Abstract
When a eukaryotic mRNA sequence specifying an amino acid motif known as 2A is directly followed by a proline codon, two nonoverlapping proteins are synthesized. From earlier work, the second protein is known to start with this proline codon and is not created by proteolysis. Here we identify the C-terminal amino acid of an upstream 2A-encoded product from Perina nuda picorna-like virus that is glycine specified by the last codon of the 2A-encoding sequence. This is an example of recoding where 2A promotes unconventional termination after decoding of the glycine codon and continued translation beginning with the 3′ adjacent proline codon.
Keywords: codon redefinition, recoding, ribosome, picorna, 2A, StopGo
INTRODUCTION
The 21st amino acid, selenocysteine, is specified by the genetic code through redefinition of the meaning of certain UGA codons that otherwise encode “stop.” In selenocysteine-containing enzymes this unusual amino acid is usually found at the active center where its chemical properties are exploited. Also, redefinition of two stop codons, UAG and UGA, can lead to specification of glutamine and tryptophan, respectively. Here, the important feature is not the identity of the amino acid inserted, but rather it is the synthesis of an extended “readthrough” product. However, in all these cases, the redefinition of codon meaning is in competition with the standard meaning: termination. Where this redefinition is utilized for gene expression, stimulatory signals, often called recoding signals, embedded in the mRNA or in its encoded nascent peptide product, either greatly facilitate or are requisite for, meaningful levels of redefinition. (Redefinition is distinct from the reassigned meaning of certain codons in all the mRNAs in specialized niches such as some mitochondria, and is one facet of reprogrammed genetic decoding, or recoding) (Gesteland and Atkins 1996).
Can the converse occur? Is the meaning of a sense codon ever redefined? This work deals with a unique case where the additional result of decoding a sense codon is unconventional chain termination. This phenomenon occurs in synthesis of a sequence of ∼18 amino acids known as “2A” (see below). It addresses the current model, formulated by Ryan and colleagues (Donnelly et al. 2001a; de Felipe et al. 2003), where a glycine codon within a 2A sequence specifies the expected amino acid but also promotes subsequent termination of translation of that chain with continued translation resulting in the next encoded amino acid becoming the N-terminal residue of a separate downstream polypeptide (Donnelly et al. 2001a; de Felipe et al. 2003). This model prompts the use of the term “StopGo” to refer to “2A” action. There is no evidence that the C terminus of the product encoded upstream and the N terminus of the downstream-encoded product are ever joined by a peptide bond.
Foot and mouth disease virus (FMDV) has an IRES-mediated translation start site followed by a single long open reading frame (ORF) from which multiple products are derived, including the leader peptide, the capsid proteins, the polymerase/replicase enzymatic functions, and three VPg proteins, which play a role in viral replication and translation (Fig. 1). The last 18 codons of the capsid-encoding region specify a sequence termed 2A, whose activity depends on the next amino acid being proline. A cassette with these 19 codons (encoding 2A plus proline), separating fused reporter-encoding sequences, functions in heterologous eukaryotic systems, including wheat germ, to efficiently generate the downstream reporter with an N-terminal proline specified by the last of the 19 codons (Ryan and Drew 1994). Generation of the N-terminal proline product is cotranslational (Ryan and Drew 1994; de Felipe et al. 2003), consistent with earlier work on a Cardiovirus (Jackson 1986), and does not occur when the cassette is expressed in prokaryotes (Donnelly, et al. 1997).
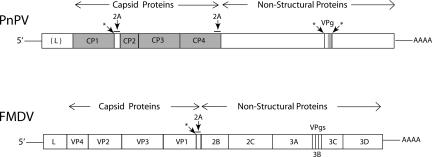
FIGURE 1.
Comparison of the genome organization of Perina nuda picorna-like virus (PnPV) and foot and mouth disease virus (FMDV). The protein products of the single open reading frame are shown as separate boxes. The shaded regions were identified by intact or digested protein analysis of PnPV virions. “L” denotes the leader peptide. Regions encoding 2A sequences are overlined and indicated by arrows. Putative virally encoded protease cleavage sites in PnPV are indicated by “*”. Only one of the virally encoded protease cleavage sites, *, is indicated in FMDV. CP1–4 denotes the four PnPV capsid proteins and VP1–4 denotes the four FMDV capsid proteins.
RNA cleavage is not involved in synthesis of 2B (Donnelly et al. 2001a). Although a cricket paralysis virus-like IRES could give methionine-independent initiation (Sasaki and Nakashima 2000; Jan et al. 2001), there is no hint of the involvement of an IRES for specific initiation of 2B. (Interestingly, in this case it is the amino acid sequence encoded before the proline rather than the nucleotide sequence itself that is important for generation of the N-terminal proline product) (Donnelly et al. 2001b).
The 3′ ends of Aphthovirus (e.g., FMDV) and Cardiovirus sequences that specifiy 2A have a consensus sequence encoding the following amino acids: D-x-E-x-N-P-G, and is immediately followed by the proline codon that encodes the first amino acid of 2B (Hahn and Palmenberg 1996; Donnelly et al. 1997). Introduction of point mutants affecting amino acid identities showed its importance for 2A activity (Donnelly et al. 2001b; Ryan et al. 2002). (2A action stands in contrast to situations where specific peptides traversing the exit tunnel cause inhibition of release factor-dependent peptidyl-tRNA hydrolysis in the peptidyl-transferase center [Cao and Geballe 1996; Cruz-Vera et al. 2006].)
