Abstract
Retroviral integrase (IN) catalyzes the integration of retroviral cDNA into host chromosome. Ini1 (integrase interactor 1) is a host protein that specifically binds and stimulates in vitro joining activity of HIV-1 IN. Ini1 has homology to yeast transcription factor SNF5 and is a component of the analogous mammalian SWI/SNF complex that can remodel chromatin. Little is known about the function of Ini1 in mammalian cells. To gain insight into the functional domains of Ini1, and to understand the details of protein–protein interactions of IN and Ini1, a structure-function analysis of Ini1 was initiated. By means of the yeast two-hybrid system, the minimal IN binding domain of Ini1 was characterized. One of the two repeat motifs present in the highly conserved regions of Ini1 was found necessary and sufficient to bind to IN in yeast as well as in vitro. Because IN binds to only one of the two repeat motifs in this conserved region of Ini1, it appears that the IN-Ini1 interaction is very specific and functionally significant. Characterization of DNA-binding properties of Ini1 revealed that Ini1 can bind to plasmid DNA, binding more readily to supercoiled DNA than to the relaxed circular DNA. The minimal domain for DNA binding was localized to a region upstream of repeat 1. The DNA binding activity of Ini1 is not required for its ability to interact with IN. The finding that the two repeat motifs of Ini1 display differential binding to HIV-1 IN and that this discrete component of mammalian SWI/SNF complex binds to DNA will help understand the role of Ini1 in HIV-1 integration and in cellular process.
Keywords: retroviral integration, protein–protein interactions, SWI, SNF complex, transcription
Integration of viral cDNA into the host chromosome is an essential part of the life cycle of all retroviruses (for reviews see refs. 1–9). During the early stages of infection, the retroviral integrase (IN) protein catalyzes a concerted integration of the two termini of the linear viral cDNA, the product of reverse transcription, into the host chromosome. Specific viral sequences at the ends of two long terminal repeats are essential for this process. Many chromosomal sequences can serve as target sites without any sequence specificity. Although retroviruses and retrotransposons are structurally related and have a similar mode of integration into the host DNA, they exhibit various degrees of bias in selecting target sites for integration in vivo. Whereas integration of Moloney murine leukemia virus retroviral DNA preferentially occur at DNase I hypersensitive regions and transcriptionally active regions (10, 11) retrotransposons such as Ty1 and Ty3 elements of yeast exhibit a greater degree of specificity in that they integrate only at RNA polymerase III-transcribed genes such as those that encode tRNA genes (12–15). The mechanism and the factors governing this bias in target site selection are not completely known.
Many lines of evidence indicate that host proteins may play a role in target site selection of retrotransposons and retroviruses. Integration of Ty3 has been demonstrated to require transcription factors TFIIIB and TFIIIC in vitro and in vivo (14). Similarly, a host factor IN interactor 1 (Ini1)/hSNF5L1 may be responsible for target site selection by HIV-1 IN. Ini1 was isolated as a binding partner for HIV-1 IN by means of the yeast two-hybrid system (16). Ini1 has amino acid sequence similarity to yeast transcriptional activator SNF5, a component of the multiprotein SWI/SNF complex (17, 18). This complex activates transcription by remodeling the chromatin (19, 20). Complexes similar to yeast SWI/SNF complex have been isolated from mammalian cells and demonstrated to have a similar set of protein components as that of yeast complex (21, 22). Ini1 is part of the mammalian SWI/SNF complex (22). Addition of complexes containing Ini1 to nucleosomal DNA results in remodeling the chromatin in vitro (22). Because Ini1 directly interacts with HIV IN and is involved in chromatin remodeling, Ini1 may target the retroviral integration machinery to open chromatin regions.
Despite Ini1 being the first mammalian homologue of SNF5 to be isolated, little is known about its function in mammalian cells. Analysis of its sequence reveals no known motifs that link it to an activity. To understand its role in HIV-1 integration, we have initiated a structure-function analysis of Ini1 to gain insight into the organization of its functional domains. Here we report the delineation of the minimal domain of Ini1 that is necessary and sufficient for interaction with HIV-1 IN, by using yeast two-hybrid based analysis as well as in vitro solution binding assays. We also have found that Ini1 is capable of binding to DNA and determined the minimal DNA binding (DB) domain of Ini1. Our results suggest that the Ini1-IN interaction is functionally significant and form a foundation for understanding the organization of the functional domains of Ini1.
MATERIALS AND METHODS
Bacterial and Yeast Strains.
Escherichia coli strains DH5α and XL1-blue were extensively used for all molecular cloning purposes. HB101 stain of E. coli was used for rescuing the plasmids from yeast. CTY10-5d containing an integrated Gal1-lacZ gene downstream of lexA operator (23), was used for the yeast two-hybrid analysis.
Generation of Ini1 Deletion Libraries.
