Abstract
The mutational response of mismatch repair-deficient animals to the alkylating agent N-methyl-N-nitrosourea was evaluated by using a transgenic lacI reporter system. Although the mutations detected in MSH2 heterozygotes were similar to those of controls, MSH2−/− animals demonstrated striking increases in mutation frequency in response to this agent. G:C to A:T transitions at GpG sites, as opposed to CpG sites, dominated the mutational spectrum of both MSH2+/+ and MSH2−/− N-methyl-N-nitrosourea -treated animals. Extrapolating to humans with hereditary non-polyposis colorectal cancer, the results suggest that MSH2 heterozygotes are unlikely to be at increased risk of mutation, even when exposed to potent DNA methylating agents. In contrast, mismatch repair-deficient cells spontaneously arising within individuals with hereditary non-polyposis colorectal cancer would likely exhibit hypermutability in response to such mutagens, an outcome predicted to accelerate the pace of tumorigenesis.
Kindreds with hereditary non-polyposis colorectal cancer (HNPCC) carry germ-line mutations in various human orthologs of the bacterial DNA mismatch repair (MMR) genes mutS and mutL (1). The gene most frequently involved in HNPCC, hMSH2, encodes a component required for DNA mismatch recognition (2, 3). Although heterozygosity for hMSH2 confers a high risk of colonic and endometrial neoplasms, malignancies at other sites, such as stomach, ovary, and urinary tract, also occur with increased frequency. Such tumors are accompanied by somatic mutations that inactivate the normal allele. The mutator phenotype arising from the combination of polymerase errors and a lack of MMR is thought to hasten the acquisition of alterations within key growth control genes, thus driving the multistep process that culminates in neoplasia (4).
To generate a model of human HNPCC, mice harboring a disruption of the MSH2 locus were generated (5, 6). However, rodents heterozygous for MSH2, unlike humans with HNPCC, failed to show an increased rate of tumor formation. Mice lacking MSH2, on the other hand, developed thymic lymphomas with high frequency. In addition, although small intestinal adenomas and adenocarcinomas were seen, colonic tumors were rarely observed in these mice (5–7). Interestingly, MMR deficiency because of a lack of MSH2 or PMS2 was compatible with normal murine growth and development (5, 6, 8), despite the presence of an elevated mutation frequency in all tissues evaluated (9, 10). Thus, the restricted spectrum of tumors in MSH2−/− animals was not attributable solely to differences in tissue-specific spontaneous mutation frequencies.
Although mice heterozygous for MSH2 did not demonstrate a mutator phenotype (10), it was conceivable that heterozygous cells might accrue mutations at a higher rate after mutagen exposure, owing to reduced levels of MSH2 protein. To determine whether specific mutagen administration would result in abnormal increases in mutation frequency in MSH2+/− and MSH2−/− tissues, we investigated the effects of an SN1 class DNA alkylating agent on MSH2-deficient animals. Such chemicals react with DNA to form various lesions, of which O6-alkylguanine (O6-alkylG) appears to be the most highly mutagenic (11). Studies of Escherichia coli and mammalian cell lines exposed to such methylating agents have shown that mispairing of O6-methylguanine (O6-meG) with thymine can result in G:C to A:T transitions (12) because of DNA polymerase base misincorporations that may occur opposite this lesion (13, 14).
MMR proteins recognize O6-meG:T base pairs, thus initiating long patch repair of these lesions (15). However, repetitive cycles of repair after DNA polymerase-mediated misincorporations of thymine opposite O6-meG sites has been postulated to result in cell death, likely as a result of the presence of excessive DNA strand breaks (16). MMR deficiency thus has been associated with tolerance to alkylation damage in terms of cell viability (16). In keeping with this, MSH2−/− embryonic stem cells demonstrated tolerance to N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), as measured by increased cell survival after exposure to this agent (6).