Single ORFs have been created comprising two reporter genes flanking 2A-encoding sequences. When such constructs are analyzed using in vitro translation systems, termination before the N-terminal proline of the downstream reporter results in a molar excess of the N-terminal reporter compared to the C-terminal reporter, further suggesting that a nonproteolytic mechanism is involved (Donnelly et al. 2001a). SDS-PAGE analyses of products produced by 2A-mediated translation are consistent with the sizes expected for products, and in particular, with each upstream encoded product having its C-terminal amino acid as glycine corresponding to the last residue of the 2A consensus sequence (in bold, above). However, size estimations using SDS-PAGE are not nearly sensitive enough to deduce the identity of the C-terminal amino acid of 2A. Whether or not this C-terminal amino acid is glycine is critical to know for assessing the StopGo model. The biotechnological importance of 2A activity (de Felipe et al. 2006), as well as its scientific interest, including the possibility that it may constitute a novel type of recoding, reprogrammed genetic decoding (Baranov et al. 2002), merits the investigation, reported here, of the identity of the C-terminal amino acid of the upstream-encoded product.
2A-encoding sequences occur in some diverse genes, although nearly all known active 2A-like sequences are from viruses (some occur in non-LTR retrotransposons found in the genomes of a range of species of the parasitic protozoan Trypanosomai) (Heras et al. 2006). The approach taken here was to identify a 2A-encoding viral sequence where the upstream- and the downstream-encoded products are present in the virion and, in theory, readily available from a natural source for accurate sequence characterization by mass spectrometry. The single-stranded RNA Perina nuda picorna-like virus, PnPV, which infects a moth, has these features and also has an additional 2A-encoding sequence where only the upstream-encoded product is present in the virion (Fig. 1; Wu et al. 2002). This virus can be readily propagated in a homologous insect cell line (Wu et al. 2002).
The PnPV genome has a single ORF that has 2986 codons and does not generate subgenomic RNAs (Wu et al. 2002). It is 89% identical at the amino acid level to Ectropis obliqua picorna-like virus (Wang et al. 2004; Lu et al. 2006), and less so (25%) to Infectious flacherie virus (Isawa et al. 1998). From the 5′ end of the PnPV ORF there are 319 codons in the position corresponding to the 202 codons in FMDV that encode its leader protease. In both viruses this is just 5′ of the sequence specifying the four capsid coat proteins, which in PnPV are termed CP 1–4. The enzymatic functions, including the polymerase, are encoded 3′ of CP4 (Fig. 1). The first PnPV 2A-encoding RNA sequence, codons 556–573, with the potential to code for QGWVPDLTVDGDVESNPG is at the 3′ end of the cp1 gene and the first codon, 574, of the adjacent cp2 gene specifies proline. This proline was confirmed by N-terminal sequence analysis (Wu et al. 2002). The second PnPV 2A-encoding RNA sequence, codons 1173–1190, with the potential to code for GGGQKDLTQDGDIESNPG, is in the vicinity of the predicted 3′ end of the cp4 gene and the 3′ adjacent codon, 1191, is a proline codon (Wu et al. 2002). N-terminal sequencing of PnPV virion proteins was previously performed by SDS-PAGE fractionation of PnPV virions followed by transfer to PVDF membranes (Wu et al. 2002). These results defined the beginnings of the four coat protein regions. The N terminus of CP2 is PFLSGLLGTV (Wu et al. 2002), consistent with the generation of a new N terminus encoded by the proline codon following the codons for the 2A sequence at the 3′ end of cp1. The primary goal of the present work, therefore, was to determine the C-terminal amino acids of either CP1 or CP4 by mass spectrometry analysis of protein digests and full-length proteins to constrain models for 2A action.
RESULTS
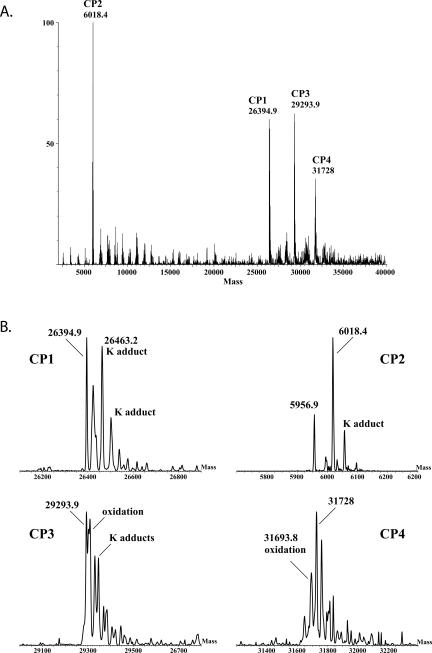
PnPV virions, purified as previously described (Wu et al. 2002), were digested with either trypsin or chymotrypsin, and the resulting peptides were analyzed by LC/MS/MS (with Fourier transform-ion cyclotron resonance [FT-ICR]). Complete sequence coverage was obtained for CP2 and CP3, and nearly complete coverage was obtained for CP1 and CP4 (Fig. 2). From the enzymatic digest analyses, all of the N-terminal and C-terminal peptides were identified for the four CP proteins. Of particular interest is the C terminus of CP1. Is it glycine encoded by codon 573, as predicted by the current model of 2A activity? By analysis of the peptide masses compared to that predicted from the RNA sequence, the C-terminal amino acid of CP1 is not glycine, but glutamine encoded by codon 556. Peptides containing amino acids encoded by codons 557–573 (GWVPDLTVDGDVESNPG) were not detected in the protein digests of isolated capsid components or in the virion sample, as analyzed by LC/MS/MS. The downstream translated protein CP2, however, begins with proline, consistent with previously described products of 2A activity. The peptide coverage for CP2 was complete.
