lilley 2002; Penedo et al. 2004). A second transition, which is spectroscopically silent but kinetically apparent, appears to take place at higher Mg2+ concentrations, as the values of [Mg2+]1/2 for catalysis range from 3.9 to ≈ 90 mM (Penedo et al. 2004; Kim et al. 2005; Roychowdhury-Saha and Burke 2006). In contrast, the folding behavior of the Schistosoma ribozyme in Na+ as the sole cation showed a two-step folding process that was similar to its minimal counterpart lacking the tertiary “loop–loop” interaction (Penedo et al. 2004). Thus, the Schistosoma ribozyme requires Mg2+ in order to form the stabilizing tertiary contacts.
FIGURE 1.
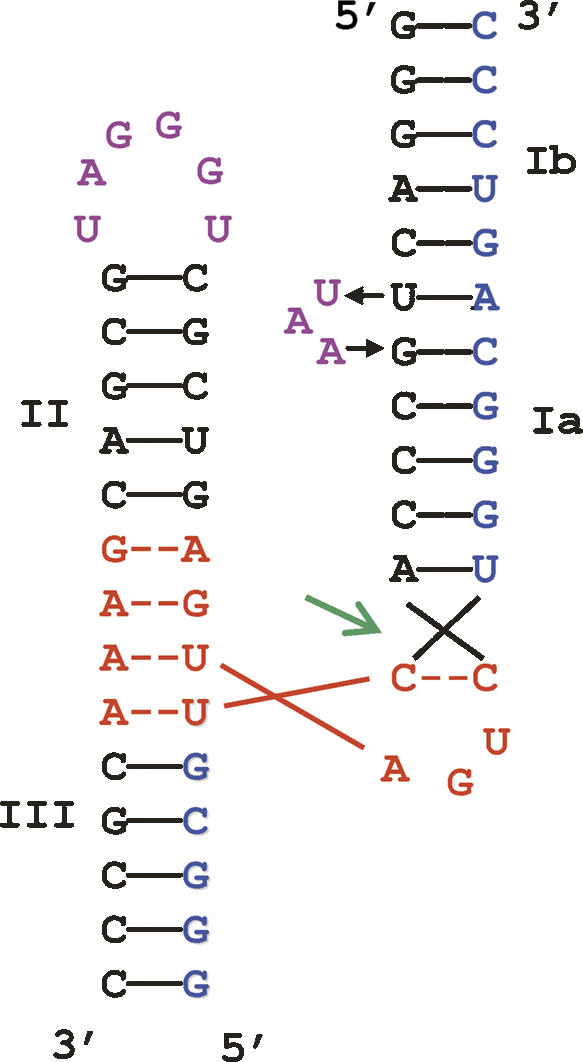
Secondary structure of RzB annealed to an 18-nt substrate (Saksmerprome et al. 2004; Roychowdhury-Saha and Burke 2006). Nucleotides of the catalytic core are shown in red, nucleotides of the substrate strand are shown in blue, and the nucleotides of the tertiary stabilization motifs in Loop II and Bulge I of the ribozyme strand are shown in purple. RzB0 sequence lacks the “UAA” loop.
The present work investigates the ability of RzB to cleave its RNA substrate in the presence of monovalent, polyamine, and exchange-inert trivalent ions. Cleavage activity for the RzB hammerhead ribozyme is improved 20–100-fold in spermidine and Na+ relative to previously published values for minimal hammerhead ribozymes (Dahm and Uhlenbeck 1991; Curtis and Bartel 2001), although catalytic activity in these metal ions was 3–4 orders of magnitude lower than the corresponding activity in Mg2+. The presence of stabilizing peripheral sequences also boosts the activity in the presence of CoHex relative to previously published values (Curtis and Bartel 2001). Together, these observations establish that the ribozyme RzB does not require inner-sphere interactions with divalent ions in order to form its productive tertiary structure, in contrast to the Schistosoma ribozyme (Nelson et al. 2005). At least one site-specific Mg2+ binding event is suggested by competition experiments with CoHex at low concentrations. Our results support the existence of an alternate folding pathway or catalytic mechanism used by the ribozyme in the presence of nondivalent cations (especially CoHex) that is distinct from that promoted by divalent cations.