DNA encoding the 130-aa-long N-terminal fragment (termed N-Ini1) was PCR-amplified by using primers 5′GTCAGGATCCCCATGGCGCTGAGCAAG3′, with a BamHI site, and 5′GTGGTGGGAGCTGTTGG3′; and by using the pINIgt clone (16) as the template DNA. The PCR-amplified fragment was cloned into BamHI–KpnI sites of pSP72 (Promega) to obtain pSP72-N-Ini1 and sequenced to demonstrate the absence of PCR-induced mutations. The full-length Ini1 clone (pSP72-Ini1) was generated by inserting a KpnI–BglII fragment isolated from clone pD2.1 (16) downstream of KpnI site of pSP72-N-Ini1.
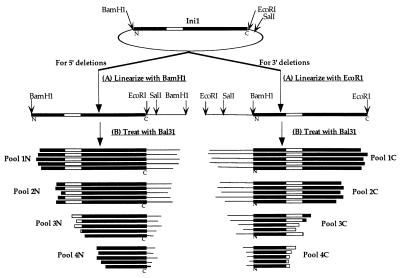
Nested deletion libraries of INI1 cDNA containing overlapping 5′ or 3′ deletions were obtained as follows (see Fig. 2). First, pSP72-Ini1 plasmid was linearized with either BamHI (to generate 5′ deletions) or EcoRI (to generate 3′ deletions). Second, linearized pSP72 DNA was treated with limiting amounts of exonuclease Bal31 for various time points. Four or five different pools of nested deletions were generated separately at each terminus of INI1 cDNA. Third, the fragments generated at each time point were treated with the Klenow fragment of DNA polymerase I and ligated to either BamHI (for 5′ deletions) or SalI (for 3′ deletions) linkers. Finally, INI1 cDNA fragments were excised by using BamHI and SalI from these deletion pools and were directionally cloned into pGADNot vector to generate 5′ and 3′ deletion libraries that express GAL4-ΔIni1 fusion proteins in yeast. Plasmid DNA from these deletion libraries, amplified in E. coli, was isolated by using a Qiagen column. The 5′ deletion libraries thus generated have Ini1 deletions fused to GAL4AD in three different reading frames and the probability of finding clones in the correct reading frame in the library is one-third.
Figure 2.
Schematic representation of the strategy for generating 5′ and 3′ deletion pools of INI1 cDNA. The plasmid carrying the INI1 cDNA was first linearized with either BamHI or EcoRI and subsequently treated with Bal31 to generate different pools of DNA fragments carrying increasing extent of deletions (pools 1N-4N and 1C-4C). The deleted pools of cDNA fragments subsequently were cloned into pGADNot for expressing GAL4AC fusion proteins in yeast.
Two-Hybrid Analysis.
A standard lithium acetate method, with slight modifications, was used to transform yeast (16). To isolate the truncations of Ini1 that retained the ability to interact with IN, either 5′ or 3′ deletion libraries of INI1 in pGADNot were individually introduced into yeast together with pSH2-IN plasmid (encoding HIV-1 IN as a fusion to LexA DNA-binding domain, LexADB). The resulting yeast transformants were subjected to 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) assay using the standard methods (16) to score for positive interaction of Ini1 truncations with IN.
Identification and Sequence Analysis of IN Binding Ini1–1 Fragments.
Subsequent to the two-hybrid analysis, DNA was isolated from several blue yeast colonies and introduced into HB101 by transformation. Because the LEU2 marker present in the pGADNot vector complements leucine auxotrophy of strain HB101, ampicillin-resistant and prototrophic colonies were selected. The pGADNot-ΔIni1 plasmids isolated from these colonies then were sequenced by using primers complementary to DNA encoding GAL4AD: 5′ GAL4 oligo (5′-CGATGATGAAGATACC-3′) and 3′GAL4 oligo (5′-GTGCACGATGCACAG-3′) to determine the truncation at the N and C terminus of Ini1, respectively. To combine the N- and C-terminal truncations into one molecular clone, a BamHI to ClaI fragment of N-terminal truncations and ClaI to SalI fragments of C-terminal truncations were isolated from plasmids obtained from the Dam− strain of E. coli CC114, and cloned together into BamHI and SalI sites of pGADNot vector in a three-fragment ligation reaction. The clones obtained were sequenced to confirm the identity.
Expression and Purification of Glutathione S-transferase (GST)-Ini1 and GST-Ini1 Truncations.
A panel of Ini1 truncations that retained the IN-binding activity in the yeast two-hybrid system were expressed as GST fusion proteins in E. coli. The DNA fragments encoding the Ini1 truncations were directionally cloned into the unique BamHI-SalI sites of pGEX3XPL (a gift from David Shore, Columbia University). GST and its fusion proteins were isolated and immobilized on glutathione-conjugated beads by using the standard procedure (16) in the lysis buffer Y (50 mM Tris-⋅Cl (pH 8.0), 1 mM EDTA, 0.5% IGEPAL, 50 mM NaCl, and protease inhibitors 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml each of leupeptin, pepstatin, and aprotinin). During these procedures we noticed that some of the truncated proteins could be extracted only on addition of NaCl to a final concentration of 0.5 M or higher. For subsequent procedures, we routinely included 0.5M NaCl in the lysis buffer for all of the deletions.