FIGURE 2.
Analysis of PnPV virion protein digests by FT-ICR. The single 2986 amino acid ORF of PnPV is depicted in sections. Underlined sequences were identified in tryptic or chymotryptic digests of virions. The two 2A sequences are italicized and the most crucial residues for “StopGo” are shown in white with black shading. Putative viral protease recognition sequences are shaded in gray. “pS” indicates phosphorylation on S 1187. The bold “Y” is the residue expected to link the putative VPg to the 5′ end of PnPV RNA.
Complete sequence coverage of CP3 was also shown, beginning at codon 638, aspartic acid, and ending at codon 906, methionine. Peptide coverage of CP4 was almost complete except for a few internal peptides (Fig. 2). The N-terminal peptide begins with codon 907, glycine. It was noted that the amino acid sequences at the ends of the leader peptide, CP1 and CP3, ending with VTAQ, VTAQ, and VTAM, respectively, are similar with a consensus sequence of VTAN. In these three cases, the next amino acid encoded is glycine, similar to recognition sequences of picorna viral cysteine proteases that generally cleave between Q or E and G, A, or S (Hellen et al. 1989).
The C-terminal region of CP4 includes the 2A-encoded amino acids and ends at codon 1190, glycine (Fig. 2). In contrast to CP1, the entire 2A-encoded sequence was found to be present at the C terminus of CP4. In addition, analysis of PnPV by LC/MS/MS of trypsin or chymotrypsin digests revealed a post-translational phosphate modification of serine residue 1187. This was confirmed from identification of several different phosphopeptides containing this same serine residue (i.e., from trypsin digestion, DLTQDGDIEpSNPG; from chymotrypsin digestion, IIGGGQKDLTQDGDIEpSNPG, NIIGGGQKDLTQDGDIEpSNPG, and RRQNIIGGGQKDLTQDGDIEpSNPG). The phosphoserine is near the C-terminal end of CP4 and within the second 2A region. The corresponding nonphosphorylated peptides were also identified, indicating that the phosphorylation is either relatively labile or the extent of phosphorylation is incomplete.
Mass spectrometry analysis of the PnPV virus digests also clearly showed the presence of a peptide encoded 3′ of the CP1–4 region (K)VNFVQTTPVVVAAK- (amino acids 2210–2223). The adjacent downstream amino acid in the polyprotein precursor is glycine. This peptide maps to a region (3B), just N terminal to the putative viral protease (3C) where VPg is found in members of the Picornaviridea (Fig. 1). In Poliovirus and other members of the Picornaviridae, VPg (3B) (Rueckert 1990) is released from the polyprotein by two cleavages at glutamine-glycine amino acid pairs (Nicklin et al. 1986). It is likely that the C terminus of the PnPV peptide was similarly generated by the action of the viral protease since it ends with VAAK and is followed by glycine, which is broadly similar to the other PnPV protease cleavage sites (VTAQ/G in both the putative leader and CP1 and VTAM/G in CP3). The N terminus of the identified peptide was generated by trypsin cleavage at (K)V. In further support, the peptide (F)VQTTPVVVAAK was identified from the chymotryptic digest LC/MS/MS data, supporting that this is the C-terminal peptide. Another putative viral protease recognition sequence is encoded by codons 2191–2195, VEAQ/A, upstream of the detected peptide. Utilization of both protease cleavage sites would generate a 29 amino acid peptide similar to, but larger than, VPgs of studied picornaviruses, that has a critical tyrosine residue near its N terminus. (The tyrosine is involved in the linkage of VPg to the 5′ end of the genomic RNA.) Similar to poliovirus VPg (Reuer et al. 1990), the putative PnPV VPg is preceded by a predicted transmembrane domain, FILKNVMLAVGAVMLAYKVY (amino acids 2162–2181). Therefore we conclude that the peptide identified by mass spectrometry most likely represents the C terminus of the VPg protein of PnPV. The LC/MS/MS data from tryptic or chymotryptic digestion of PnPV was also searched for other possible modifications: uridylylation (on serine, threonine, and tyrosine residues) (Richards et al. 2006) and myristoylation (on lysine and N-terminal glycine residues) (Moscufo et al. 1991) in the entire PnPV sequence, and neither of these modifications was identified.