The mutational response to DNA alkylation of tissues from MSH2-deficient animals has not been reported. Furthermore, although MSH2 heterozygous cells do not appear to demonstrate resistance to alkylation-induced cell death (6), the specific mutational response of normal MSH2+/− tissues to DNA alkylation has not been explored. The experiments with N-methyl-N-nitrosourea (MNU) described herein provide an in vivo assessment of the mutational consequences of alkylation tolerance. Furthermore, because alkylating agents have the potential to be either formed endogenously or ingested, it was important to establish whether MSH2+/− and/or MSH2−/− animals demonstrated increased sensitivity to such agents when compared with controls.
To examine the response of MMR-deficient animals to DNA alkylating agent exposure, a transgenic lacI-bearing lambda shuttle phage line (BC-1), previously developed for in vivo mutation detection (17), was crossed onto the MSH2−/− background (5). This enabled us to evaluate the effects of specific mutagen exposure on the tissue-specific lacI mutation frequency and spectrum. In this study, the consequences of MNU administration to MSH2+/+, MSH2+/−, and MSH2−/− mice were assessed.
MATERIALS AND METHODS
Transgenic Mice.
MSH2+/− heterozygotes generated by gene targeting (5) were bred with BC-1 transgenic mice (17) to obtain MSH2−/−/BC-1, MSH2+/−/BC-1, and MSH2+/+/BC-1 mice. Genotypes were determined as previously described (7, 10).
One male and two female animals, approximately 21 days of age, were chosen from each of the MSH2+/+, MSH2+/−, and MSH2−/− genotypes. Mice were given intraperitoneal injections of 50 mg/kg MNU (Sigma) on 2 consecutive days, followed by a 3-week recovery period to ensure that the repair of DNA lesions had occurred and that mutations had become fixed. MNU was selected as a prototypic SN1 alkylating agent, a class known to efficiently generate O6-alkylguanine DNA lesions. Animals were sacrificed by carbon dioxide inhalation, and tissues were flash-frozen in liquid N2. DNA was extracted from small intestine, thymus, and heart and then packaged and plated as previously described (18).
Determination of lacI Gene Mutation Frequency and Spectrum.
Tissue isolation and transgenic lambda phage rescue were carried out as described (18, 19). Briefly, phage genomes within high-molecular-weight BC-1 transgenic mouse DNA were excised and packaged by using a highly efficient phage packaging extract, Transpack (Stratagene). Rescued phage were then plated in the presence of 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) on an SCS-8 (Stratagene) bacterial cell lawn, and lacI mutant frequency was established by determining the ratio of mutant (blue) to nonmutant (colorless) plaques. LacI mutations were verified as previously described (17). Following phage rescue and the isolation of single mutant clones, lacI genes were amplified by PCR of phage templates from randomly selected mutants. Templates were then directly sequenced by using primers spanning the lacI gene (17) and an ABI 388 sequencer (Applied Biosystems).
RESULTS
Mutation Frequencies.
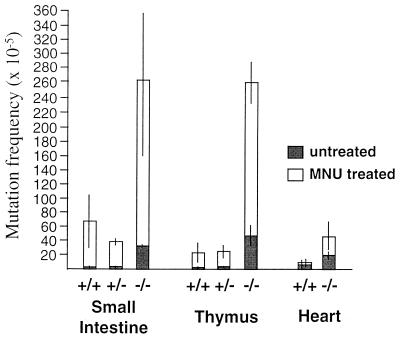
Mutation frequencies were determined for lacI genes rescued from small intestine, thymus, and heart DNA of MSH2+/+, MSH2−/+, and MSH2−/− animals. The number of lacI mutant (blue) plaques and the total number of plaque forming units (pfu) counted for the tissues of each animal are presented in Table 1. In repair proficient animals, MNU treatment resulted in increased lacI mutation frequencies in small intestine (74 × 10−5) and thymus (22 × 10−5), but much less so in heart (12 × 10−5). Treated MSH2 heterozygotes demonstrated mutation frequency inductions similar to those of the controls for small intestine (39 × 10−5) and thymus (24 × 10−5). MSH2-deficient animals, in contrast, revealed striking lacI mutation frequency increases above the already elevated backgrounds in all three tissues, with the highest increases occurring in small intestine (265 × 10−5) and thymus (260 × 10−5). A comparison of the mean mutation frequencies for each tissue from the three MSH2 genotypes is illustrated in Fig. 1, where the excess mutations resulting specifically from MNU treatment can be seen superimposed on the spontaneous background frequencies of MSH2−/−, MSH2+/−, and MSH2+/+ mice. Thus, the induction of mutations by MNU was greatest in MSH2 nullizygous mice. Mutation frequency inductions were tissue-specific, with heart being far lower than tissues containing substantial proliferative cell populations, as exemplified by small intestine and thymus. Furthermore, with respect to the latter two tissues, the inclusion of nonproliferative cell populations such as thymic stromal cells or intestinal smooth muscle cells in the DNA preparation suggests that the mutation frequency increases measured underestimate the inductions within the proliferating cell populations.