In Vitro Binding of GST-ΔIni1 to Recombinant IN.
The GST and GST-ΔIni1 proteins immobilized onto G beads were added to E. coli extract containing bacterially expressed HIV-1 IN, under buffer conditions described previously (16). After the binding reaction, G beads were washed several times, and the bound proteins were eluted by boiling in sample buffer containing SDS and DTT and subjected to SDS/PAGE followed by immunoblotting. Monoclonal anti-IN antibodies (a gift of Dug Helland, University of Bergen, Norway) were used to visualize the IN bound by GST-Ini1 proteins by the chemiluminiscence detection method (Pierce).
DNA Binding Studies.
The plasmid pBluescript SK− DNA, purified by CsCl-ethidium bromide equilibrium density gradient centrifugation, was incubated with various concentrations of GST and GST-fusion proteins in buffer (25 mM Hepes, pH 7.2/50 mM NaCl/0.1 mM EDTA/1 mM DTT/10% glycerol/3 mM MnCl) for 1 hr at 30°C or 37°C. After the reaction, the DNA molecules were separated by electrophoresis on agarose gels containing ethidium bromide and were visualized under UV light.
RESULTS
Structural Motifs of Ini1 and its Homologues.
Comparison of Ini1 with known amino acid sequences reveals no known structural motifs indicative of its function. There are at least five proteins and ORFs that have amino acid sequence similarity to Ini1 (Fig. 1a). The yeast SNF5 protein shares homology with Ini1 in its central charged region (16). The N-terminal domain of SNF5 is a glutamine- and proline-rich region and C-terminal domain is a proline-rich region (18). These two regions are absent from: (i) Ini1; (ii) the Drosophila protein, SNR1 (SNF5 related gene 1; ref. 24); (iii) the Caenorhabditis elegans protein, CeSNF5 (R07E5.3, PIR accession no. S43599; ref. 25); and (iv) an ORF from Saccharomyces cerevisiae, SFH1 (SNF Five Homologue 1, L8543.4; PIR accession no. S53399; ref. 22). Another ORF C2F7.08, encoding a hypothetical 71.9-kDa protein from Schizosaccharomyces pombe that has close similarity to Ini1 (BLASTP value: 3.5 × 10−29 with Ini1; SP accession no. Q09699 and GenBank accession no. YA28SCHPO) has been recently identified (we refer to this ORF as SpSNF5). Ini1 is more similar to SpSNF5 than to that of the two yeast homologues (BLASTP value of Ini1: with L8543.4 is 5.9 × 10−27; and with SNF5 is 4.3 × 10−20). The C2F7.08 protein lacks the N-terminal glutamine- and proline-rich region of SNF5. However, it has a C-terminal region that is somewhat proline-rich, containing 7% of prolines as compared with 18% prolines present in SNF5 (17 of 244-aa C-terminal region of SpSNF5 are prolines).
Figure 1.
Alignment of proteins/ORFs with homology to Ini1 protein. (a) Alignment of highly conserved regions of six protein/ORFs, Drosophila protein SNR1, C. elegans ORF R07E5.3 (CeSNF5 protein), S. pombe ORF encoding C2F7.08C protein (SpSNF5), and the two yeast proteins, SNF5 and SFH1. The alignment was performed by using the program multialign. The three highly conserved regions are indicated by horizontal arrows. Consensus sequence is indicated at the bottom. The first amino acid residue of Ini1 in each lane is numbered. (b) Graphical representation of six Ini1 homologues displaying the two repeat motifs. The name of the protein is indicated on the left. The two repeats are indicated by thick arrows, and the highly conserved regions that contain the two repeats are indicated by boxes. Empty box, repeat 1 (Rpt1) and shaded box, repeat 2 (Rpt2). The numbers at the beginnings and ends of the boxes represent the position of the amino acid residue at each edge of the region. Percentage identity of primary sequence between Ini1 and a given protein within the particular repeat is indicated inside the boxed area. The N-terminal and C-terminal glutanine- and proline-rich regions of SNF5 and SpSNF5 are indicated by dashed lines. (c) Alignment of the sequence of two repeat motifs with each other and their consensus sequence. The repeats, 1 and 2 of six Ini1-related proteins, were aligned by using pileup program in GCG and displayed by using the pretty program. Consensus sequences of the repeat motif, determined by using the plurality of 6 of 12 identical or conserved residues, is indicated at the bottom. The invariant residues are indicated by bold letter. Highly conserved and invariant residues are indicated by shading.