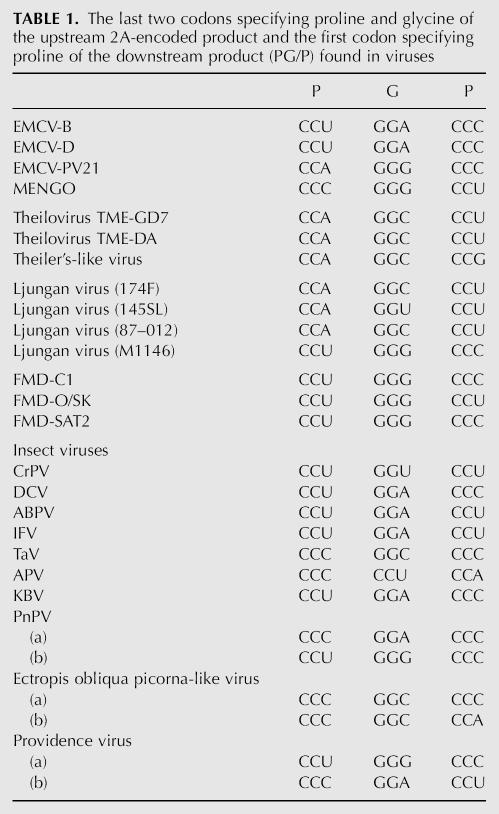
The intact proteins of PnPV were also analyzed by electrospray ionization mass spectrometry (ESI/MS). Figure 3 shows the neutral molecular-mass spectrum obtained by electrospray ionization of the entire virus capsid (i.e., not digested), in which molecular masses corresponding to intact CP1, CP2, CP3, and CP4 proteins are indicated. The molecular mass of intact CP1 at 26,394.9 Da is consistent with the CP1 sequence beginning with glycine at the N terminus (amino acid 320) and ending with glutamine at the C terminus (amino acid 556). This supports the conclusion from the protein digest analysis that the predicted amino acids 557–573 are not found in the mature CP1 protein. The measured mass of 26,394.9 Da is ∼33 Da larger than the theoretical mass of 26,361.7 Da. Correspondingly, the observed complexity of higher mass molecular peaks in roughly 32 Da increments suggests that minor-level oxidation or other modifications existed in the protein as analyzed.
FIGURE 3.
Electrospray molecular-mass spectra. (A) The mass spectrum of denatured virions showing the four major coat proteins, CP1–4. (B) Expanded views of the regions around the major peaks for CP1–4. Potassium (K) adducts and oxidation products are indicated. Oxidation of cysteine, proline, methionine, or other residues in the protein may have occurred from storage of the virus sample, exposure to reagents, reaction with degraded β-mercaptoethanol in solution, or may be due to post-translational modifications.
The molecular mass of intact CP2, determined to be 6018.4 Da, is in agreement with the theoretical mass of 6019.0 Da (average-isotope composition), with no apparent post-translational modifications present. There is also a protein at 5956.9 Da present that does not result from simple cleavage of any of the CP proteins, and its source was not determined. CP3 yielded an intact molecular mass of 29,293.9 Da, which is consistent with the theoretical mass of 29,294.2 Da, and some degree of oxidation or minor modification of the protein is apparent from the presence of higher mass peaks (e.g., 29,345 Da).
The analysis of intact CP4 shows a molecular mass at 31,693.8 Da (theoretical mass of 31,662.9) that is consistent with an oxidized form of the protein. However, similar to CP1, the complexity of molecular species indicates other modifications of the protein may also be present. Taken together with the unequivocal protein digest data, we conclude that CP4 ends with glycine encoded by codon 1190 at the end of the 2A sequence.
DISCUSSION
One of the goals of this work was to identify the carboxy-terminus of a protein containing a 2A site. There were two candidates in the PnPV virion, CP4 and CP1. Identification of peptides by LC/MS/MS with FT-ICR showed that the coding region of CP4 extends to the terminal glycine, encoded by codon 1190, of the 2A sequence. Analysis of full-length virion proteins by ESI/MS indicated that CP4 is encoded by codons 907–1190. The peptide and full-length protein data for CP1 showed that it is encoded by codons 320–556 rather than the expected 320–573. There would have been 17 additional amino acids if all of 2A had been present. PnPV cp1 specifies a potential virally encoded cysteine protease cleavage site ending 17 codons before the 3′ end of a sequence specifying 2A, which likely accounts for the observed truncation.
What is the role of the 2A?
It could be that the selective advantage of a 2A-encoding sequence at the 3′ end of cp1 is to permit a step-down in the level of products encoded 3′ as has been observed with reporter constructs using the FMDV 2A sequence (Donnelly et al. 2001a,b). However, Donnelly et al. did not observe this step-down of expression in the native FMDV polyprotein context, implying that there are important sequence features in addition to 2A that are contained in the 14 amino acids preceding the 2A sequence (Donnelly et al. 2001b). A step-down is not evident from the proportion of CP1 and CP3 from SDS-PAGE analysis of PnPV virions (data not shown; Wu et al. 2002). It could be that before the viral protease is synthesized, precursor CP1 containing the C-terminal 2A sequence has a nonstructural role, e.g., a role in translation/replication regulation. If so, this form of CP1 would not be detected in this analysis since it would not be expected to be incorporated into virions. Yet another possibility for 2A function is to cause a pause in translation (Donnelly et al. 2001a) allowing relevant RNA or protein folding(s) to occur. Resolution of this issue will require experimental investigation.
Interestingly, analysis of FMDV capsids showed a corresponding viral protease cleavage that removed most of the 2A-encoded sequence at the 3′ end of the terminal coat protein gene with the protein ending with Q (Ryan et al. 1989). Extrapolating from the now established C terminus and the viral genome sequence encoding 2A, the released peptide is predicted to be (Q)LLNFDLLKLAGDVESNPG(P), i.e., one amino acid longer than its PnPV counterpart (M.D. Ryan, unpubl.).
