Abstract
Interphasic nuclear organization has a key function in genome biology. We demonstrate that p21WAF-1, by influencing gene expression and inducing chromosomal repositioning in tumor suppression, plays a major role as a nuclear organizer. Transfection of U937 tumor cells with p21WAF-1 resulted in expression of the HUMSIAH (human seven in absentia homologue), Rb, and Rbr-2 genes and strong suppression of the malignant phenotype. p21WAF-1 drastically modified the compartmentalization of the nuclear genome. DNase I genome exposure and fluorescence in situ hybridization show, respectively, a displacement of the sensitive sites to the periphery of the nucleus and repositioning of chromosomes 13, 16, 17, and 21. These findings, addressing nuclear architecture modulations, provide potentially significant perspectives for the understanding of tumor suppression.
Keywords: cancer genetics, chromatin architecture, genome organization, p53
The concept of specific topological and functional architecture in the organization of the genome in the nuclei of eukaryotic cells is supported by accumulating evidence (1–3). Remodeled chromatin has been initially detected through previous work on DNase I hypersensitivity sites associated with gene activation (4–6). Factors involved in transcript processing are identified within discrete foci in nuclei (7, 8). Spatial rearrangement of chromatin was recently observed during the cell cycle (9, 10). Studies in Drosophila demonstrate a position effect variegation with striking consequences on gene expression depending upon the gene’s distance from the centromere and its association with a block of heterochromatin (11–13).
We recently designed model systems to study tumor suppression. We derived cells with a strongly suppressed malignant phenotype from the malignant human erythroleukemic cell line K562 and from the monocytic cell line U937 (14, 15). We used the H-1 parvovirus as a tool to isolate the cells with a suppressed phenotype from a population of malignant cells. Although the parental cells are tumorigenic both in vitro and in vivo, the H-1 parvovirus-selected clones (US3 and US4) display a strongly reduced tumorigenicity. For example, U937 cells form tumors in 80% of injected sites in scid/scid mice; in contrast, the US3 do not form any tumors at all and the US4 cells form one tumor in 20 injected sites (15). We have identified some of the molecular events underlying tumor suppression in these models. Importantly, these cells with a suppressed malignant phenotype show reactivation of the molecular pathways of tumor suppression. Whereas the K562-derived KS clones reexpress the p53, the US3 and US4 cells reexpress p21WAF-1 independently from the p53 pathway (14, 15).
Because p21WAF-1 is activated in different model systems of tumor suppression and apoptosis, and considering its association with p53 signaling and its role as an inhibitor of cyclin dependent kinases (16–19) as well as its absence in knock-out cells resulting in chromosomal endoreduplication (20), we investigated its potential role—as a reorganizer of the nucleus—in tumor suppression.
Previously, chromosomal translocations, point mutations, and loss of genetic material have been identified as the major genetic alterations in cancer. In the present work, we have identified an event associated with cancer—global disorganization of chromosomal positioning in the nucleus. This altered nuclear architecture is shown to be reversible by p21WAF-1 during tumor suppression, including a repositioning of the chromosomes.
MATERIALS AND METHODS
Transfections.
The full-length cDNA encoding human p21WAF-1 (generously provided by B. Weinstein, Columbia University, New York) has been cloned into the EcoRI site of the pBK–RSV phagemid vector (Stratagene). The constructs were transfected using Lipofectin (GIBCO/BRL). U937 cells (3.5 × 106) were incubated 8 h with the Lipofectin (30 μg)–DNA (20 μg) complex. Selection was done in the presence of 1.5 mg/ml of G418.
Northern Blot Analysis.
RNA extraction and Northern blot analysis were performed using standard procedures. p21WAF-1 probe corresponds to the 2.1-kb cDNA. For HUMSIAH (human seven in absentia homologue), a 1.4-kb fragment of the cDNA was used. The Rb probe is HP126 (Medgene). The Rbr-2 probe is a 393-bp PCR fragment obtained with the following primers: 5′-TGGGACAGCTACCGCAGCATGAGCGA-3′ and 5′-CACAGTACAGGGCTGTCGCCGCTGTT-3′.