RESULTS
Spermidine and Na+ promote RzB catalysis in the absence of divalent cations
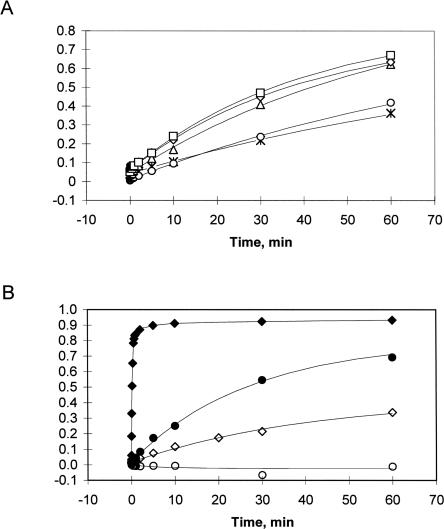
Single-turnover trans-cleavage of a 13-nt RNA substrate was monitored for hammerhead ribozyme RzB as a function of spermidine. The observed rate of cleavage increased with spermidine at low concentrations and saturated at ∼10 mM spermidine with k max = 0.025 ± 0.001 min−1 (Fig. 2A). Previous studies of catalysis by a minimal hammerhead ribozyme in polyamines (Dahm and Uhlenbeck 1991) reported a rate of 0.00024 min−1 in 0.5 mM spermine (a tetra-amine) at 25°C, which is ∼100-fold slower than the rate we observe here in spermidine (a tri-amine). The tertiary stabilization afforded by the peripheral sequence elements likely accounts for these differences in polyamine utilization. This interpretation is supported by the absence of cleavage activity in the control ribozyme lacking tertiary stabilization (ribozyme RzB0), even at 50 mM spermidine after 2 h (data not shown). These kinetic measurements were performed in the presence of 10 μM EDTA to sequester any trace contaminating divalent ions; increasing EDTA concentration to 10 mM did not alter the cleavage activity of RzB. Interestingly, the net cleavage rate (k obs = k cleave + k lig) observed in 1 M Na+ (k obs = 0.037 ± 0.002 min−1) is comparable (Fig. 2B) to the rate observed in 5 mM spermidine. The control ribozyme, RzB0, did not cleave the substrate in 1 M Na+ in 2 h (Fig. 2B). These observations are in contrast to those obtained using extended hammerhead ribozymes, where both the extended version (with tertiary stabilization) and the minimal version (without) show equivalent cleavage rates in 2 M Li+ (Nelson et al. 2005). These differences in ion sensitivities may reflect differences in the nature of the tertiary interactions present in the two different stabilized ribozymes (RzB versus Schistosome natural ribozyme).
FIGURE 2.
(A) Plot of cleavage kinetics of RzB in spermidine: 0.5 mM (asterisks); 1 mM (circles); 5 mM (triangles); 10 mM (diamonds); 20 mM (squares). (B) Plot of cleavage kinetics of RzB (filled symbols) and RzB0 (open symbols) in 1 mM Mg2+ (diamonds) or in 1 M Na+ (circles). Data for Mg2+ are from Roychowdhury-Saha and Burke (2006).
The maximal extent of cleavage by RzB at long incubation times is similar in 5–20 mM spermidine (70%), 1 M Na+ (80%), and 1 mM Mg2+ (>90%) (Fig. 2). Thus, the RzB ribozyme is capable of accessing a catalytically competent conformation in each of these cations. However, the maximal cleavage rate observed in spermidine or Na+ is 10,000-fold lower than the maximal rate of 357 ± 23 min−1 in Mg2+ (10 mM) under otherwise identical conditions (Roychowdhury-Saha and Burke 2006). This dramatic difference in rate may potentially reflect a different mechanism or rate-limiting step. Multichannel reaction pathways involving divalent-dependent and divalent-independent cleavage mechanisms have been noted previously for the HDV ribozyme and for a minimal hammerhead ribozyme (Nakano et al. 2001, 2003; Zhou et al. 2002). Analogous multiple pathways for hammerhead ribozymes may reflect different local architectures of the active site—such as the presence or absence of specific coordination of a metal ion to the scissile phosphate or to a nucleobase—even if the global folded structures of the ribozyme–substrate complex are similar. Such scenarios could affect the energetics of the cleavage reaction and result in dissimilar rates, particularly if only a subset of the cations can play a role that extends significantly beyond gross structural stabilization (see Discussion).