Mechanism of 2A action
Our results, together with published results, support the model postulated by Ryan and colleagues (Donnelly et al. 2001a) and here termed “StopGo.” The model is that a sense codon specifies the C-terminal amino acid of one protein and a 3′ adjacent sense codon specifies a non-methionine N-terminal amino acid of a separate protein. Previous work with a 2A sequence flanked by reporter genes showed that proteolysis is not involved (Ryan and Drew 1994) and that, in a nascent peptide sequence-dependent manner, cotranslational generation of the two products results in relatively more of the upstream-encoded product. The possibility that peptidyl-tRNA containing the upstream encoded product “drops off” from the ribosome prior to hydrolysis has not been directly tested, but how this could happen with retention of tRNAGly in the A-site for subsequent synthesis of the downstream-encoded product is far from obvious. We will address here the intraribosomal peptidyl-tRNA hydrolysis model of Donnelly et al. (2001a).
There is only one directly encoded protein residue, proline, which is an imino acid. Its φ dihedral angle is constrained by its pyrrolidine ring to ∼60°. In the 2A sequence, it is the penultimate amino acid in the nascent chain and would be expected to distort the positioning of the relevant part of the nascent peptide. The C-terminal amino acid is glycine, which forms a flexible peptide bond and is expected to allow the peptide at this position to be less constrained. An understanding of the significance of the properties of these two amino acids and how the key amino acids encoded upstream affect the phenomenon will require detailed structural knowledge. In addition to the importance of these amino acids and the ribosome, the CCA end of tRNA is doubtless also critical (Weinger et al. 2004; Feinberg and Joseph 2006) and can have effects on codon:anticodon dissociation (O'Connor et al. 1993).
Protein synthesis is dependent on reaction of the α-amino group of aminoacyl-tRNA bound to the ribosomal A-site with the ester carbon of peptidyl-tRNA bound in the P-site (Green and Lorsch 2002). Pro-tRNAPro acts as a particularly poor nucleophile (as does the glycine counterpart) (Nathans and Neidle 1963; Rychlík et al. 1970). Continuation of standard protein synthesis in response to an accepted A-site aminoacyl-tRNA involves ribosomal RNA undergoing movements that reorient the ester linkage, making it accessible for attack (Schmeing et al. 2005a,b). Before the next tRNA is accepted into the A-site, the peptidyl-transfer center positions the ester linkage so that it is not accessible to nucleophilic attack by water with resulting deacylation of peptidyl-tRNA as happens when a release factor is accepted in an A-site containing a stop codon. The exact features that can reprogram the ribosome so there is hydrolysis of peptidyl-tRNAGly, but with continued synthesis resulting in the proline acylated to the next A-site tRNA and becoming the N-terminal amino acid of a separate chain, are not understood.
Despite the rRNA differences between prokaryotic and eukaryotic ribosomes, other studies in Escherichia coli are pertinent. A strong regulatory pause occurs during translation of E. coli secM mRNA until the N terminus of the nascent chain is “pulled” by the secretory apparatus. The pause is mediated by specific amino acids encoded upstream of the pause site interacting with specific 23S rRNA nucleotides and ribosomal protein L22 that are located at the constriction of the nascent peptide exit tunnel through the large ribosomal subunit (Nakatogawa and Ito 2002). This interaction signal is transduced to the peptidyl-transfer center (Mitra et al. 2006; Woolhead et al. 2006). The strong pause occurs with cognate peptidyl-tRNAGly at the ribosomal P-site and cognate aminoacyl-tRNAPro accepted in the ribosomal A-site (Muto et al. 2006). Although the block occurs before transfer of the peptide chain to Pro tRNAPro, having that tRNA in the A-site is critical for the process (Muto et al. 2006) (and, in addition, its presence inhibits tmRNA accessibility) (Garza-Sánchez et al. 2006). The specific ribosomal changes in response to the nascent peptide signal in this case do not remove the protection of the ester linkage of the peptidyl-tRNAGly from P-site nucleophilic attack by water with resulting deacylation. The distinctions that cause deacylation with 2A remain to be determined (uncharged tRNA in the A-site triggers peptidyl-tRNA deacylation but the tRNA in this case is, of course, acylated). The relevant glycine codon in the first 2A-encoding sequence in PnPV RNA is GGA, and in the second it is GGG. In 2A sequences in other genomes (Table 1) or in tested mutants, there is no preferred glycine codon. Since no restriction to GGG codons is evident, the situation is different from drop-off in E. coli, which has been reported to occur preferentially at NGG sequences (Gonzalez de Valdivia and Isaksson 2005). Although some detailed studies of ribosome binding with esterified amino acids have been performed (Fahlman and Uhlenbeck 2004), and there are also early reports on preferential hydrolysis (Hentzen et al. 1972), we are not aware of data directly relevant to the 2A issue.
TABLE 1.
The last two codons specifying proline and glycine of the upstream 2A-encoded product and the first codon specifying proline of the downstream product (PG/P) found in viruses
Studies in E. coli have shown that, in a cooperative manner, the identity of the last two amino acids in the nascent chain influence A-site decoding, with proline and glycine, in particular, permitting elevated suppressor reading of stop codons (Björnsson et al. 1996; Mottagui-Tabar and Isaksson 1997). This effect is sensitive to the characteristics of the A-site tRNA (Mottagui-Tabar et al. 1994). Supportive results come from studies of tmRNA tagging. In certain contexts, with either P-site peptidyl-tRNAPro or peptidyl-tRNAGly, there are elevated levels of tmRNA tagging on full-length proteins with an A-site stop codon (Hayes et al. 2002; Sunohara et al. 2002). Formally, this could be due directly to facilitated tmRNA entry, but all the evidence points to partial inhibition of release factor access, thereby favoring tmRNA in its competition with the release factor.