Alignment of the amino acid sequences of all six proteins/ORFs reveals that the C-terminal halves of Ini1, SNR1, CeSNF5, and SFH1 and the C-terminal half of the central regions of SNF5 and SpSNF5 proteins have three highly conserved regions (Fig. 1). The first two conserved regions are imperfect repeat motifs (amino acids 186–245 and 259–319 of Ini1, Fig. 1b). The degree of similarity among the six related proteins is highest in the first two regions (for example, as high as 85% identity in the first conserved region of Ini1 and SNR1). Alignment of sequences of repeats 1 and 2 from all the six proteins together revealed the presence of several invariable residues, suggesting that these residues may be functionally important. The consensus sequence for these 12 regions from six different proteins is shown in Fig. 1c. There are two invariant residues L and D that are separated by 10 amino acid residues in these repeats from all Ini1-related proteins except repeat 1 of SNF5, where these residues are separated by 12 amino acids. The third region of similarity is more conserved among Ini1, SNR1, and SpSNF5 proteins than among the remaining three proteins. Searching for the presence of coils by using the program coils (26) indicated that the region downstream of repeat 2, including the third conserved region in all six proteins, potentially can form coiled coil structures (data not shown). No functional information is available about any of these regions. In our effort to delineate the minimal domain of Ini1 necessary for interaction with IN, we found that one of the three conserved regions was necessary and sufficient for interaction with IN, indicating that at least one of these regions is a protein–protein interaction motif.
Deletion Mutagenesis of Ini1 to Determine the IN-Interaction Domain.
In the absence of information about functional domains or motifs, we used two-hybrid analysis to determine the IN-interaction domain of Ini1. Our previous experience with the yeast two-hybrid system indicated that examination of individual truncated proteins for their ability to interact in this system is likely to give misleading results (16). Therefore, instead of screening for truncations that show absence of interaction and hence lack of an interaction domain, we screened for libraries of N- and C-terminal truncations of Ini1 for those that retained the interaction domain as illustrated in Fig. 2. First, several pools of deletions were generated at both 5′ and 3′ end of INI1 cDNA by using Bal31. After the digestion, the pools of deletion fragments were directionally cloned into a yeast vector for the expression of GAL4AD-ΔIni1 proteins in the yeast. This procedure resulted in several deletion libraries of INI1 cDNA, each representing successive pools of heterogeneous but progressively larger truncations at each terminus. Restriction analysis of DNA from each pool confirmed that the average number of nucleotides deleted increased with longer exposure to Bal31.
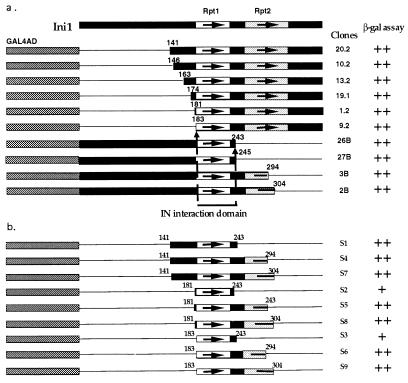
Each of the GAL4AD-Ini1 libraries with N- or C-terminal truncations were individually expressed in yeast strain CTY10–5d along with LexADB-IN fusion proteins for two-hybrid analysis. In this strain, interaction of LexADB-IN and GAL4AD-Ini1 results in the transactivation of the lacZ gene (27). If the GAL4AD-Ini1 truncations retain the ability to interact with IN, they will be scored positive in the X-Gal assay. The Ini1 truncation clones that either lost the interaction domain or resulted in an unstable protein or unfavorable protein fusion will be scored negative. Several thousand transformants were obtained with each of the deletion pools for X-Gal staining (Table 1). The percentage of blue clones decreased as the extent of deletion increased either from 5′ or 3′ end of INI1 cDNA. Deletion pools tested at 5′ end have a maximum of 35% of blue colonies because of random cloning of INI1 deletions in all three reading frames. Truncation of average of 173 amino acid residues at the N terminus of Ini1 did not decrease the percentage of blue colonies (pools 1N and 2N). However, truncation beyond 173 residues decreased the percentage of interacting clones to below 35% (3N and 4N). At the C terminus, truncation of an average of 150 amino acids (pool 2C) reduced the percentage of blue colonies. However, deleting further beyond 150 amino acids from the C terminus completely eliminated the interaction (pools 3C and 4C). We rescued plasmids from about 20 blue colonies derived from pools 2N, 3N, and 4C and sequenced the junction to determine the extent of truncation (Fig. 3a). The shortest clones retaining IN interaction were missing 182 residues at the N terminus (clone 9.2) or 142 amino acids at the C terminus (clone 26B). These two clones define the N- and C-terminal borders of IN-interaction domain. The common overlapping region in all of the interacting clones therefore spanned the residues 183–243.
Table 1.