CoHex promotes RzB catalysis
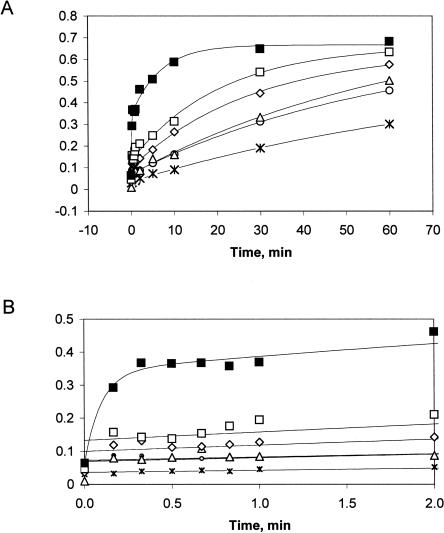
Both inner- and outer-sphere mechanisms have been proposed for metal-ion-mediated catalysis by hammerhead ribozymes. To discriminate between these two possibilities, substrate cleavage by RzB was monitored in the presence of CoHex. The kinetic profile has two distinct phases, with a rapid burst that is complete within the first 10 sec, followed by a slower phase (Fig. 3A,B). In previous work, we (Roychowdhury-Saha and Burke 2006) and others (Canny et al. 2004; Zamel et al. 2004) have used a quench-flow strategy to measure rapid ribozyme cleavage mediated by Mg2+ or transition metal ions. A critical aspect of these experiments is the ability to quench the reaction on millisecond time scales by sequestering the metal ions with EDTA. This approach is not available for assessing RzB cleavage in CoHex, as this exchange-inert metal ion complex cannot be sequestered by EDTA. An alternative quench strategy employing strong denaturing conditions was explored (3 volumes of 99% formamide); however, this strategy was ineffective, as CoHex-promoted cleavage was faster than denaturation of the RzB active structure. Burst phase rates therefore represent lower-limit estimates, and a Hill coefficient for CoHex was not explicitly determined.
FIGURE 3.
(A) Cleavage kinetics of RzB in CoHex: 0.1 mM (asterisks); 0.5 mM (circles); 1 mM (triangles); 5 mM (diamonds); 10 mM (open squares); 20 mM (filled squares). The time courses were fit to a single-exponential equation, except for the data collected at 20 mM, which were fit to a double-exponential equation. (B) Initial time points of the data shown in A through the first 2 min. Single-exponential fit of the kinetic trace in 20 mM CoHex using only the data through the first minute gave a lower limit estimate of ∼8.9 ± 0.9 min−1.
Both the rate of cleavage during the slow phase (assumed to reflect rearrangement from an inert to an active conformation) and the magnitude of the burst fraction (cleavage occurring during the first 10 sec, assumed to reflect the fraction that is prefolded in a catalytically competent conformation) are stimulated by increasing concentrations of CoHex (Fig. 3A,B). In 20 mM CoHex, the burst phase rate (≥8.9 ± 0.9 min−1) is higher than any previously reported for a hammerhead ribozyme in the presence of CoHex. Cleavage rates during the slow phase ranged from 0.012 ± 0.002 min−1 (0.1 mM CoHex) to 0.149 ± 0.027 min−1 (20 mM CoHex). The low end of this range is similar to values obtained previously (0.009 min−1) for the extended Schistosoma hammerhead in 0.1 mM CoHex (Osborne et al. 2005). RzB0 does not show detectable cleavage after 2 h even at CoHex concentrations as high as 20 mM (data not shown), similar to the nonreactivity or poor reactivity observed previously for minimal hammerhead ribozymes (Curtis and Bartel 2001; Osborne et al. 2005).
Competition between Mg2+ and CoHex
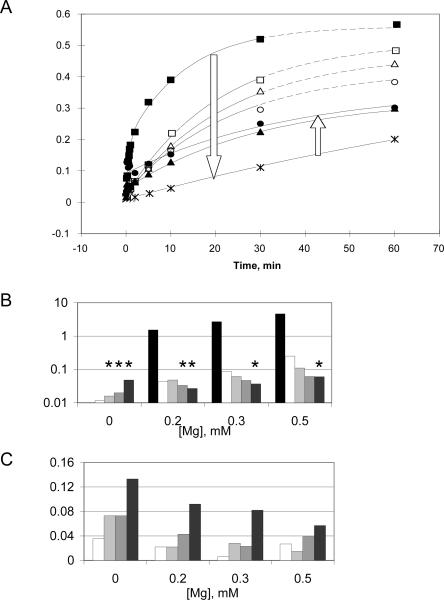
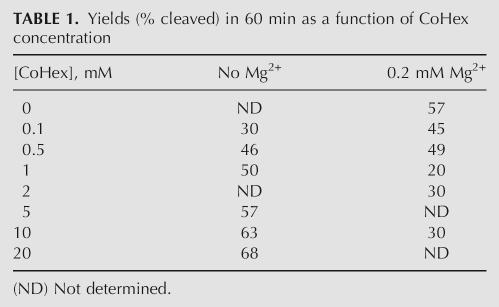
To the extent that Co(NH3)6 3+ acts as a true structural analog of Mg(H2O)6 2+ (similar geometry and size), both ions together should have additive effects, whereas if they promote catalysis via different mechanistic pathways or promote alternative folds, they are expected to exhibit competitive behavior. Therefore, competition assays were performed to address the potential synergy or antagonism in the cleavage mechanisms stimulated by Mg2+ versus CoHex (Fig. 4; Table 1). Cleavage kinetics in a constant background of 0.2 mM Mg2+ were monitored in various concentrations of CoHex (Fig. 4A). The initial rate decreased as CoHex concentration was increased to ∼1.0 mM (Fig. 4B), but at still higher concentrations, cleavage increased markedly and exhibited burst kinetics (Fig. 4A,C). Throughout this series of reactions, increases in the CoHex concentration yielded a concomitant decrease in the fraction cleaved at equilibrium (F ∞) (Fig. 4A). Time courses at 0.3 and 0.5 mM constant Mg2+ showed similar decreases in F ∞ with increasing CoHex (data not shown). In contrast, when no Mg2+ was present, increasing the CoHex concentration increased both the burst phase population and the final equilibrium fraction, F ∞ (cf. Fig. 3). Both of these results suggest differences in the mechanistic details of catalysis by ribozyme RzB in the presence of CoHex versus in the presence of Mg2+ (see Discussion).