Table 1.
Spontaneous and MNU-induced mutation frequencies for BC-1 controls, BC-1/MSH2+/− and BC-1/MSH2−/−
| Tissue | Animal | BC-1 controls
|
|||||
|---|---|---|---|---|---|---|---|
| Untreated control animals
|
MNU-treated animals
|
||||||
| Total pfu | No. of mutants | Mutation frequency, ×10−5 | Total pfu | No. of mutants | Mutation frequency, ×10−5 | ||
| Small intestine | a | 286,860 | 11 | 3.8 | 211,384 | 106 | 50.1 |
| b | 261,540 | 4 | 1.5 | 191,800 | 102 | 53.2 | |
| c | 265,320 | 11 | 4.1 | 179,380 | 210 | 117.1 | |
| Mean | 3.1 ± 1.4 | 74 ± 38 | |||||
| Thymus | a | 292,890 | 13 | 4.4 | 206,580 | 13 | 6.3 |
| b | 264,180 | 8 | 3.0 | 203,720 | 65 | 31.9 | |
| c | 285,680 | 5 | 1.8 | 195,680 | 54 | 27.6 | |
| Mean | 3.1 ± 1.3 | 22 ± 14 | |||||
| Heart | a | 244,960 | 10 | 4.1 | 202,620 | 29 | 14.3 |
| b | 229,400 | 30 | 13.1 | 158,220 | 14 | 8.8 | |
| Mean | 8.6 ± 6.4 | 12 ± 4 | |||||
| Tissue | Animal | BC-1/MSH2+/−
|
|||||
|---|---|---|---|---|---|---|---|
| Untreated control animals
|
MNU-treated animals
|
||||||
| Total pfu | No. of mutants | Mutation frequency, ×10−5 | Total pfu | No. of mutants | Mutation frequency, ×10−5 | ||
| Small intestine | a | 253,560 | 7 | 2.8 | 217,044 | 91 | 41.9 |
| b | 299,340 | 7 | 2.3 | 217,660 | 78 | 35.8 | |
| Mean | 2.6 ± 0.4 | 39 ± 4 | |||||
| Thymus | a | 259,080 | 5 | 1.9 | 201,550 | 61 | 30.3 |
| b | 257,900 | 17 | 6.6 | 214,040 | 39 | 18.2 | |
| Mean | 4.3 ± 3.3 | 24 ± 9 | |||||
| Tissue | Animal | BC-1/MSH2−/−
|
|||||
|---|---|---|---|---|---|---|---|
| Untreated control animals
|
MNU-treated animals
|
||||||
| Total pfu | No. of mutants | Mutation frequency, ×10−5 | Total pfu | No. of mutants | Mutation frequency, ×10−5 | ||
| Small intestine | a | 282,640 | 93 | 32.9 | 221,340 | 330 | 149 |
| b | 269,980 | 99 | 36.7 | 200,420 | 687 | 343 | |
| c | 246,440 | 80 | 32.5 | 202,640 | 613 | 303 | |
| Mean | 34 ± 2 | 265 ± 102 | |||||
| Thymus | a | 300,460 | 89 | 29.6 | 203,420 | 394 | 194 |
| b | 295,020 | 166 | 56.3 | 214,920 | 652 | 303 | |
| c | 281,640 | 155 | 55.0 | 200,840 | 566 | 282 | |
| Mean | 47 ± 15 | 260 ± 58 | |||||
| Heart | a | 215,740 | 34 | 15.8 | 252,360 | 64 | 25.4 |
| b | 96,080 | 16 | 16.7 | 88,660 | 42 | 47.4 | |
| c | 196,980 | 54 | 27.4 | 94,480 | 60 | 63.5 | |
| Mean | 20 ± 6 | 45 ± 19 | |||||
pfu, plaque forming units.