Deletion mutagenesis of INI1 cDNA to determine the minimal IN-interaction domain
| Bal31 deletion pools | Average no. of bases deleted | % Blue colonies IN-interaction |
|---|---|---|
| (Full-length INI 1) | (1,150 bp) (385 a.a.) | (100) |
| 5′ deletions | ||
| 1N | 400 (∼133 a.a.) | >35 |
| 2N | 520 (∼173 a.a.) | >35 |
| 3N | 650 (∼217 a.a.) | ∼30 |
| 4N | 770 (∼250 a.a.) | ∼2 |
| 5N | 1,000 (∼333 a.a.) | 0 |
| 3′ deletions | ||
| 1C | 150 (∼50 a.a.) | ∼94 |
| 2C | 450 (∼150 a.a.) | ∼28 |
| 3C | 750 (∼250 a.a.) | 0 |
| 4C | 950 (∼317 a.a.) | 0 |
First column represents INI1 deletion libraries: 1N-5N are 5′ deletions; and 1C-4C are 3′ deletions. Second column represents average number of bases (and amino acids) deleted in each pool. The third column represents % blue colonies obtained by scoring 2,000-3,000 yeast transformants for each deletion library in the yeast two-hybrid assay.
Figure 3.
Interaction of HIV-1 IN with Ini1 truncations in the two-hybrid system. (a) Various truncations of Ini1 that retained the ability to interact with IN in the two-hybrid system. (b) Repeat 1 of Ini1 is sufficient for the interaction with IN. The top bar represents the full-length Ini1. The two highly conserved regions containing the two repeats are represented by open and shadowed boxes with arrows. Ini1 deletions obtained in the two-hybrid analysis are indicated as fusion to GAL4AD below the top bar. The thin lines represent the region of the protein deleted. The numbers on the right of deletions represent the clone number. The amino acid residue at the junction of both N- and C-terminal deletions are represented for each clone. The two shortest clones (9.2, and 26B) define the N- and C-terminal borders of IN-interaction domain. Relative strengths of interaction of Ini1 mutants with IN is indicated by ++, strong blue, and +, light blue.
Repeat 1 Region of Ini1 Is Necessary and Sufficient for Interaction with IN.
The 183–243-aa region of Ini1 coincides with the first of the two highly conserved repeats present in all the proteins/ORFs with similarity to Ini1, suggesting that repeat 1 is the IN-interaction domain. In addition, even though thousands of transformants were obtained with the deletion clones in pools 4N and 3C (which were devoid of the first repeat) none were blue in the X-Gal assay, indicating that repeat 1 of Ini1 is necessary for IN interaction.
Because all of the truncations we tested still retained either N- or C-terminal extensions around repeat 1, to determine whether these regions are contributing to IN interaction, we combined several of the N- and C-terminal truncations to generate smaller clones as indicated in Fig. 3b. These smaller clones were expressed as fusion to GAL4AD and tested for their ability to interact with LexADB-IN in the two-hybrid system. All of these clones were able to interact with IN. However, the smallest clones, S2 and S3, which are about 60 amino acids in length (amino acids 181–243 and amino acids 183–243), exhibited weaker X-Gal staining compared with the slightly larger clones. However, it is clear that the smallest clones containing minimal repeat 1 region still can interact with IN.
The above conclusion was supported by testing the interaction of clones that retained only the second repeat for their interaction with IN. We isolated a clone from pool 5N (Table 1) that expressed a truncated protein as a fusion to GAL4AD, that had lost the first repeat but retained the second repeat (A.M. and G.V.K., unpublished data) and tested its ability to interact with LexADB-IN. Although this clone expresses stable protein (data not shown), it did not result in transactivation of LacZ gene in the two-hybrid system, indicating that the protein containing repeat 2 region is unable to interact with IN. These data together demonstrate that the first but not the second repeat of Ini1 is necessary and sufficient for interaction with HIV-1 IN.
Repeat 1 of Ini1 Is Sufficient for Binding to IN in Vitro.
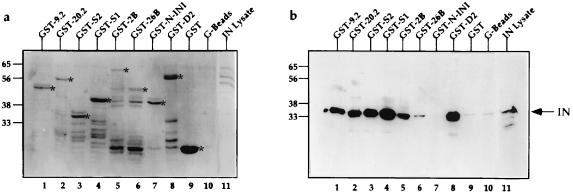
The above analyses indicate that the minimal IN binding domain of Ini1 must contain repeat 1. To demonstrate that the interactions as defined in the two-hybrid system truly reflect protein–protein interactions between IN and Ini1 truncation, we carried out in vitro binding studies by using GST fusion proteins. A panel of Ini1 truncations were expressed as fusions to GST (Fig. 4a) and used for binding to HIV-1 IN expressed in bacteria. The IN bound to GST fusion proteins was visualized with monoclonal anti-IN antibodies in a Western analysis (Fig. 4b). The results indicated that all of the deletions that retained the repeat 1 were able to bind to IN in solution (Fig. 4b, lanes 1–6 and 8). The glutathione beads (G beads) bound to either the truncation mutant containing only the N terminal 100 amino acids of Ini1 (GST-N-Ini1, Fig. 4b, lane 7), or GST (Fig. 4b, lane 9) or no protein (beads alone, Fig. 4b, lane 10) were unable to bind to IN. Interestingly, the mutant S2 (amino acids 183–243) containing the minimal portion of Ini1 with repeat 1 alone was able to bind to IN equally well, as GST-D2 (107–385 amino acids), a near full-length clone (Fig. 4b, compare lane 3 to lane 8). Based on these results we conclude that repeat 1 is sufficient for binding to IN.