FIGURE 4.
(A) Plot of competition assays at 37°C, pH 7.4 in the presence of 0.2 mM Mg2+ only (filled squares, 1.54 min−1) or in the presence of 0.2 mM Mg2+ supplemented with CoHex at concentrations of 0.1 mM (open triangles, 0.043 min−1), 0.5 mM (open squares, 0.048 min−1), 0.75 mM (open circles, 0.042 min−1), 1 mM (asterisks, 0.0071 min−1), 2 mM (filled triangles, 0.033 min−1), or 10 mM (filled circles, 0.031 min−1). k obs was calculated by fitting each time course to a single-exponential equation, except the one in Mg2+ alone, for which a double-exponential equation was used; burst phase noticed in 5 mM (not shown) and 10 mM [Co (NH3)6]3+. Dashed lines are used for the best-fit curves when CoHex concentration was below 1 mM; solid lines are used for 1 mM and above. Arrows show progression of curves as CoHex concentration increased. (B,C) Bar graphs of competition assays under four different Mg2+ concentrations (indicated below the graphs) supplemented with CoHex at concentrations of 0 mM (black); 0.1 mM (white); 0.5 mM (light gray); 2 mM (gray); 10 mM (dark gray). (B) k obs values calculated under different metal conditions. Asterisks (*) indicate conditions under which detectable burst was observed and single-exponential curve fitting was used to estimate the rate during the slow phase. (C) The burst magnitude (F 0) was calculated from curve fitting the kinetic data with a single-exponential equation. A background value of approximately 0.04 was observed, arising from the background radioactivity of the gels and representing the noise in the burst fraction obtained from the curve fit.
TABLE 1.
Yields (% cleaved) in 60 min as a function of CoHex concentration
DISCUSSION
Multichannel catalysis by ribozyme RzB
The catalytic activities reported here for hammerhead ribozyme RzB in the presence of sodium, spermidine, and CoHex confirm and extend previous findings of divalent-ion-independent hammerhead ribozyme catalysis. For ribozyme RzB, the difference between reaction rates observed in divalent ions (Roychowdhury-Saha and Burke 2006) and those observed in Na+ or spermidine (Fig. 2) is likely due to tertiary stabilization in RzB, which could then result in differing kinetic pathways. Previous modeling of minimal hammerhead ribozyme kinetics suggested a multichannel reaction mechanism with Mg2+-dependent, Li+-dependent, and cooperative pathways (Takagi et al. 2004). In those studies, the reaction in divalent ions was ∼20-fold faster than in monovalent ions (Curtis and Bartel 2001). Previous studies of the Mg2+ dependence of substrate cleavage by RzB yielded a Hill coefficient (for catalysis) of 2.0 ± 0.3 (Roychowdhury-Saha and Burke 2006), suggesting a requirement for two metal ions to access the catalytic transition state. CoHex may act in some respects as a true analog of Mg(H2O)n 2+ by replacing its outer-sphere functions of charge neutralization and hydrogen bond donating. However, there are significant differences in the kinetic behavior of Mg2+-mediated and CoHex-mediated reactions that suggest the CoHex-mediated pathway may differ in mechanistic detail from the pathway promoted by Mg(H2O)n 2+. First, the maximal rate attained by RzB in the presence of Mg2+ alone (Roychowdhury-Saha and Burke 2006) is as much as 40-fold higher than that attained in CoHex (357 ± 23 versus ≥ 8.9 ± 0.9 min−1). A caveat here is that because the CoHex rate is a lower-limit value, the actual difference in initial rate could be significantly less. Second, the calculated values of F ∞ at 55 ± 5% for reactions carried out in the presence of CoHex (Fig. 3A) are significantly lower than those seen in Mg2+ or other divalent ions at >90% or in sodium or spermidine at 70%–80% (Fig. 2B; Roychowdhury-Saha and Burke 2006). Thus, the ribozyme is less able to access a catalytically competent conformation in CoHex even at long times. Third, CoHex and Mg2+ compete for binding the ribozyme. Cleavage rates in a constant Mg2+ background initially decrease with increasing CoHex concentrations as CoHex displaces Mg2+ and interferes with Mg2+-mediated catalysis. Still higher concentrations lead to increases in the initial rates and final cleavage extents and to burst kinetics, but the rates and yields do not return to those observed at elevated CoHex in the absence of Mg2+ (Table 1). Thus, CoHex interferes with Mg2+-mediated catalysis and Mg2+ interferes with CoHex-mediated catalysis.