Tumorigenicity Analysis.
Injection of scid/scid mice was done as described (14). Cells (107) were injected per site. Animals were followed for 3 months. The Mann–Whitney test was used for the statistical analysis.
DNase I Genome Exposure.
The DNase I genome exposure assay was modified from Puck et al. (5). Cells were fixed in 4% paraformaldehyde/PBS, dehydrated, and stored at −80°C. In situ nick-translation (NT) was performed using 30 units of DNase I (Boehringer Mannheim) in NT buffer (50 mM Tris⋅HCl, pH 8/5 mM MgCl2/10 mM 2β-mercaptoethanol/50 ng/ml BSA) at room temperature and 40 units of DNA polymerase I (Boehringer Mannheim), 100 mM dNTP digoxigenin labeling mix (Boehringer Mannheim). DNA in situ repair was revealed using anti-digoxigenin peroxidase conjugated antibodies (Boehringer Mannheim) and tyramide–tetramethylrhodamine B isothiocyanate (DuPont/NEN) as substrate. Slides were counterstained with chromomycin A3 (Sigma). Analysis was done by confocal scanning laser microscopy (MRC 600; Bio-Rad) in fluorescence mode.
Fluorescent In Situ Hybridization (FISH).
FISH was performed as described (21). The following probes (Oncor) were used: chromosome 16 centromeric alphoid probe (D16Z2), chromosome 17 centromeric alphoid probe (D17Z1), chromosomes 13/21 centromeric alphoid probes (D13Z1/D21Z1), p53, and Rb. Analysis was done by epifluorescence microscopy [with a cooled three-charged coupled device (3CCD) camera (Lhesa France) equipped with a triple band pass] using low numeric aperture (N.A.) lenses (×40; 0.75 N.A.), digitalized in a matrix of 768 × 512 pixels. Each pixel corresponds to a 55 × 55 nm of the object.
Image Analysis.
Distance measurements were performed using a samba ips image analysis system (Unilog, Meylan, France). The markers were extracted by top-hat transformation, and the nuclei were segmented by thresholding the 4′,6-diamidino-2-phenylindole fluorescence image. Two binary images were obtained, respectively, for markers and nuclei. Successive erosion was used to measure the distance between the markers and the nuclear border. The markers’ positions were determined by the intersection of their image with the distance one. This method allows to analyze the totality of nuclei, independently of their shape. To compensate the differences in nuclear size, the measurements were systematically normalized by dividing each distance value by the corresponding nuclear radius.
Model Distribution of Distances.
Model curves used for comparison with experimentally derived distances were obtained by stereological simulation. The nuclei in three-dimensional representation were assimilated to a spherical model formed by a set of cylinders. The volume of one elementary cylinder (vi) is related to the probability (Pi) of markers presence by Pi = vi/Σ vi.
If R is the nuclear radius, ri the cylinder radius, ΔR the thickness of cylinder, and hi the height, then vi = Π ΔR (2 ri − ΔR) hi.
This equation allows to trace the random distribution in a sphere and the peripheral distribution in shells of different thickness. With this model, the function obtained is independent of elliptical spreading of nuclei. FISH spots at one setting of the focus adjustment (equatorial plan) were analyzed. More than 100 nuclei were evaluated per case.
Statistical Analysis.
The nuclear radius was divided into eight classes. Each measured distance between the nuclear border, and the FISH signals was categorized into one of these eight classes. The χ2 test was used to probe two hypotheses. (i) The measured distances in p21WAF-1 U937 cells were distributed in a similar way as these measured for the U937 cells transfected with the vector alone. (ii) The measured distances in both cell lines were distributed in a similar way as are random points in a sphere. P < 0.001 was considered to indicate a significant difference.
RESULTS AND DISCUSSION
p21WAF-1 Induces Specific Gene Expression and Tumor Suppression.