Specification of the C-terminal amino acid of a protein by a codon that is in-frame, and 5′ adjacent to, a codon that specifies the N terminus of a separate protein is the opposite of tmRNA action where adjacent amino acids in a protein are specified from two different mRNAs. Both phenomena broaden the repertoire of recoding and exemplify the sophisticated layers of decoding revealed in the past decade. Once the exciting characteristics of tmRNA became known (Keiler et al. 1996), its name changed from 10Sa, which was not informative (Atkins and Gesteland 1996; Jentsch 1996). We propose that the “2A” phenomenon (minimally requiring 2A followed by a proline codon) be named “StopGo,” as has been adopted in this paper, or some appealing alternative that reflects function. This is especially relevant since “extra-ribosomal” proteases of other picornaviruses (coxsackievirus, rhinoviruses, enteroviruses, and poliovirus) that mediate post-translational polyprotein cleavages, as well as a cellular serine–threonine phosphatase (Cho and Xu 2007), are also known as 2A.
MATERIALS AND METHODS
For analysis by LC/MS/MS, virus samples were digested with TPCK-modified trypsin (Promega) or chymotrypsin (Princeton). Most digests were performed for 2–4 h, but different digest time periods were performed, ranging from 5 min to overnight. Proteolytic enzymes (in 50 mM ammonium bicarbonate) were added to virus protein in ∼1:25 ratio (enzyme to virus). LC/MS/MS data were acquired using a LTQ-FT hybrid mass spectrometer (ThermoElectron Corp). Peptide molecular masses were measured by FT-ICR, and all peptides assigned in this work had mass errors <3 ppm. Peptide sequencing was performed by collision-induced dissociation (CID) in the linear ion trap of this hybrid instrument. Digest samples were introduced by nanoLC (Eksigent, Inc.) with nano-electrospray ionization (ThermoElectron Corp). NanoLC was performed using a homemade C18 nanobore column (75 μm i.d. × 10 cm; Atlantis C18 [Waters Corp.]; 3 μm particle). Peptides were eluted from a 50-min linear gradient run from 4% acetonitrile (with 0.1% formic acid) to 70% acetonitrile (with 0.1% formic acid).
All identified peptides from protein digests were assigned from MSBD, NCBI, or custom PnPV protein database searches, using in-house processing with the MASCOT search engine (Matrix Science, Inc.). All identified peptides (including the phosphopeptides) showed MASCOT scores >20 and had mass errors <3 ppm to be acceptable.
To achieve successful molecular weight measurements of intact viral capsid proteins by ESI/MS, several methods were attempted to denature the viral capsid prior to ESI/MS analysis. The most effective method of denaturing the capsid was a brief exposure to chymotrypsin (5 min in 50 mM ammonium bicarbonate, 1:25 ratio of enzyme to protein). Following the chymotrypsin treatment, the protein sample was immediately purified using a C18 Ziptip (Millipore), with elution of the proteins into three 1 μL aliquots of 60% acetonitrile and 2% formic acid and one aliquot of 98% acetonitrile and 2% formic acid. ESI/MS analysis of the intact proteins was performed using a Quattro-II mass spectrometer (Micromass, Inc.). The eluent from Ziptip purification was infused at 3 μL/min for ESI/MS. Data were acquired with a cone voltage of 50 eV, a spray voltage of 2.8 kV, and the instrument was scanned from 800 to 1400 m/z in 4 sec. Scans were accumulated for ∼1 min, the spectra were combined, then deconvoluted into molecular-mass spectra (i.e., processed into neutral molecular weight) using MaxEnt software (Micromass, Inc.).
ACKNOWLEDGMENTS
We thank Dr. P.V. Baranov for earlier discussions, Dr. R.F. Gesteland for prompting our interest in viral capsid proteomics, and both for comments on the manuscript. This work was supported by an award from Science Foundation Ireland, The National Science Council of the Republic of China under Grant 95N 1302 (2), NIH Grants GM071853 and GM079523, and support from the BBSRC and Wellcome Trust.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.487907.