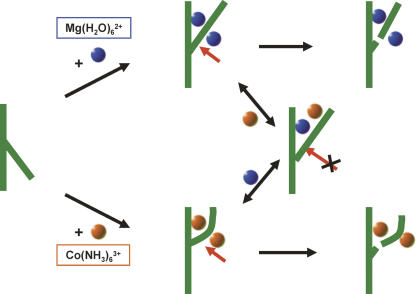
We interpret these data to indicate that hammerhead ribozyme RzB can fold into a Mg2+-dependent or a Mg2+-independent active state in the presence of either metal ion alone, but that simultaneous binding by both metal ions creates an intermediate state with little or no catalytic activity (Fig. 5). Competition between CoHex and Mg2+ ions, with accompanying structural changes, has also been observed in a minimal hammerhead ribozyme (Horton and DeRose 2000).
FIGURE 5.
Multichannel reaction mechanism in tertiary stabilized hammerhead ribozyme in cobalt hexammine and hexahydrated magnesium. Two Mg2+ ions have been shown in accordance with the determination of the Hill coefficient for this ribozyme (Roychowdhury-Saha and Burke 2006). Two CoHex ions are shown as an extrapolation based on the precedent of the Mg(H2O)n 2+ ions.
Mechanistic model for reduced activity in the absence of divalent ions
The “alternate” reaction pathways promoted by Na+, spermidine, and CoHex could be either structural or mechanistic in nature. For example, photo-cross-linking studies (Lambert et al. 2006a,b) suggest a different active site geometry in the hairpin riboyzme in CoHex than in the structures proposed from crystallographic studies in Ca2+ (Rupert and Ferre-D'Amare 2001) or in CoHex (Alam et al. 2005). To the extent that the conformation captured by cross-linking is on a path to catalysis, these results suggest a conformational mobility that may be ion dependent. In addition, mutations near the hairpin catalytic core induced distinctive metal ion dependencies (Alam et al. 2005). In contrast, Neurospora VS ribozyme shows similar conformational changes in both Mg2+ and CoHex during chemical modification structure probing assays, although Mg2+ is more effective than CoHex in inducing important structural changes (Maguire and Collins 2001).
For the present work an attractive alternative is that the “alternate pathways” involve removal of a specific catalytic mechanism that is available to Mg2+ and to transition metal ions but not to monovalent ions, polyamines, or exchange-inert cations. There is significant debate regarding the functional and structural roles of Mg2+ in hammerhead ribozyme catalysis. Magnesium can act as a Lewis acid via inner-sphere contacts or by any of several outer-sphere interactions, such as hydrogen bonding, diffuse Coulombic interactions, or proton transfer. Its potential role in proton transfer has been called into question by recent studies in which simultaneous substitution of G8 and G12 with 2, 6-diaminopurine (diAP; pKa 5.1 versus pKa 9.6 for G) in a minimal hammerhead ribozyme yielded a bell-shaped pH-rate profile, implicating these two nucleobases in both proton transfer steps (Han and Burke 2005). Furthermore, our recent analysis of the pH dependence of substrate cleavage by RzB in various metal ions suggests that the metal ion hydrate is not acting as the general base for the reaction (Roychowdhury-Saha and Burke 2006). However, the pKa of the general acid correlates with the pKa of the metal ion hydrate, displaced to lower values by 1.0–1.7 units (Roychowdhury-Saha and Burke 2006), suggesting that the metal ion may perturb one of the guanosine's pKa by a direct (inner-sphere) coordination of the metal ion to the nucleobase or by proximity of the metal ion's positive charge. The slower catalysis observed in Na+, spermidine, and cobalt hexammine may therefore stem from the decreased ability for general acid protonation of the 5′ oxyanion leaving group due to the inability of these cations to form strong inner-sphere coordination with an active site guanosine.