Figure 1.
LacI mutation frequencies (and standard errors) from tissues from MSH2+/+, MSH2+/−, and MSH2−/− animals, untreated and treated with MNU.
Spectrum of Alkylation-Induced Mutations.
The spectrum of mutations within small intestinal and thymic DNA obtained from MNU-treated MSH2+/+ and MSH2−/− animals are presented in Table 2. The types of mutations observed were similar for both small intestine and thymus (data not shown) and thus were combined. Mutations isolated more than once per tissue per animal were included only once in the mutation frequency and spectrum data presented. This was done to eliminate any potential bias arising from the clonal expansion of cells harboring a particular mutation.
Table 2.
Mutation spectra from small intestine and thymus of MNU-treated and untreated control and MSH2−/− animals corrected for clonality
| Mutation | Untreated MSH2+/+
|
MNU-treated MSH2+/+
|
Untreated MSH2−/−
|
MNU-treated MSH2−/−
|
||||
|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | |
| Transitions | ||||||||
| G:C −> A:T | 16 | 50 | 29 | 74 | 15 | 42 | 31 | 82 |
| A:T −> G:C | 4 | 13 | 0 | 0 | 9 | 25 | 0 | 0 |
| Transversions | 9 | 28 | 7 | 18 | 6 | 17 | 1 | 3 |
| Deletions | 1 | 3 | 0 | 0 | 1 | 3 | 2 | 5 |
| Frameshifts | 2 | 6 | 3 | 8 | 5 | 14 | 4 | 11 |
| Total | 32 | 100 | 39 | 100 | 36 | 100 | 38 | 100 |
In untreated controls, G:C to A:T and A:T to G:C transitions were both present (50% and 13%, respectively) and together formed the most frequent class of mutation, followed by transversions (28%) (Table 2). MNU treatment of controls strikingly altered the distribution of lacI transition mutations, these being restricted to G:C to A:T mutations (74%).
The spontaneous lacI mutation spectrum of MMR-deficient mice was similar to that of controls, in that transitions, both G:C to A:T and A:T to G:C, predominated (42% and 25%, respectively) (Table 2). The proportion of frameshift mutations was increased and transversions were decreased in MSH2-deficient mice as compared with controls. Similar to controls treated with MNU, the spectrum shifted, with 82% of all mutations being G:C to A:T transitions. LacI mutants sampled from MNU-treated MMR-deficient mice also showed no A:T to G:C transitions.
Sequence Specificity of Mutations.
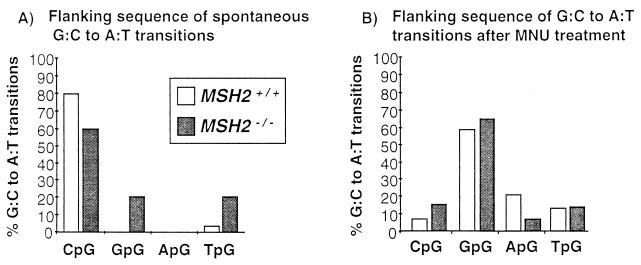
In addition to a shift in mutation type, the site specificity of G:C to A:T transitions was altered in MNU-treated mouse tissues (Fig. 2). In untreated controls, 80%, and in untreated MSH2−/− animals, 60%, of G:C to A:T transitions occurred at CpG sites. In contrast, after MNU treatment, the majority of G:C to A:T transitions were at GpG sites in both MSH2+/+ and MSH2−/− mice (59% and 65%, respectively).
Figure 2.
Flanking sequence of G:C to A:T transitions observed in untreated MSH2+/+ and MSH2−/− animals (A) and MNU-treated MSH2+/+ and MSH2−/− animals (B).
The dramatic alteration in mutation spectrum of MNU-treated animals confirmed that mutation frequency inductions observed were mutagen-specific and also eliminated the possibility that high endogenous background mutation frequencies had been present in some of the animals.