REFERENCES
- Atkins, J.F., Gesteland, R.F. A case for trans translation. Nature. 1996;379:769–771. doi: 10.1038/379769a0. [DOI] [PubMed] [Google Scholar]
- Baranov, P.V., Gesteland, R.F., Atkins, J.F. Recoding: Translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- Björnsson, A., Mottagui-Tabar, S., Isaksson, L.A. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J. 1996;15:1696–1704. [PMC free article] [PubMed] [Google Scholar]
- Cao, J., Geballe, A.P. Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell. Biol. 1996;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, U.S., Xu, W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- Cruz-Vera, L.R., Gong, M., Yanofsky, C. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. 2006;103:3598–3603. doi: 10.1073/pnas.0600082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe, P., Hughes, L.E., Ryan, M.D., Brown, J.D. Co-translational, intraribosomal cleavage of polypeptides by the foot-and-mouth disease virus 2A peptide. J. Biol. Chem. 2003;278:11441–11448. doi: 10.1074/jbc.M211644200. [DOI] [PubMed] [Google Scholar]
- de Felipe, P., Luke, G.A., Hughes, L.E., Gani, D., Halpin, C., Ryan, M.D. E unum pluribus: Multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006;24:68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Donnelly, M.L.L., Gani, D., Flint, M., Monoghan, S., Ryan, M.D. The cleavage activity of aptho- and cardiovirus 2A proteins. J. Gen. Virol. 1997;78:13–21. doi: 10.1099/0022-1317-78-1-13. [DOI] [PubMed] [Google Scholar]
- Donnelly, M.L.L., Luke, G., Mehrotra, A., Li, X., Hughes, L.E., Gani, D., Ryan, M.D. Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction but a novel translational effect: A putative ribosomal “skip.”. J. Gen. Virol. 2001a;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Donnelly, M.L.L., Hughes, L.E., Luke, G., Mendoza, H., ten Dam, E., Gani, D., Ryan, M.D. The “cleavage” activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring “2A-like” sequences. J. Gen. Virol. 2001b;82:1027–1041. doi: 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- Fahlman, R.P., Uhlenbeck, O.C. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P and A sites. Biochemistry. 2004;43:7575–7583. doi: 10.1021/bi0495836. [DOI] [PubMed] [Google Scholar]
- Feinberg, J.S., Joseph, S. A conserved base pair between tRNA and 23S rRNA in the peptidyl transferase center is important for peptide release. J. Mol. Biol. 2006;364:1010–1020. doi: 10.1016/j.jmb.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Garza-Sánchez, F., Janssen, B.D., Hayes, C.S. Prolyl-tRNAPro in the A site of secM-arrested ribosomes inhibits the recruitment of transfer-messenger RNA. J. Biol. Chem. 2006;281:34258–34268. doi: 10.1074/jbc.M608052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland, R.F., Atkins, J.F. Recoding: Dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- Gonzalez de Valdivia, E.I., Isaksson, L.A. Abortive translation caused by peptidyl-tRNA drop-off at NGG codons in the early coding region of mRNA. FEBS J. 2005;272:5306–5316. doi: 10.1111/j.1742-4658.2005.04926.x. [DOI] [PubMed] [Google Scholar]
- Green, R., Lorsch, J.R. The path to perdition is paved with protons. Cell. 2002;110:665–668. doi: 10.1016/s0092-8674(02)00965-0. [DOI] [PubMed] [Google Scholar]
- Hahn, H., Palmenberg, A.C. Mutational analysis of the encephalomyocarditis virus primary cleavage. J. Virol. 1996;70:6870–6875. doi: 10.1128/jvi.70.10.6870-6875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, C.S., Bose, B., Sauer, R.T. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J. Biol. Chem. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- Hellen, C.U., Krausslich, H.G., Wimmer, E. Proteolytic processing of polyproteins in the replication of RNA viruses. Biochemistry. 1989;28:9881–9890. doi: 10.1021/bi00452a001. [DOI] [PubMed] [Google Scholar]
- Hentzen, D., Mandel, P., Garel, J.-P. Relation between aminoacyl-tRNA stability and the fixed amino acid. Biochim. Biophys. Acta. 1972;281:228–232. doi: 10.1016/0005-2787(72)90174-8. [DOI] [PubMed] [Google Scholar]
- Heras, S.R., Thomas, M.C., Garcia-Canadas, M., de Felipe, P., Garcia-Pérez, J.L., Ryan, M.D., López, M.C. L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain a functional viral-like self-cleaving 2A sequence in frame with the active proteins they encode. Cell. Mol. Life Sci. 2006;63:1449–1460. doi: 10.1007/s00018-006-6038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isawa, H., Asano, S., Sahara, K., Iizuka, T., Bando, H. Analysis of genetic information of an insect picorna-like virus, infectious flacherie virus of silkworm: Evidence for evolutionary relationships among insect, mammalian and plant picorna (-like) viruses. Arch. Virol. 1998;143:127–143. doi: 10.1007/s007050050273. [DOI] [PubMed] [Google Scholar]
- Jackson, R.J. A detailed kinetic analysis of the in vitro synthesis and processing of encephalomyocarditis virus products. Virology. 1986;149:114–127. doi: 10.1016/0042-6822(86)90092-9. [DOI] [PubMed] [Google Scholar]
- Jan, E., Thompson, S.R., Wilson, J.E., Pestova, T.V., Hellen, C.U., Sarnow, P. Initiator Met-tRNA-independent translation mediated by an internal ribosome entry site element in cricket paralysis-like insect viruses. Cold Spring Harb. Symp. Quant. Biol. 2001;66:285–292. doi: 10.1101/sqb.2001.66.285. [DOI] [PubMed] [Google Scholar]
- Jentsch, S. When proteins receive deadly messages at birth. Science. 1996;271:955–956. doi: 10.1126/science.271.5251.955. [DOI] [PubMed] [Google Scholar]