Figure 4.
In vitro binding assay of GST-Ini1 truncations to HIV-1 IN. (a) Coomassie-stained PAGE of bound proteins. (b) Western analysis using monoclonal anti-IN antibodies. Lanes are identical in a and b. Numbers on the left represent the position of molecular weight markers in kDa. GST-fusion proteins used for binding experiment is indicated on top of each lane. G beads, glutathione agarose beads not bound to any proteins. IN lysate in lane 11 represents the bacterial lysate expressing IN, before binding experiment. The bands of the expected size length GST or GST-fusion proteins are indicated by ∗ on the right side of each band.
Ini1 Has DNA Binding Activity.
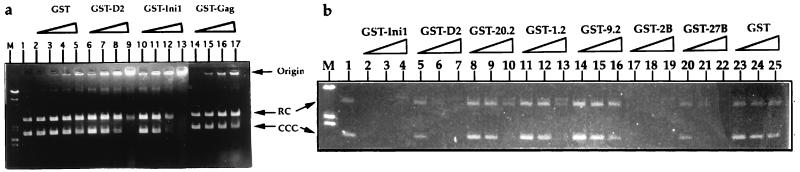
During the isolation of GST-Ini1 fusion proteins and GST-Ini1 truncation derivatives, we noticed that full-length GST-Ini1 and some of the truncation derivatives (clones 27B, 3B, 2B, and 26B) could be extracted only in the presence of high salt such as 0.5 M NaCl, whereas other truncation derivatives could be extracted with low salt such as 50 mM NaCl. This finding suggested to us that Ini1 and some of the truncation derivatives may have DNA binding ability. To determine whether Ini1 exhibited DNA binding activity, increasing concentrations of GST-Ini1 proteins were incubated with plasmid DNA prepared by CsCl and ethidium bromide equilibrium density gradient centrifugation, which contains a mixture of supercoiled and nicked circular DNAs. The plasmid DNA then was analyzed by electrophoresis in an agarose gel, and the position of migration of the DNA was determined by ethidium bromide staining (Fig. 5a). GST fusion proteins containing either the full-length Ini1 sequence (GST-Ini1) or the C-terminal fragment of Ini1 from residues 107 to 385 (GST-D2) showed DNA binding activity. As 3-fold increasing concentrations of the proteins were added, bands corresponding to the supercoiled DNA, followed by that corresponding to the relaxed circular DNA, decreased in intensity. This decrease in intensity of bands was accompanied by the appearance of ethidium bromide-stained DNA at the origin, suggesting that DNA was retained by binding to protein. The supercoiled DNA was bound preferentially over the relaxed circular DNA by both proteins. The extent of binding increased sharply over a very narrow range of protein concentrations, suggesting strong cooperativity in the binding (Fig. 5a, see lanes 7–9 and 11–13). The full-length Ini1 fusion bound DNA at lower protein concentrations than the fragment of Ini1; binding of the complete Ini1 fusion first was observed at a protein/DNA ratio of approximately 1:1 by weight, indicating that many protein molecules may be bound to each DNA molecule and that the binding is nonspecific. Treatment of the complexes with proteinase K before electrophoresis released both the supercoiled and relaxed circles, indicating that the DNA was not nicked or degraded during the incubations (data not shown). Control experiments with several-fold molar excess of GST protein alone, or with a GST-Gag fusion, showed very little or no binding under these conditions. We conclude that the Ini1 protein in these fusions can bind and retain DNA.
Figure 5.
DNA binding activity of Ini1. Ethidium bromide-stained gel of electrophoresis of plasmid DNA incubated with GST or GST-fusion proteins. (a) DNA binding was carried out by incubating 200 ng of plasmid pBluescript with increasing concentrations of GST or GST-fusion proteins in a 20-μl reaction. After the incubation, the plasmid DNAs were separated on agarose gels and stained with ethidium bromide. Lane M, lambda HindIII markers. Lane 1, pBluescript SK− plasmid DNA. Plasmid DNA incubated with 5, 10, 20, and 40 pMols of GST (lanes 2–5); 1.4, 2.8, 5.6, and 11.2 pMols of GST-D2 (lanes 6–9); 0.4, 0.8, 1.6, and 3.2 pMols of GST-Ini1 (lanes 10–13); or GST-gag (lanes 14–17). Origin, wells in the agarose gel indicating the retention of protein–DNA complexes. RC, relaxed circle DNA. CCC, covalently closed circular DNA. (b) Deletion analysis to determine the DNA binding domain of Ini1. DNA binding assay was carried out as before with uniformly increasing concentrations (1.6, 3.2, and 6.4 pMols respectively) of all the GST and GST-fusion proteins and using 100 ng of pBluescript DNA. Lane M, lambda HindIII marker. Lane 1, pBluescript SK− DNA. Lanes 2–25, plasmid DNA incubated with increasing concentrations of GST-Ini1 (lanes 2–4) GST-D2 (lanes 5–7), GST-20.2 (lanes 8–10), GST-1.2 (lanes 11–13), GST-9.2 (lanes 14–16), GST-2B (lanes 17–19), GST-27B (lanes 20–22), or GST (lanes 23–25) proteins as indicated on top of the gel. RC, relaxed circular DNA. CCC. covalently closed circular DNA.