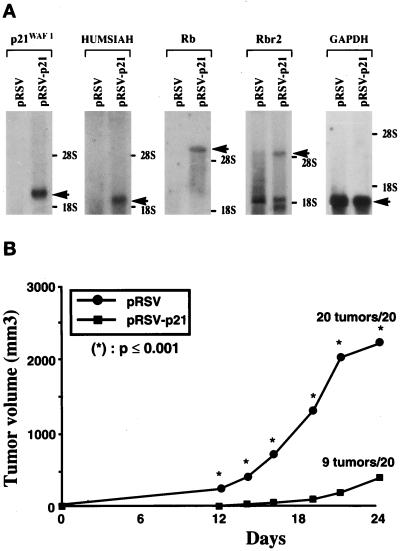
Parental U937 tumor cells from monocytic origin barely express any p21WAF-1 at the mRNA or protein level. In contrast, the daughter cell lines US3 and US4 that we derived with a suppressed malignant phenotype express constitutively high levels of p21WAF-1 (15). These observations led us to investigate the effect of p21WAF-1 in stable transfected U937 cells on specific gene expression and tumor suppression. These transfected cells (pRSV-p21) expressed high levels of p21WAF-1 at the protein level (data not shown). As shown in Fig. 1A, the p21-transfected cells exhibit high levels of HUMSIAH. This gene (or TSAP 3; tumor suppressor activated pathway 3) is activated by wild-type p53 at the early onset of programmed cell death, during physiological apoptosis and tumor suppression (15, 22). The retinoblastoma susceptibility gene Rb (23) and the gene encoding the Rb-related p130 (Rbr-2) (24) are also strongly activated in the p21 transfectants of U937 cells (Fig. 1A). The expression of the p53 gene at the mRNA and protein levels remained undetectable in both the control and the transfectants (data not shown). We analyzed the expression of the Rb gene because of its established tumor suppressive properties (23) and Rbr-2 (24), for its implication in the cell cycle. However, the cell cycle analysis by fluorescence-activated cell sorter in three independent experiments (using propidium iodide) did not reveal any significant difference between the U937 and the p21WAF-1 transfectants (typical experiment: for U937 cells transfected with the vector alone, in G1 65%/S 29%/G2 6% and for the p21WAF-1 transfectants G1 64%/S 30%/G2 6%). The doubling time was also similar for both cell lines, with 33 h for the U937 cells and 36 h for the p21WAF-1 transfectants, respectively. Importantly, these p21WAF-1 stable transfected U937 cells have a significantly suppressed malignant phenotype (Fig. 1B). After 24 days, the scid/scid mice injected with the U937 cells containing the control vector alone had tumors at all sites (20/20) and of such a volume that they had to be killed. In contrast, only 9/20 injection sites with pRSV-p21 U937 cells formed tumors. These tumors were significantly delayed in appearance and of much smaller volume. No new tumors were detected after an additional 6 weeks of observation. This suppression of the malignant phenotype corroborates previous findings where it was demonstrated that p21WAF-1 acts as a tumor suppressor in vivo (25). We suggest that the specific induction of gene expression by p21WAF-1 in the present case, without important cell cycle variations, is part of the activation of the “molecular engine of tumor suppression” (14, 22).
Figure 1.
(A) Differential expression of mRNA in control pRSV-transfected U937 cells and p21WAF-1-expressing cells (pRSV-p21). Northern blot analysis indicated activation of p21WAF-1, HUMSIAH, Rb, and Rbr-2 in pRSV-p21-transfected cells compared with cells transfected with the vector alone. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is used as control probe. (B) Tumorigenicity of U937 cells transfected with the vector alone compared with the pRSV-p21-transfected cells after subcutaneous injection of 107 cells in scid/scid mice. ∗, P value that for each point was significantly different (P ≤ 0.001) between the pRSV and pRSV-p21-transfected U937 cells.
p21WAF-1 Modulates Genomic Exposure to DNase I.
