Abstract
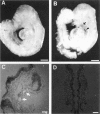
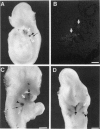
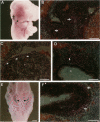
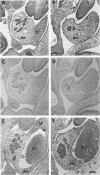
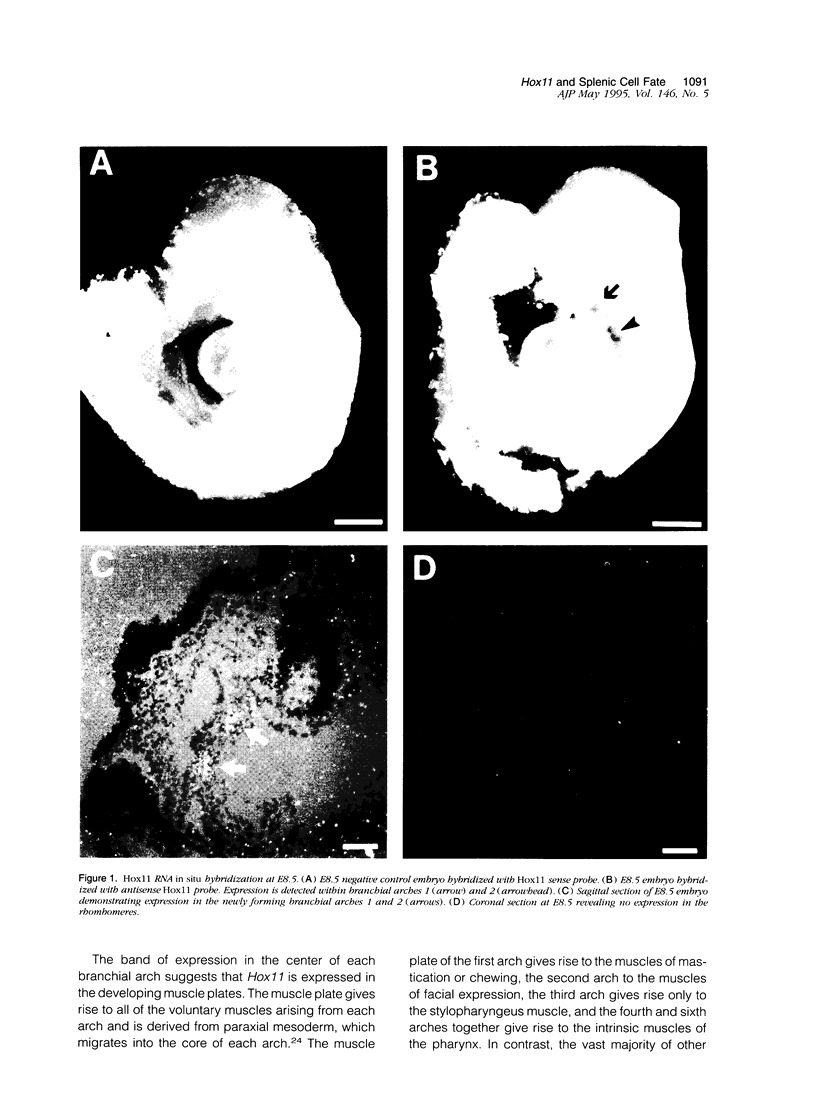
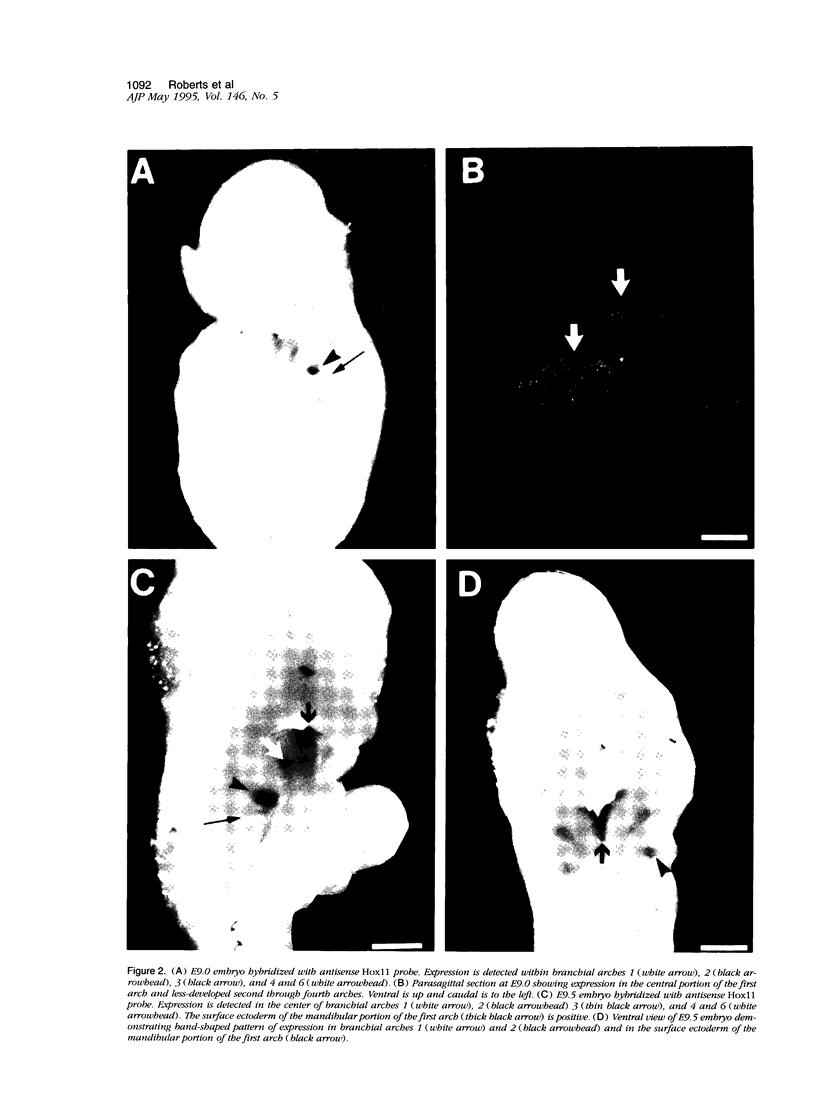
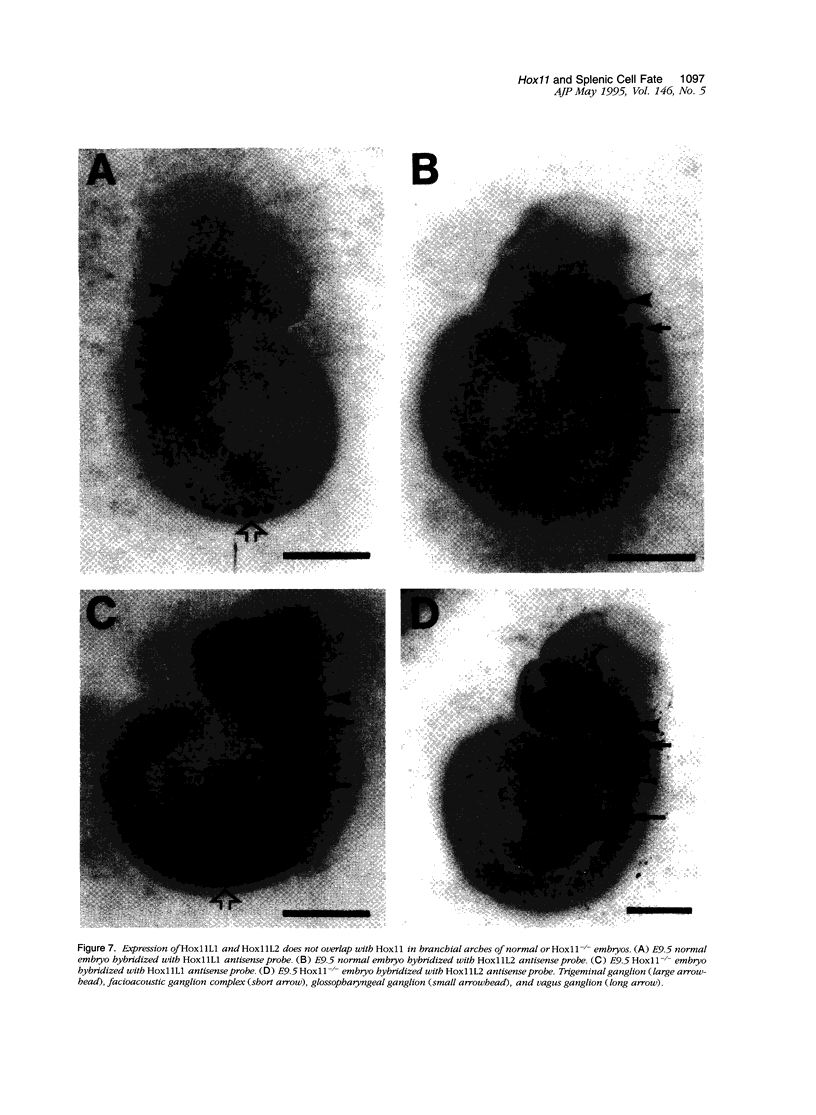
Hox11 is the first member of a novel class of orphan homeobox genes. We report that Hox11 is expressed in a discrete temporal and spatially segmented pattern during embryonic development and appears critical for the specification of splenic cell fate. Expression is first observed in the developing muscle plates of branchial arches 1, 2, 3 and 4/6, and subsequently within motor neurons of cranial nerves V, VII, IX, and X, which innervate these muscles. Hox11 serves as a molecular maker distinguishing branchial from somatic motor nuclei. Additionally, Hox11 is expressed in the surface ectoderm of the first branchial arch in the region destined to become the tongue and teeth and then in ganglia innervating this area. However, Hox11-deficient mice have no apparent morphological of functional defects within these structures. Notably the closely related homeobox genes, Hox11L.1 and Hox1L1.2, were not expressed in a redundant pattern. Neither Hox11L1 nor Hox11L2 was expressed in the branchial arches or their motor nuclei within wild-type or Hox11-/- mice. Beginning at E11.5, Hox11 is normally expressed at a single site in the abdomen within splanchnic mesoderm destined to form the spleen, and Hox11-/- mice have no spleen. We noted no increase in cell death within the dorsal mesogastrium of Hox11-deficient mice. Instead the dorsal mesogastrium fails to separate from the stomach. Hox11-/- mice display a larger stomach and possibly pancreas, suggesting that these mesodermal cells now contribute to other organs.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aplan P. D., Lombardi D. P., Ginsberg A. M., Cossman J., Bertness V. L., Kirsch I. R. Disruption of the human SCL locus by "illegitimate" V-(D)-J recombinase activity. Science. 1990 Dec 7;250(4986):1426–1429. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- Blochlinger K., Jan L. Y., Jan Y. N. Transformation of sensory organ identity by ectopic expression of Cut in Drosophila. Genes Dev. 1991 Jul;5(7):1124–1135. doi: 10.1101/gad.5.7.1124. [DOI] [PubMed] [Google Scholar]