DISCUSSION
To determine the mutability of mice heterozygous or homozygous for a mutation in the MSH2 mismatch repair gene, MSH2−/−, MSH2+/−, and MSH2+/+ animals carrying a mutational reporter system were challenged with the alkylating agent MNU. We show that mice heterozygous for MSH2 demonstrated lacI mutation frequency inductions similar to those of controls, whereas mice deficient in MSH2 were hypersensitive to mutation induction by this agent.
Prior studies on alkylation-induced mutations in MMR-deficient systems have been limited to analyses of tumor cell lines such as MT-1, which is deficient in hMSH6/GTBP (20), a subunit of the hMSH2α complex (21, 22). MNNG-induced mutations of hprt genes in MT-1 cells were predominantly G:C to A:T transitions (20). The use of lacI transgenic mice, however, has permitted an in vivo assessment of mutations in a passive reporter gene within MMR-deficient hosts after exposure to an alkylating agent.
Previously, we demonstrated that spontaneous mutation frequencies in MSH2+/− mice were similar to those of controls, whereas mutation frequencies in MSH2−/− mice demonstrated 5- to 15-fold elevations as compared with controls in the tissues examined (10). Herein, we have shown that MNU administration led to an increase in mutation frequency in MSH2+/− animals that was similar to that of the MSH2+/+ animals. The responses of these animals were also in keeping with studies employing “Big Blue” lacI transgenic mice, as well as rat cell lines (23–25). The lower inductions seen in heart, as compared with small intestine and thymus, were consistent with the reduced mutation frequency increases observed in brain (23), another tissue having relatively low levels of cell turnover in the adult animal.
In contrast to the results obtained in the MSH2+/− and MSH2+/+ mice, MNU treatment of MSH2−/− mice resulted in a dramatic mutation frequency increase over the already elevated spontaneous backgrounds, reaching lacI mutation frequencies of >250 × 10−5 for small intestine and thymus (Fig. 1). Thus, when compared with controls, MSH2−/− animals proved exquisitely sensitive to mutation induction by this DNA methylating agent. Alkylation tolerance is a likely factor contributing to the greatly elevated mutation frequencies observed in MSH2−/− tissues, because MSH2−/− cells experiencing high levels of DNA damage would be predicted not to undergo apoptosis, thus contributing to the tissue-specific mutation frequency data obtained.
When background MSH2−/− mutation frequencies were subtracted from MNU-induced frequencies, the resulting increase because of alkylation varied from tissue to tissue, with thymus and small intestine demonstrating much greater inductions (5- to 8-fold) than were present in heart. Although these differences might have resulted from multiple factors, such as differences in drug distribution or differences in O6-methylguanine-DNA-methyltransferase levels in the different tissues (16), they likely reflect primarily differences in tissue-specific mitotic activity (23).
MNU-induced mutations in repair-proficient cells (Table 2, Fig. 2) were similar to those obtained with “Big Blue” transgenic mice, where 91% of all MNU mutations sequenced were G:C to A:T transitions, with 71% occurring at the 3′ G of GpG sequences (23). This was also in keeping with studies in bacteria that indicated that mispairing of O6-meG with T was the principal mutagenic mechanism of MNU-induced DNA base alkylation (12, 26–32). MNU-induced mutations of hprt were also dominated by G:C to A:T transitions at A/GpG sites, further suggesting O6-meG as the principal premutagenic lesion stemming from MNU exposure (33, 34).
The increased sensitivity of the MSH2−/− animals to alkylating agents and the similarity of the induced lacI mutant spectrum to that of MNU-treated controls suggested that MSH2−/− mice may prove to be useful adjuncts in evaluating the mutagenic potential of chemicals with potential DNA alkylating activity.