We further assessed whether p21WAF-1 has a more global role as “reorganizer” of the nucleus. The rationale to investigate nuclear organization stems from a series of experiments during the past decade (1, 2, 5, 11, 12) indicating that chromosome positions within the nucleus may contribute to phenotypic changes. Transcriptionally active chromatin is preferentially sensitive to digestion by nucleases. Such chromatin usually contains the DNase I hypersensitive sites (4). However, these nucleosome-free DNA segments are also associated with a series of regulatory functions in gene expression. For the purpose of this study, we investigated the hypersensitivity to DNase I in situ because it is a powerful tool for a global analysis of the genome organization. The initial experiments of Puck et al. (5) already suggested a difference between normal and malignant cells in compartmentalization of chromatin containing DNase I sensitive sites. Nevertheless, no molecular mechanism has been hitherto suggested for these observations. We hypothesized that p21WAF-1 might play such a role of nuclear organizer based upon its above mentioned properties (16–18). The most striking result is the chromosomal endoreduplication in p21WAF-1 knock-out cells that may represent the first step in aneuploidy, giving rise to the chromosomal variability found in tumors (20). Studies describing the overexpression of p21WAF-1 in transgenic mice and transfection of human breast carcinoma cell lines indicate its involvement in the formation of polyploid nuclei and giant cells (26, 27).
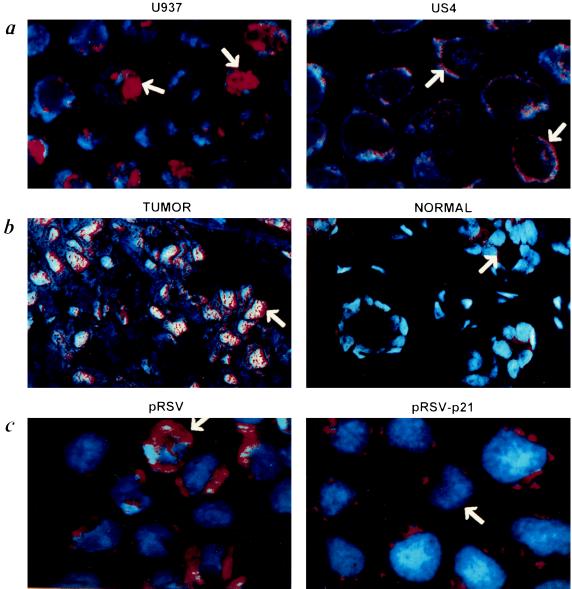
Genomic exposure studies were carried out using a nick translation technique so as to make visible the exposed DNase I-sensitive nuclear DNA in situ. Genomic exposure was examined by epifluorescence and confocal microscopy (Fig. 2). The parental U937 cancer cells (Fig. 2a; U937) or those transfected with the control vector alone (Fig. 2c; pRSV) display a diffuse pattern such as the one observed in breast carcinoma biopsies, specifically in the malignant cells (Fig. 2b; TUMOR). p21WAF-1 transfected U937 cells (Fig. 2c; pRSV-p21) have radical changes in their genome exposure to DNase I, displaying a pattern similar to the one of suppressed phenotype US3 and US4 cells (Fig. 2a; US4), and epithelial cells of normal breast tissue (Fig. 2b; NORMAL). p21WAF-1 induces redistribution of DNase I hypersensitivity sites in a shell at the periphery of the nucleus. This nuclear reorganization might be associated with the activation of genes in tumor suppression that are silenced in the tumoral process. Although it is outside the scope of this study to investigate the mechanisms by which p21WAF-1 induces a reorganization of the genome, at least two hypotheses could be considered. This nuclear reorganization, induced by p21WAF-1 is either a direct effect or a consequence of its function as inhibitor of cdks activity. One of the characterized targets for mitotic cdks is the group of nuclear lamins (28). Changes in the phosphorylation status of these nuclear lamins might result in nuclear modifications. Because we observe an important change in the distribution of DNase I exposure during tumor suppression, one possible mechanism underlying these events is the triggering, initiated by p21, of chromosomal movements in the nucleus. We addressed this question next.
Figure 2.