- Bodmer R., Barbel S., Sheperd S., Jack J. W., Jan L. Y., Jan Y. N. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell. 1987 Oct 23;51(2):293–307. doi: 10.1016/0092-8674(87)90156-5. [DOI] [PubMed] [Google Scholar]
- Boehm T., Greenberg J. M., Buluwela L., Lavenir I., Forster A., Rabbitts T. H. An unusual structure of a putative T cell oncogene which allows production of similar proteins from distinct mRNAs. EMBO J. 1990 Mar;9(3):857–868. doi: 10.1002/j.1460-2075.1990.tb08183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Cheng J. T., Tasi L. H., Schneider N., Buchanan G., Carroll A., Crist W., Ozanne B., Siciliano M. J., Baer R. The tal gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix-loop-helix protein. EMBO J. 1990 Feb;9(2):415–424. doi: 10.1002/j.1460-2075.1990.tb08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisaka O., Capecchi M. R. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature. 1991 Apr 11;350(6318):473–479. doi: 10.1038/350473a0. [DOI] [PubMed] [Google Scholar]
- Chisaka O., Musci T. S., Capecchi M. R. Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature. 1992 Feb 6;355(6360):516–520. doi: 10.1038/355516a0. [DOI] [PubMed] [Google Scholar]
- Dear T. N., Sanchez-Garcia I., Rabbitts T. H. The HOX11 gene encodes a DNA-binding nuclear transcription factor belonging to a distinct family of homeobox genes. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4431–4435. doi: 10.1073/pnas.90.10.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé I. D., Kamel-Reid S., Yuan C. C., Lu M., Wu X., Corpus G., Raimondi S. C., Crist W. M., Carroll A. J., Minowada J. A novel human homeobox gene lies at the chromosome 10 breakpoint in lymphoid neoplasias with chromosomal translocation t(10;14). Blood. 1991 Dec 1;78(11):2996–3003. [PubMed] [Google Scholar]
- Freyd G., Kim S. K., Horvitz H. R. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature. 1990 Apr 26;344(6269):876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y., Sherman Y., Ben-Sasson S. A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992 Nov;119(3):493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg J. M., Boehm T., Sofroniew M. V., Keynes R. J., Barton S. C., Norris M. L., Surani M. A., Spillantini M. G., Rabbitts T. H. Segmental and developmental regulation of a presumptive T-cell oncogene in the central nervous system. Nature. 1990 Mar 8;344(6262):158–160. doi: 10.1038/344158a0. [DOI] [PubMed] [Google Scholar]