Do alkylating agents play a role in the genesis of colonic tumors in individuals with HNPCC? These compounds may be endogenously produced in the intestine from dietary nitrates that are reduced to nitrites in the proximal colon by bacteria, resulting in N-nitroso compounds with alkylating activity (35–37). In addition, amines and amides arising from protein catabolism are substrates for nitrosation by nitric oxide, with the resulting alkylating N-nitroso compounds being capable of DNA alkylation (38, 39). Other sources of methylating agents in the colon potentially include activated subepithelial macrophages (40) or nitrosated bile acids in the bowel lumen (41). Indeed, O6-meG-containing DNA has been demonstrated in both normal and malignant colonic epithelia of humans (42), indicating that sources of alkylating compounds exist in the normal colon. Also, MNU-induced murine lymphomas typically contain activating mutations of K-ras within the GpG of codon 12 (43, 44), a site frequently involved in human colorectal cancers, including HNPCC (45, 46). Although the K-ras mutations could arise by other mechanisms, their presence suggests a potential role for agents capable of alkylating O6-G in this process. It could be speculated that an alkylating agent(s) gradient exists in the human colon, with the highest concentrations being present in the proximal colon, as a way of explaining the tendency of HNPCC lesions to arise in the proximal bowel.
Whether MMR-deficient cells arise spontaneously in the normal colonic mucosa of HNPCC carriers, and whether these mutational events tend to predominate in the proximal colon or are distributed equally throughout the colon, is not known. However, given the presence of a mutagen gradient as suggested above, it might be predicted that the second MSH2 allele would be at increased risk for mutational inactivation in the proximal colon, with such cells subsequently undergoing the increased mutation rates typical of MMR deficiency. There are very few reports on the nature of the second somatic MMR gene mutation in HNPCC colon tumors, and therefore there is insufficient evidence at present to exclude DNA alkylation damage as an etiologic factor in these mutations (2, 3, 47–49). If MMR deficient cells were to occur sporadically throughout the colon, our results suggest that in the presence of the putative alkylating agent gradient these cells would be at a greatly increased risk for G:C to A:T transition mutations within the proximal colon.
MSH2 nullizygous but not heterozygous mice demonstrated an increased incidence of small intestinal tumors but were not predisposed to lesions of the large bowel, even on a Min−/− background (7). This difference from human HNPCC likely stems from a host of interspecies variables, including differences in life span, diet composition and mutagen content, intestinal flora and level of gut colonization, as well as the ability of the rodent to generate endogenous ascorbic acid (50).
We have determined the in vivo mutational frequencies and spectra of three tissues from MSH2+/+, MSH2+/−, and MSH2−/− animals. On exposure to MNU, heterozygosity for MSH2 resulted in mutation frequency inductions similar to those of control animals. In contrast, MSH2-deficient mice were highly sensitive to mutation induction by this agent. MNU treatment resulted almost exclusively in G:C to A:T transitions in both MSH2+/+ and MSH2−/− animals, in keeping with the miscoding properties of O6-meG (13). These results suggest that HNPCC carriers may not be at increased risk for alkylation-induced mutations. However, spontaneously arising MMR-deficient cells would be predicted to accumulate mutations at a greatly accelerated rate on exposure to specific types of alkylating agents.
Acknowledgments
We are indebted to P. Glazer and S. Baker for their critical reviews of this manuscript. We also thank David Spear of the Canadian Human Genetic Diseases Network Sequencing Core Facility. This work was funded by the National Cancer Institute of Canada with funds from the Canadian Cancer Society. F.R.J. is the recipient of a Research Scientist Award from the Canadian Arthritis Society. S.E.A. holds a Medical Research Council of Canada Postdoctoral Fellowship award.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: MMR, mismatch repair; HNPCC, hereditary non-polyposis colorectal cancer; MNU, N-methyl-N-nitrosourea.