DNase I genome exposure analysis using confocal microscopy. (a) The U937 cells have a diffuse pattern for the DNase I genome exposure, whereas the US4 cells with a suppressed malignant phenotype exhibit a rim-like pattern (arrow). (b) Difference between breast cancer cells and normal cells. The normal cells have a weak signal with a rim-like pattern (arrow). (c) U937 cells transfected with pRSV exhibit a diffuse pattern while those expressing the p21WAF-1 (pRSV-p21) have a rim-like pattern for the DNase I sensitivity.
p21WAF-1 Regulates Chromosome Positioning Within the Nucleus.
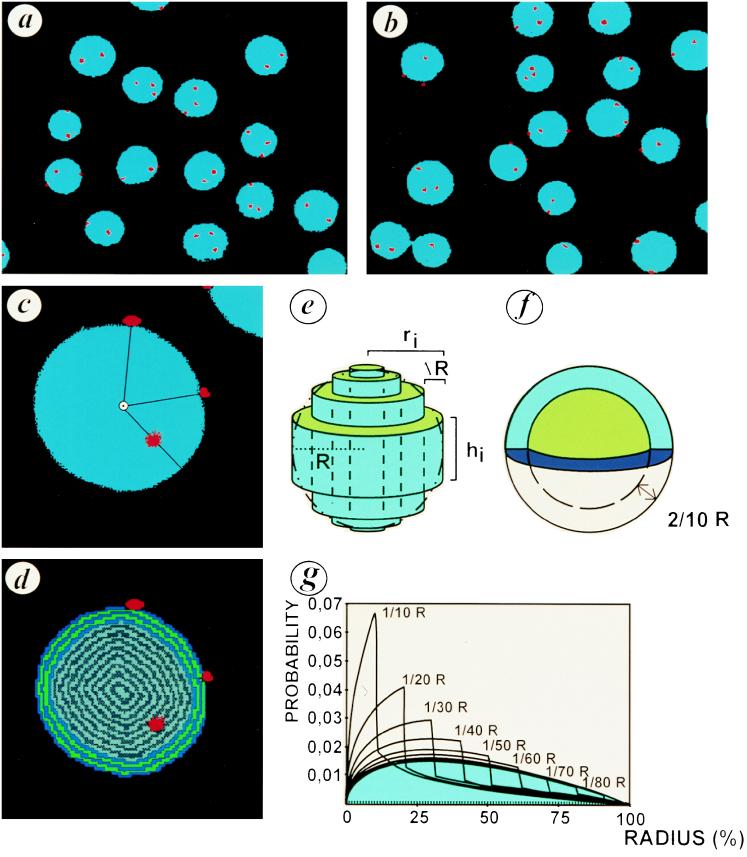
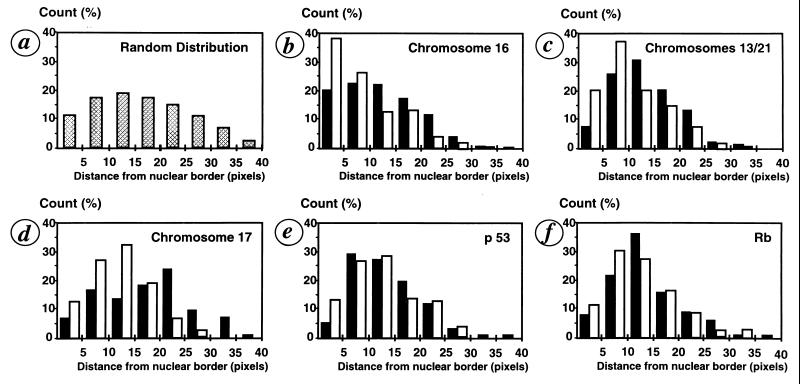
For every probe used, the distance was measured from the border of the nucleus and integrated in a model, as exemplified in Fig. 3 for a chromosome 16 centromeric probe. We compared U937 cells transfected with the vector alone and the p21WAF-1 U937 transfectants regarding chromosome and specific gene positioning within the nucleus. HUMSIAH and Rbr-2 are colocalized on chromosome 16q12 (15, 24) whereas Rb is located on chromosome 13q14 (29) and p53 on chromosome 17p13.1. Using FISH with alphoïd centromeric probes corresponding to chromosomes 16, 13/21, and 17 (Fig. 4), we found that the p21WAF-1-transfected U937 cells display a global redistribution of the signals with a clear shift to the peripheral region of the nucleus. This distribution of the FISH signals corresponding to the centromeric probes is statistically different between the tumor cells and those with a suppressed phenotype. The χ2 test is highly significant (P < 0.001). By using specific probes for the Rb and p53 genes, we did not observe a statistically significant difference, using the χ2 test, in distribution between the FISH spots in the pRSV alone and the p21WAF-1-transfected cells. From a physiological point of view, these observations are complementary to the growing evidence for a territorial interphase chromosome organization (1, 2, 5). Specific chromatin domains occupy distinct, nonrandom, spatial positions as reported previously (9, 10). Whether there is a correlation between gene expression and chromosome repositioning remains to be investigated further because some results indicate that both active and inactive genes localize preferentially in the periphery of chromosome territories (30). Our study suggests that in the specific case of p21WAF-1-induced tumor suppression, centromeric regions containing noncoding genetic material are repositioned. This repositioning of the centromeric regions of the chromosomes analyzed above may underlie the mechanism by which the architecture of the chromosomal territory is modified. Such territorial modifications might influence the spatial environment of specific genes and modulate their expression. Obviously, the precise topological conformation of genes within the nucleus may have functional repercussions. The recent work describing the position-effect variegation in Drosophila has clearly demonstrated the functional importance of a specific nuclear architecture on gene expression (11, 12).