DNA Binding Domain and IN-Interaction Domains of Ini1 Are Distinct from Each Other.
Because Ini1 has no motifs that suggest DNA binding activity, we set out to determine the minimal domain of Ini1 that can bind to DNA. We tested a panel of Ini1 truncations isolated in our two-hybrid analysis, expressed as GST fusions, in agarose gel-based gel shift assay as described above. The results indicated that the two C-terminal truncation mutants (clones 2B and 27B) retained the DNA binding ability (Fig. 5b, lanes 17–22). However, many of the N-terminal truncations (clones 20.2, 1.2, and 9.2) demonstrated either decreased or no DNA binding activity. These results indicated that the DNA binding domain of Ini1 lies in the overlapping region that is common to the two truncated proteins, D2 (amino acids 106–385) and 27B (amino acids 1–243). Because the truncations 1.2 and 9.2 have reduced DNA binding activity, it appears that amino acids 106–183 of Ini1, situated adjacent to repeat 1, are most important for DNA binding. Interestingly, several of the truncations that retained the IN-interaction domain in the repeat 1 region did not retain the DNA binding activity, suggesting that DNA binding is not a requirement for IN binding.
DISCUSSION
Ini1/hSNF5L1 is a component of mammalian SWI/SNF complex (16). Amino acid sequence analysis of Ini1 reveals no structural motifs. However, alignment of six proteins/ORFs with amino acid similarity to Ini1 reveals the presence of two regions that are highly conserved. These two regions are imperfect repeats with each other (25). In addition, a third conserved region that can potentially form a coiled structure was noticed. Although the functional significance of any of these regions is unclear, we present here evidence to indicate the role of one of the conserved regions of Ini1 in HIV IN interaction.
We carried out a deletion analysis of Ini1 to define the IN-interaction domain by using two-hybrid system. While mapping the domain of HIV-1 IN involved in oligomerization, we had found that examination of specific deletion mutants is not an efficient means to map the minimal interaction domain as instability of the mutant protein or the geometric constraints posed by the fusion of deletions to DNA binding or AC domains may affect the normal functioning of GAL4 and falsely result in noninteractive phenotype (16, 28). We therefore developed the strategy of using nested deletion libraries to screen thousands of deletions to identify the largest deletions that retain the ability to interact. This strategy allowed us to demonstrate that the repeat 1, but not repeat 2, is both necessary and sufficient for binding to IN. In vitro binding studies confirmed our two-hybrid analysis. All of the deletions, including the smallest Ini1 fragment (clone S2, amino acids 181–143), containing repeat 1, efficiently bound to IN. The weaker interactions exhibited by the smaller clone S2 in the two-hybrid system probably is not caused by its decreased ability to bind to IN, but could be because of decreased protein stability.
Defining the IN-interaction domain of Ini1 gives a clue to the function of the highly conserved region. Although it previously has been demonstrated that the C-terminal half containing all three homology regions of Ini1 is necessary for coimmunoprecipitation with BRG1 (mammalian homologue of SWI2 protein), these studies did not distinguish between various repeats of Ini1 (25). Thus it was not clear if repeat 1, 2, or both are necessary for interaction with BRG1. Moreover, there was no evidence to suggest that BRG1-Ini1 interaction is direct (25). Our results indicate that repeat 1 is capable of direct heteromeric protein–protein interactions. The lack of IN-binding capacity of repeat 2 suggests that the two repeat motifs, despite their similarity, may interact with different sets of proteins in the cell, possibly involved in diverse functions such as transcription and recombination. On the other hand, repeat 1 may be involved in protein–protein interactions and repeat 2 may have diverged to participate in or take on a different function. It will be interesting to investigate these questions as well as to study the differential regulation of cellular function by Ini1 or SWI/SNF complex depending on the type of proteins that may interact with each of the two repeats.