References
- 1.Kolodner R D. Trends Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 2.Fishel R A, Lescoe M K, Rao M R S, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 3.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 4.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 5.Reitmair A H, Schmits R, Ewel A, Bapat B, Redston M, Mitri A, Waterhouse P, Mittrucker H-W, Wakeham A, Liu B, Thomason A, Griesser H, Gallinger S, Ballhausen W G, Fishel R, Mak T W. Nat Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- 6.deWind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 7.Reitmair A H, Redston M, Cai J-C, Chuang T C Y, Bjerknes M, Cheng H, Hay K, Gallinger S, Bapat B V, Mak T W. Cancer Res. 1996;56:3842–3849. [PubMed] [Google Scholar]
- 8.Baker S, Bronner C E, Zhang L, Plug A W, Robatzek M, Warren G, Elliott E A, Yu J, Ashley T, Arnheim N, Flavell R A, Liskay R M. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 9.Narayanan L, Fritzell J A, Baker S M, Liskay R M, Glazer P M. Proc Natl Acad Sci USA. 1997;94:3122–3127. doi: 10.1073/pnas.94.7.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrew S E, Reitmair A H, Fox J, Hsiao L, Francis A, McKinnon M, Mak T W, Jirik F R. Oncogene. 1997;15:123–129. doi: 10.1038/sj.onc.1201180. [DOI] [PubMed] [Google Scholar]
- 11.Loveless A. Nature (London) 1969;223:206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- 12.Horsfall M J, Gordon A J E, Burns P A, Zielenska M, van der Vliet M E, Glickman B W. Environ Mol Mutagen. 1990;15:107–122. doi: 10.1002/em.2850150208. [DOI] [PubMed] [Google Scholar]
- 13.Swann P F. Mutat Res. 1990;233:81–94. doi: 10.1016/0027-5107(90)90153-u. [DOI] [PubMed] [Google Scholar]
- 14.Griffin S, Karran P. Biochemistry. 1993;32:13032–13039. doi: 10.1021/bi00211a012. [DOI] [PubMed] [Google Scholar]
- 15.Fishel R, Kolodner R D. Curr Opin Gen Dev. 1995;5:382–395. doi: 10.1016/0959-437x(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 16.Karran P, Bignami M. Nucleic Acids Res. 1992;20:2933–2940. doi: 10.1093/nar/20.12.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrew S E, Pownall S, Fox J, Hsiao L, Hambleton J, Penney J E, Kohler S W, Jirik F R. Mutat Res. 1996;357:57–66. doi: 10.1016/0027-5107(96)00080-2. [DOI] [PubMed] [Google Scholar]
- 18.Kohler S W, Provost G S, Kretz P L, Fieck A, Sorge J A, Short J M. GATA. 1990;7:212–218. doi: 10.1016/0735-0651(90)90003-x. [DOI] [PubMed] [Google Scholar]
- 19.Kohler S W, Provost G S, Fieck A, Kretz P L, Bullock W O, Sorge J A, Putman D L, Short J M. Proc Natl Acad Sci USA. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kat A, Thilly W G, Fang W-H, Longley M J, Li G-M, Modrich P. Proc Natl Acad Sci USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drummond J T, Li G-M, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 22.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan J J, Jiricny J. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 23.Provost G S, Kretz P L, Hamner R T, Matthews C D, Rogers B J, Lundberg K S, Dycaico M J, Short J M. Mutat Res. 1993;288:133–149. doi: 10.1016/0027-5107(93)90215-2. [DOI] [PubMed] [Google Scholar]
- 24.Allay E, Koc O N, Gerson S L. Proc Am Assoc Cancer Res. 1994;35:115. (abstr.). [Google Scholar]
- 25.Wyborski D L, Malkhosyan S, Moores J, Perucho M, Short J M. Mutat Res. 1995;334:161–165. doi: 10.1016/0165-1161(95)90007-1. [DOI] [PubMed] [Google Scholar]
- 26.Suri A, deBoer J, Kusser W, Glickman B W. Mutat Res. 1996;372:23–31. doi: 10.1016/S0027-5107(96)00105-4. [DOI] [PubMed] [Google Scholar]