MATERIALS AND METHODS
Quantification of the ribozyme reaction
Kinetic measurements were performed under ribozyme-saturated, single-turnover conditions with two RNA constructs (RzB and RzB0), each of which was assembled from separate ribozyme and substrate strands. This design ensures that the conversion of the ribozyme–substrate complex to the ribozyme–product complex could be monitored without the complications arising from kinetic processes associated with annealing of the complex. Ribozyme strands were synthesized by in vitro transcription using T7 RNA polymerase from the Epicentre Ampliscribe kit following the supplier's protocol. A chemically synthesized substrate strand (Integrated DNA Technologies) was 5′ radiolabeled using [γ-32P] ATP and polynucleotide kinase. All cleavage reactions were performed at 37°C, pH 7.4. Reactions were carried out with a 2 μM ribozyme strand and a 50 nM substrate strand in 50 mM Tris•HCl and 10 μM EDTA, as described previously (Saksmerprome et al. 2004). Briefly, each reaction was initiated by adding 5 μL of a cation solution to 70 μL of preannealed ribozyme–substrate solution at 37°C, pH 7.4. Aliquots (5 μL) were withdrawn at each time point and quenched with 1–2 volumes of 90% formamide, 50 mM EDTA, and 0.005% each of xylene cyanol and bromophenol blue. Quenched samples were stored at −80°C prior to denaturing (8 M urea) polyacrylamide gel electrophoresis. Metal ion competition experiments were initiated by addition of 5 μL of a mixture of MgCl2 and [Co(NH3)6]Cl3 to 70 μL of preannealed ribozyme–substrate solution at 37°C, pH 7.4. The extent of cleavage in each reaction was quantified using ImageQuant software (Molecular Dynamics). The fraction cleaved (Ft) at time t was fit using KaleidaGraph 3.5 either to a single-exponential equation:
or to a biexponential equation:
where F 0 is the zero point correction, F ∞ is the estimated plateau value at infinite time, and α and (1 − α) are the fractions of the cleaved population that have rate constants of k obs,1 and k obs,2, respectively. Reported errors reflect uncertainties of the Kaleidagraph fit of the data to these equations.
ACKNOWLEDGMENTS
We thank Steven Rhee and Carla Da Costa for their insightful comments on this work. This work was supported by Exobiology award NAG5-12360 from NASA and by an Interdisciplinary Science award from the David and Lucille Packard Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.339207.
REFERENCES
- Akashi, H., Kawasaki, H., Kim, W.J., Akaike, T., Taira, K., Maruyama, A. Enhancement in the cleavage activity of a hammerhead ribozyme by cationic comb-type polymers and an RNA helicase in vitro. J. Biochem. 2002;131:687–692. doi: 10.1093/oxfordjournals.jbchem.a003152. [DOI] [PubMed] [Google Scholar]
- Alam, S., Grum-Tokars, V., Krucinska, J., Kundracik, M.L., Wedekind, J.E. Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn. Biochemistry. 2005;44:14396–14408. doi: 10.1021/bi051550i. [DOI] [PubMed] [Google Scholar]
- Basolo, F., Pearson, R.G. Mechanisms of inorganic reactions. Wiley; New York: 1988. [Google Scholar]
- Butcher, S.E., Allain, F.H.T., Feigon, J. Determination of metal ion binding sites within the hairpin ribozyme domains by NMR. Biochemistry. 2000;39:2174–2182. doi: 10.1021/bi9923454. [DOI] [PubMed] [Google Scholar]
- Canny, M.D., Jucker, F.M., Kellogg, E., Khvorova, A., Jayasena, S.D., Pardi, A. Fast cleavage kinetics of a natural hammerhead ribozyme. J. Am. Chem. Soc. 2004;126:10848–10849. doi: 10.1021/ja046848v. [DOI] [PubMed] [Google Scholar]
- Cho, B.R., Burke, D.H. Topological rearrangement yields structural stabilization and interhelical distance constraints in the Kin.46 self-phosphorylating ribozyme. RNA. 2006;12:2118–2125. doi: 10.1261/rna.173506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, E.A., Bartel, D.P. The hammerhead cleavage reaction in monovalent cations. RNA. 2001;7:546–552. doi: 10.1017/s1355838201002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm, S.C., Uhlenbeck, O.C. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- De la Pena, M., Gago, S., Flores, R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw, D.J., Gait, M.J. Hairpin ribozyme cleavage catalyzed by aminoglycoside antibiotics and the polyamine spermine in the absence of metal ions. Nucleic Acids Res. 1998;26:5551–5561. doi: 10.1093/nar/26.24.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, R.L., Tinoco, I. Solution structure and thermodynamics of a divalent metal ion binding site in an RNA pseudoknot. J. Mol. Biol. 1999;289:1267–1282. doi: 10.1006/jmbi.1999.2841. [DOI] [PubMed] [Google Scholar]