- Guthrie S., Muchamore I., Kuroiwa A., Marshall H., Krumlauf R., Lumsden A. Neuroectodermal autonomy of Hox-2.9 expression revealed by rhombomere transpositions. Nature. 1992 Mar 12;356(6365):157–159. doi: 10.1038/356157a0. [DOI] [PubMed] [Google Scholar]
- Hatano M., Roberts C. W., Minden M., Crist W. M., Korsmeyer S. J. Deregulation of a homeobox gene, HOX11, by the t(10;14) in T cell leukemia. Science. 1991 Jul 5;253(5015):79–82. doi: 10.1126/science.1676542. [DOI] [PubMed] [Google Scholar]
- Hunt P., Wilkinson D., Krumlauf R. Patterning the vertebrate head: murine Hox 2 genes mark distinct subpopulations of premigratory and migrating cranial neural crest. Development. 1991 May;112(1):43–50. doi: 10.1242/dev.112.1.43. [DOI] [PubMed] [Google Scholar]
- Karlsson O., Thor S., Norberg T., Ohlsson H., Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990 Apr 26;344(6269):879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- Kennedy M. A., Gonzalez-Sarmiento R., Kees U. R., Lampert F., Dear N., Boehm T., Rabbitts T. H. HOX11, a homeobox-containing T-cell oncogene on human chromosome 10q24. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):8900–8904. doi: 10.1073/pnas.88.20.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T., Ishimaru S., Higashijima S., Takayama E., Akimaru H., Sone M., Emori Y., Saigo K. Identification of a different-type homeobox gene, BarH1, possibly causing Bar (B) and Om(1D) mutations in Drosophila. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4343–4347. doi: 10.1073/pnas.88.10.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Gong Z. Y., Shen W. F., Ho A. D. The tcl-3 proto-oncogene altered by chromosomal translocation in T-cell leukemia codes for a homeobox protein. EMBO J. 1991 Oct;10(10):2905–2910. doi: 10.1002/j.1460-2075.1991.tb07840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lufkin T., Dierich A., LeMeur M., Mark M., Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991 Sep 20;66(6):1105–1119. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Krumlauf R. Homeobox genes and axial patterning. Cell. 1992 Jan 24;68(2):283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- McGuire E. A., Hockett R. D., Pollock K. M., Bartholdi M. F., O'Brien S. J., Korsmeyer S. J. The t(11;14)(p15;q11) in a T-cell acute lymphoblastic leukemia cell line activates multiple transcripts, including Ttg-1, a gene encoding a potential zinc finger protein. Mol Cell Biol. 1989 May;9(5):2124–2132. doi: 10.1128/mcb.9.5.