- 27.Burns P A, Gordon A J E, Glickman B W. Carcinogenesis. 1988;9:1607–1610. doi: 10.1093/carcin/9.9.1607. [DOI] [PubMed] [Google Scholar]
- 28.Richardson K K, Richardson F C, Crosby R M, Swenberg J A, Skopek T R. Proc Natl Acad Sci USA. 1987;84:344–348. doi: 10.1073/pnas.84.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DuBridge R B, Tang P, Hsia H C, Lelong P M, Miller J H, Calos M P. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed J, Hutchinson F. Mol Gen Genet. 1987;208:446–449. doi: 10.1007/BF00328137. [DOI] [PubMed] [Google Scholar]
- 31.Glickman B W, Horsfall M J, Gordon A J, Burns P A. Environ Health Perspect. 1987;76:29–32. doi: 10.1289/ehp.877629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon A J E, Glickman B W. Mutat Res. 1988;208:105–108. doi: 10.1016/s0165-7992(98)90008-2. [DOI] [PubMed] [Google Scholar]
- 33.Akagi T, Hiromatsu K, Iyehara-Ogawa K, Kimura H, Kato T. Carcinogenesis. 1993;14:725–729. doi: 10.1093/carcin/14.4.725. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L-H, Jenssen D. Carcinogenesis. 1991;12:1903–1909. doi: 10.1093/carcin/12.10.1903. [DOI] [PubMed] [Google Scholar]
- 35.Kang H-I, Konishi C, Kuroki T, Huh N-H. Carcinogenesis. 1995;16:1277–1280. doi: 10.1093/carcin/16.6.1277. [DOI] [PubMed] [Google Scholar]
- 36.Bartsch H, Oshima H, Shuker D E G, Pignatelli B, Calmels S. Mutat Res. 1990;238:255–267. doi: 10.1016/0165-1110(90)90017-6. [DOI] [PubMed] [Google Scholar]
- 37.Bingham S A, Pignatelli B, Pollock J R A, Ellul A, Malaveille C, Gross G, Runswick S, Cummings J H, O’Neill I K. Carcinogenesis. 1996;17:515–523. doi: 10.1093/carcin/17.3.515. [DOI] [PubMed] [Google Scholar]
- 38.Rowland I R, Granli T, Bockman O C, Key P E, Massey R C. Carcinogenesis. 1991;12:1359–1401. doi: 10.1093/carcin/12.8.1395. [DOI] [PubMed] [Google Scholar]
- 39.Ward F W, Coates M E, Walker R. Food Chem Toxicol. 1989;27:445–449. doi: 10.1016/0278-6915(89)90030-6. [DOI] [PubMed] [Google Scholar]
- 40.Marletta M A. Chem Res Toxicol. 1988;1:249–257. doi: 10.1021/tx00005a001. [DOI] [PubMed] [Google Scholar]
- 41.Puju S, Shuker D E G, Bishop W W, Falchuk K R, Tannenbaum S R, Thilly W G. Cancer Res. 1982;42:2601–2604. [PubMed] [Google Scholar]
- 42.Hall C N, Badawi A F, O’Connor P J, Saffhill R. Br J Cancer. 1991;64:59–63. doi: 10.1038/bjc.1991.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diamond L E, Guerrero I, Pellicer A. Mol Cell Biol. 1988;8:2233–2236. doi: 10.1128/mcb.8.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newcomb E W, Ateinber J J, Pellicer A. Cancer Res. 1988;48:5514–5521. [PubMed] [Google Scholar]
- 45.Forrester K, Almoguera C, Han K, Grizzle W E, Perucho M. Nature (London) 1987;327:298–303. doi: 10.1038/327298a0. [DOI] [PubMed] [Google Scholar]
- 46.Bos J L. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 47.Nicolaides N C, Papadopoulos N, Liu B, Wei Y-F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter J C, Dunlop M G, Hamilton S R, Petersen G M, de la Chapelle A, Vogelstein B, Kinzler K W. Nature (London) 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 48.Lu S-L, Akiyama Y, Nagasaki H, Nomizu T, Ikeda E, Baba S, Ushio K, Iwama T, Maruyama K, Yuasa Y. Jpn J Cancer Res. 1996;87:279–287. doi: 10.1111/j.1349-7006.1996.tb00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Onda A, Okumara Y, Kishi N, Iwama T, Mori T, Koike M, Ushio K, Chiba M, Nomizu S, Konishi F, Utsunomiya J, Miyaki M. Gastroentology. 1996;111:307–317. doi: 10.1053/gast.1996.v111.pm8690195. [DOI] [PubMed] [Google Scholar]
- 50.Meister A. J Biol Chem. 1994;269:9397–9400. [PubMed] [Google Scholar]