- Hammann, C., Lilley, D.M.J. Folding and activity of the hammerhead ribozyme. ChemBioChem. 2002;3:691–700. doi: 10.1002/1439-7633(20020802)3:8<690::AID-CBIC690>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Hampel, A., Cowan, J.A. A unique mechanism for RNA catalysis: The role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol. 1997;4:513–517. doi: 10.1016/s1074-5521(97)90323-9. [DOI] [PubMed] [Google Scholar]
- Hampel, K.J., Burke, J.M. Solvent protection of the hammerhead ribozyme in the ground state: Evidence for a cation-assisted conformational change leading to catalysis. Biochemistry. 2003;42:4421–4429. doi: 10.1021/bi020659c. [DOI] [PubMed] [Google Scholar]
- Han, J., Burke, J.M. Model for general acid–base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- Heckman, J.E., Lambert, D., Burke, J.M. Photo-cross-linking detects a compact, active structure of the hammerhead ribozyme. Biochemistry. 2005;44:4148–4156. doi: 10.1021/bi047858b. [DOI] [PubMed] [Google Scholar]
- Horton, T.E., DeRose, V.J. Cobalt hexammine inhibition of the hammerhead ribozyme. Biochemistry. 2000;39:11408–11416. doi: 10.1021/bi001141g. [DOI] [PubMed] [Google Scholar]
- Hunsicker, L.M., DeRose, V.J. Activities and relative affinities of divalent metals in unmodified and phosphorothioate-substituted hammerhead ribozymes. J. Inorg. Biochem. 2000;80:271–281. doi: 10.1016/s0162-0134(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Lescoute, A., Westhof, E., Jayasena, S.D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Kieft, J.S., Tinoco, I. Solution structure of a metal-binding site in the major groove of RNA complexed with cobalt (III) hexammine. Structure. 1997;5:713–721. doi: 10.1016/s0969-2126(97)00225-6. [DOI] [PubMed] [Google Scholar]
- Kim, N.K., Murali, A., DeRose, V.J. Separate metal requirements for loop interactions and catalysis in the extended hammerhead ribozyme. J. Am. Chem. Soc. 2005;127:14134–14135. doi: 10.1021/ja0541027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koculi, E., Lee, N.K., Thirumalai, D., Woodson, S.A. Folding of the Tetrahymena ribozyme by polyamines: Importance of counterion valence and size. J. Mol. Biol. 2004;341:27–36. doi: 10.1016/j.jmb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Lambert, D., Heckman, J.E., Burke, J.M. Cation-specific structural accommodation within a catalytic RNA. Biochemistry. 2006a;45:829–838. doi: 10.1021/bi0513709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, D., Heckman, J.E., Burke, J.M. Three conserved guanosines approach the reaction site in native and minimal hammerhead ribozymes. Biochemistry. 2006b;45:7140–7147. doi: 10.1021/bi052457x. [DOI] [PubMed] [Google Scholar]
- Maderia, M., Hunsicker, L.M., DeRose, V.J. Metal-phosphate interactions in the hammerhead ribozyme observed by P-31 NMR and phosphorothioate substitutions. Biochemistry. 2000;39:12113–12120. doi: 10.1021/bi001249w. [DOI] [PubMed] [Google Scholar]
- Maguire, J.L., Collins, R.A. Effects of cobalt hexammine on folding and self-cleavage of the Neurospora VS ribozyme. J. Mol. Biol. 2001;309:45–56. doi: 10.1006/jmbi.2001.4625. [DOI] [PubMed] [Google Scholar]
- Martick, M., Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky, P.J., Takach, J.C., Feig, A.L. Entropy-driven folding of an RNA helical junction: An isothermal titration calorimetric analysis of the hammerhead ribozyme. Biochemistry. 2004;43:5870–5881. doi: 10.1021/bi0360657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, J.B., Seyhan, A.A., Walter, N.G., Burke, J.M., Scott, W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- Nakano, S., Chadalavada, D.M., Bevilacqua, P.C. General acid–base catalysis in the mechanism of a hepatitis δ virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- Nakano, S., Proctor, D.J., Bevilacqua, P.C. Mechanistic characterization of the HDV genomic ribozyme: Assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- Nakano, S., Cerrone, A.L., Bevilacqua, P.C. Mechanistic characterization of the HDV genomic ribozyme: Classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- Nelson, J.A., Shepotinovskaya, I., Uhlenbeck, O.C. Hammerheads derived from sTRSV show enhanced cleavage and ligation rate constants. Biochemistry. 2005;44:14577–14585. doi: 10.1021/bi051130t. [DOI] [PubMed] [Google Scholar]