- Keiler, K.C., Waller, P.R., Sauer, R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Lu, J., Zhang, J., Wang, X., Jiang, H., Liu, C., Hu, Y. In vitro and in vivo identification of structural and sequence elements in the 5′ untranslated region of Ectropis obliqua picorna-like virus required for internal initiation. J. Gen. Virol. 2006;87:3667–3677. doi: 10.1099/vir.0.82090-0. [DOI] [PubMed] [Google Scholar]
- Mitra, K., Schaffitzel, C., Fabiola, F., Chapman, M.S., Ban, N., Frank, J. Elongation arrest by secM via a cascade of ribosomal RNA rearrangements. Mol. Cell. 2006;22:533–543. doi: 10.1016/j.molcel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Moscufo, N., Simons, J., Chow, M. Myristoylation is important at multiple stages in poliovirus assembly. J. Virol. 1991;65:2372–2380. doi: 10.1128/jvi.65.5.2372-2380.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottagui-Tabar, S., Isaksson, L.A. Only the last amino acids in the nascent peptide influence translation termination in Escherichia coli genes. FEBS Lett. 1997;414:165–170. doi: 10.1016/s0014-5793(97)00978-2. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar, S., Björnsson, A., Isaksson, L.A. The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J. 1994;13:249–257. doi: 10.1002/j.1460-2075.1994.tb06255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto, H., Nakatogawa, H., Ito, K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for secM-mediated ribosomal stall. Mol. Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Nakatogawa, H., Ito, K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Nathans, D., Neidle, A. Structural requirements for puromycin inhibition of protein synthesis. Nature. 1963;197:1076–1077. doi: 10.1038/1971076a0. [DOI] [PubMed] [Google Scholar]
- Nicklin, M.J.H., Toyoda, H., Murray, M.G., Wimmer, E. Proteolytic processiong in the replication of polio and related viruses. Biotechnology (N. Y.) 1986;4:33–42. [Google Scholar]
- O'Connor, M., Wills, N.M., Bossi, L., Gesteland, R.F. Functional tRNAs with altered 3′ ends. EMBO J. 1993;12:2559–2566. doi: 10.1002/j.1460-2075.1993.tb05911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuer, Q., Kuhn, R.J., Wimmer, E. Characterization of poliovirus clones containing lethal and nonlethal mutations in the genome-linked protein VPg. J. Virol. 1990;64:2967–2975. doi: 10.1128/jvi.64.6.2967-2975.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, O.C., Spagnolo, J.F., Lyle, J.M., Vleck, S.E., Kuchta, R.D., Kirkegaard, K. Intramolecular and intermolecular uridylyation by poliovirus RNA-dependent RNA polymerase. J. Virol. 2006;80:7405–7415. doi: 10.1128/JVI.02533-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert, R.R. In: Virology. 2nd ed. Fields B.M., et al., editors. Raven Press; New York: 1990. [Google Scholar]
- Ryan, M.D., Drew, J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994;13:928–933. doi: 10.1002/j.1460-2075.1994.tb06337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, M.D., Belsham, G.J., King, A.M.Q. Specificity of enzyme-substrate interactions in foot-and-mouth disease virus polyprotein processing. Virology. 1989;173:35–45. doi: 10.1016/0042-6822(89)90219-5. [DOI] [PubMed] [Google Scholar]
- Ryan, M.D., Luke, G., Hughes, L.E., Cowton, V.M., Dam, E.T., Gani, D., Xuejun, L., Donnelly, M.L.L., Mehrotra, A., Gani, D. The Aphto- and Cardiovirus “Primary” 2A/2B polyprotein “cleavage.”. In: Semler B.L., Simmer E., editors. Molecular biology of picornaviruses. ASM Press; Washington, DC: 2002. pp. 213–223. [Google Scholar]
- Rychlík, I., Černá, J., Chládek, S., Pulkrábek, P., Žemlička, J. Substrate specificity of ribosomal peptidyl transferase. Eur. J. Biochem. 1970;16:136–142. doi: 10.1111/j.1432-1033.1970.tb01064.x. [DOI] [PubMed] [Google Scholar]
- Sasaki, J., Nakashima, N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing, T.M., Huang, K.S., Strobel, S.A., Steitz, T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005a;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Schmeing, T.M., Huang, K.S., Kitchen, D.E., Strobel, S.A., Steitz, T.A. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell. 2005b;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Sunohara, T., Abo, T., Inada, T., Aiba, H. The C-terminal amino acid sequence of nascent peptide is a major determinant of SsrA tagging at all three stop codons. RNA. 2002;8:1416–1427. doi: 10.1017/s1355838202020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Zhang, J., Lu, J., Yi, F., Liu, C., Hu, Y. Sequence analysis and genomic organization of a new insect picorna-like virus, Ectropis obliqua picorna-like virus, isolated from Ectropis obliqua . J. Gen. Virol. 2004;85:1145–1151. doi: 10.1099/vir.0.19638-0. [DOI] [PubMed] [Google Scholar]
- Weinger, J.S., Parnell, K.M., Dorner, S., Green, R., Strobel, S.A. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- Woolhead, C.A., Johnson, A.E., Bernstein, H.D. Translation arrest requires a two-way communication between a nascent polypeptide and the ribosome. Mol. Cell. 2006;22:587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Wu, C.Y., Lo, C.F., Huang, C.J., Yu, H.T., Wang, C.H. The complete genome sequence of Perina nuda picorna-like virus, an insect-infecting RNA virus with a genome organization similar to that of the mammalian picornaviruses. Virology. 2002;294:312–323. doi: 10.1006/viro.2001.1344. [DOI] [PubMed] [Google Scholar]