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellentin J. D., Smith S. D., Cleary M. L. lyl-1, a novel gene altered by chromosomal translocation in T cell leukemia, codes for a protein with a helix-loop-helix DNA binding motif. Cell. 1989 Jul 14;58(1):77–83. doi: 10.1016/0092-8674(89)90404-2. [DOI] [PubMed] [Google Scholar]
- Noden D. M. Interactions and fates of avian craniofacial mesenchyme. Development. 1988;103 (Suppl):121–140. doi: 10.1242/dev.103.Supplement.121. [DOI] [PubMed] [Google Scholar]
- Noden D. M. The embryonic origins of avian cephalic and cervical muscles and associated connective tissues. Am J Anat. 1983 Nov;168(3):257–276. doi: 10.1002/aja.1001680302. [DOI] [PubMed] [Google Scholar]
- Raju K., Tang S., Dubé I. D., Kamel-Reid S., Bryce D. M., Breitman M. L. Characterization and developmental expression of Tlx-1, the murine homolog of HOX11. Mech Dev. 1993 Nov;44(1):51–64. doi: 10.1016/0925-4773(93)90016-q. [DOI] [PubMed] [Google Scholar]
- Roberts C. W., Shutter J. R., Korsmeyer S. J. Hox11 controls the genesis of the spleen. Nature. 1994 Apr 21;368(6473):747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Tamkun J. W., Hartzell G. W., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989 Jul 28;989(1):25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Wanaka A., Johnson E. M., Jr, Milbrandt J. Localization of FGF receptor mRNA in the adult rat central nervous system by in situ hybridization. Neuron. 1990 Sep;5(3):267–281. doi: 10.1016/0896-6273(90)90164-b. [DOI] [PubMed] [Google Scholar]
- Way J. C., Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988 Jul 1;54(1):5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- Wilkinson D. G., Bhatt S., Cook M., Boncinelli E., Krumlauf R. Segmental expression of Hox-2 homoeobox-containing genes in the developing mouse hindbrain. Nature. 1989 Oct 5;341(6241):405–409. doi: 10.1038/341405a0. [DOI] [PubMed] [Google Scholar]
- Wilkinson D. G., Krumlauf R. Molecular approaches to the segmentation of the hindbrain. Trends Neurosci. 1990 Aug;13(8):335–339. doi: 10.1016/0166-2236(90)90145-z. [DOI] [PubMed] [Google Scholar]