- Nixon, P.L., Theimer, C.A., Giedroc, D.P. Thermodynamics of stabilization of RNA pseudoknots by cobalt(III) hexaammine. Biopolymers. 1999;50:443–458. doi: 10.1002/(SICI)1097-0282(19991005)50:4<443::AID-BIP9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- O'Rear, J.L., Wang, S., Feig, A.L., Beigelman, L., Uhlenbeck, O.C., Herschlag, D. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA. 2001;7:537–545. doi: 10.1017/s1355838201002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, E.M., Schaak, J.E., DeRose, V.J. Characterization of a native hammerhead ribozyme derived from schistosomes. RNA. 2005;11:187–196. doi: 10.1261/rna.7950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo, J.C., Wilson, T.J., Jayasena, S.D., Khvorova, A., Lilley, D.M. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracchi, A., Beigelman, L., Scott, E.C., Uhlenbeck, O.C., Herschlag, D. Involvement of a specific metal ion in the transition of the hammerhead ribozyme to its catalytic conformation. J. Biol. Chem. 1997;272:26822–26826. doi: 10.1074/jbc.272.43.26822. [DOI] [PubMed] [Google Scholar]
- Pley, H.W., Flaherty, K.M., McKay, D.B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Roychowdhury-Saha, M., Burke, D.H. Extraordinary rates of transition metal ion-mediated ribozyme catalysis. RNA. 2006;12:1846–1852. doi: 10.1261/rna.128906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudisser, S., Tinoco, I. Solution structure of cobalt(III)hexammine complexed to the GAAA tetraloop, and metal-ion binding to G center dot A mismatches. J. Mol. Biol. 2000;295:1211–1223. doi: 10.1006/jmbi.1999.3421. [DOI] [PubMed] [Google Scholar]
- Rueda, D., Wick, K., McDowell, S.E., Walter, N.G. Diffusely bound Mg2+ ions slightly reorient stems I and II of the hammerhead ribozyme to increase the probability of formation of the catalytic core. Biochemistry. 2003;42:9924–9936. doi: 10.1021/bi0347757. [DOI] [PubMed] [Google Scholar]
- Rupert, P.B., Ferre-D'Amare, A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Saksmerprome, V., Roychowdhury-Saha, M., Jayasena, S., Khvorova, A., Burke, D.H. Artificial tertiary motifs stabilize trans-cleaving hammerhead ribozymes under conditions of submillimolar divalent ions and high temperatures. RNA. 2004;10:1916–1924. doi: 10.1261/rna.7159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Suga, H., Cowan, J.A., Szostak, J.W. Unusual metal ion catalysis in an acyl-transferase ribozyme. Biochemistry. 1998;37:10118–10125. doi: 10.1021/bi980432a. [DOI] [PubMed] [Google Scholar]
- Suh, Y.A., Kumar, P.K.R., Taira, K., Nishikawa, S. Self-cleavage activity of the genomic Hdv ribozyme in the presence of various divalent metal ions. Nucleic Acids Res. 1993;21:3277–3280. doi: 10.1093/nar/21.14.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, Y., Inoue, A., Taira, K. Analysis on a cooperative pathway involving multiple cations in hammerhead reactions. J. Am. Chem. Soc. 2004;126:12856–12864. doi: 10.1021/ja031991u. [DOI] [PubMed] [Google Scholar]
- Tuschl, T., Gohlke, C., Jovin, T.M., Westhof, E., Eckstein, F. A three-dimensional model for the hammerhead ribozyme based on fluorescence measurements. Science. 1994;266:785–789. doi: 10.1126/science.7973630. [DOI] [PubMed] [Google Scholar]
- Vogt, M., Hunsicker, L., Cosper, N., Scott, R., DeRose, V.J. Spectroscopic probes for metals and catalysis in the hammerhead ribozyme. Biophys. J. 2003;84:180A–181A. [Google Scholar]
- Wang, S.L., Karbstein, K., Peracchi, A., Beigelman, L., Herschlag, D. Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry. 1999;38:14363–14378. doi: 10.1021/bi9913202. [DOI] [PubMed] [Google Scholar]
- Wedekind, J.E., McKay, D.B. Crystallographic structures of the hammerhead ribozyme: Relationship to ribozyme folding and catalysis. Annu. Rev. Biophys. Biomol. Struct. 1998;27:475–502. doi: 10.1146/annurev.biophys.27.1.475. [DOI] [PubMed] [Google Scholar]
- Yen, L., Svendsen, J., Lee, J.S., Gray, J.T., Magnier, M., Baba, T., D'Amato, R.J., Mulligan, R.C. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- Zamel, R., Poon, A., Jaikaran, D., Andersen, A., Olive, J., De Abreu, D., Collins, R.A. Exceptionally fast self-cleavage by a Neurospora Varkud satellite ribozyme. Proc. Natl. Acad. Sci. 2004;101:1467–1472. doi: 10.1073/pnas.0305753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M., Zhou, D.M., Takagi, Y., Kasai, Y., Inoue, A., Baba, T., Taira, K. Existence of efficient divalent metal ion-catalyzed and inefficient divalent metal ion-independent channels in reactions catalyzed by a hammerhead ribozyme. Nucleic Acids Res. 2002;30:2374–2382. doi: 10.1093/nar/30.11.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]