The yeast SWI/SNF complex has been demonstrated to have DNA binding activity (29). Its DNA binding property is similar to that of high mobility group box proteins in that it binds to DNA in a nonspecific and length-dependent manner and shows affinity to cruciform DNAs. In that study it also was demonstrated that three polypeptides of the yeast SWI/SNF complex, corresponding to 150K SWI1, and two other components p68 and p78 can crosslink with DNA in the presence of UV. However, that study does not exclude the possibility that other components of SNF/SWI complex can bind to DNA in vivo. Our studies indicate that Ini1 can bind plasmid DNA, binding supercoiled DNA with a higher preference than relaxed circular DNA. At this time it is not clear if Ini1 recognizes specific DNA sequences. It may bind to nonspecific DNA. The differential binding of Ini1 to supercoiled versus relaxed circle DNA may indicate a preference for structures such as cruciform and four-way junctions that may be present in supercoiled DNA. More experiments are needed to address whether Ini1 contributes to the ability of SWI/SNF complex to bind to DNA in a manner similar to that of high mobility group proteins. The DNA binding property of Ini1 is intriguing and understanding of the details of this process will shed light on the role of Ini1 in HIV-1 integration as well as transcriptional activity of SWI/SNF complex in mammalian cells. The DNA binding domain of Ini1, defined by the deletion analysis, does not contain any known DNA binding motif. No significant conservation was observed in this region among various proteins with similarity to Ini1. It needs to be determined if this DNA binding motif is unique to Ini1 or if other proteins share a similar three-dimensional structure in this region despite the lack of primary sequence conservation.
Analysis of DNA binding property and IN interaction of Ini1 will lay a foundation for understanding Ini1’s role in HIV-1 integration and in cellular processes. Although it is still not clear what role Ini1 may play in integration, because IN interacts with a highly conserved region suggests that its association is functionally significant. Knowledge of the minimal IN-interaction domain of Ini1 will enable studies aimed at modulation of HIV integration in vivo. For example, the minimal repeat 1 of Ini1 may act as a dominant negative inhibitor of HIV-1 integration. Our results provide a basis for a detailed understanding of protein–protein interactions of IN and Ini1 in the hope that inhibitors to HIV-1 replication may be developed by using this information.
Acknowledgments
We thank S. Goff for helpful discussions, V. Prasad, S. Emmons, G. Childs, K. Davies, and V. Mehra for critically reading the manuscript, S. Marmon for technical support in constructing pSP72-Ini1 clone, and Cancer Center Sequencing facilities for their services. This work was supported by National Institutes of Health Grant RO1 AI39951-01 to G.V.K.
ABBREVIATIONS
- IN
integrase
- Ini1
integrase interactor 1
- GST
glutathione S-transferase
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
References
- 1.Goff S P. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 2.Kulkosky J, Skalka A M. Pharmacol Ther. 1994;61:185–203. doi: 10.1016/0163-7258(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 3.Lieber M. Curr Biol. 1996;6:134–136. doi: 10.1016/s0960-9822(02)00443-8. [DOI] [PubMed] [Google Scholar]
- 4.Skalka A M. In: Integrative Recombination of Retroviral DNA. Kucherlapati R, Smith G R, editors. Washington, D.C.: Am. Soc. Microbiol. Publ.; 1989. pp. 701–724. [Google Scholar]
- 5.Skalka A M. Gene. 1993;135:175–182. doi: 10.1016/0378-1119(93)90063-9. [DOI] [PubMed] [Google Scholar]
- 6.Rice P, Craigie R, Davies D R. Curr Opin Struct Biol. 1996;6:76–83. doi: 10.1016/s0959-440x(96)80098-4. [DOI] [PubMed] [Google Scholar]
- 7.Andrake M D, Skalka A M. J Biol Chem. 1996;271:19633–1966. doi: 10.1074/jbc.271.33.19633. [DOI] [PubMed] [Google Scholar]
- 8.Polard P, Chandler M. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 9.Vink C, Plasterk R H A. Trends Genet. 1994;9:433–437. doi: 10.1016/0168-9525(93)90107-s. [DOI] [PubMed] [Google Scholar]
- 10.Vijaya S, Steffen D L, Robinson H L. J Virol. 1986;60:683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohdewohld H, Weiher H, Reik W, Jaenisch R, Breindl M. J Virol. 1987;61:336–343. doi: 10.1128/jvi.61.2.336-343.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalker D L, Sandmeyer S B. Genes Dev. 1992;6:117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- 13.Ji H, Moore D P, Blomberg M A, Braiterman L T, Voytas D F, Natsoulis G, Boeke J D. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 14.Kirchner J, Connolly C M, Sandeyer S B. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 15.Sandmeyer S B, Hansen L J, Chalker D L. Annu Rev Genet. 1990;24:491–518. doi: 10.1146/annurev.ge.24.120190.002423. [DOI] [PubMed] [Google Scholar]
- 16.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 17.Carlson M, Laurent B C. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 18.Laurent B C, Treitel M A, Carlson M. Mol Cell Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirschhorn J N, Brown S A, Clark C D, Winston F. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 20.Cote J, Quinn J, Workman J L, Peterson C L. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 21.Imbalzano A N, Know H, Green M R, Kingston R E. Nature (London) 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Cote J, Zhou S, Muchardt C, Khavari P, Bigger S R, Xue Y, Kalpana G V, Goff S P, Yaniv M, Tjian R, Workman J, Crabtree G. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 23.Kalpana G V, Goff S P. Proc Natl Acad Sci USA. 1993;90:10593–10597. doi: 10.1073/pnas.90.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dingwall A, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 27.Chien C-T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golemis E A, Brent R. Mol Cell Biol. 1992;12:3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. Nature (London) 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]