Figure 3.
Repositioning of chromosome 16 in p21WAF-1 transfectants of U937 cells. (a and b) FISH using a chromosome 16 alphoid centromeric probe. The nuclei of the control transfectants U937 cells (a; pRSV) and the p21 transfectants (b; pRSV-p21) are counterstained with 4′,6-diamidino-2-phenylindole (blue). The FISH signal is revealed by tyramide–tetramethylrhodamine B isothiocyanate (red). Because of a trisomy 16, three spots are detected. (c) Magnification of a p21 transfected U937 with the distance code image of a nucleus (d). The successive strata (one pixel in size) were labeled as a function of the border distance. (e) Reconstruction of nuclear volume from a 2-D image. The sphere (R, radius) was fitted by a set of cylinders (radius = ri, height = hi, thickness = ΔR). (f) Peripheral distribution was modelized by random distribution in a shell whose thickness was 2/10 R. (g) Model of signal distribution in shells of different thicknesses according to the distance from the nuclear border. The blue area corresponds to the distribution of random points in a sphere.
Figure 4.
Histograms of the signals distribution as measured from the extreme border of the nuclei. (a) Distribution of random points in a sphere. (b–f) Distribution of the FISH signals as measured from the border of the nuclei for chromosome 16 (b), chromosomes 13/21 (c), chromosome 17 (d), p53 (e), and Rb gene (f), respectively. Comparison between the control (▪) U937 pRSV transfectants and U937 pRSV-p21 transfectants (□).
In conclusion, using both model systems for the analysis of tumor suppression and stable transfectants, our data indicate that p21WAF-1 induces repositioning of genetic material within the nucleus that is associated with potentially important regulatory functions for achieving the suppressed phenotype. These results suggest the central role played by p21WAF-1 in nuclear organization, chromosomal positioning, and activation of pathways of tumor suppression.
Acknowledgments
A.T. and R.B.A. dedicate this work to Daniel Cohen, Georges Schnek, and Marc Van Montagu for constant encouragement. G.L.-C. and H.B.-G. have contributed equally as first authors. V.A. and J.-P.R. are equal as second authors. R.B.A., F.C., and A.T. share equally the contribution to this work. We thank Gilles Thomas for his support. We thank Lucien Cazes and Fabienne Dufour for the cell cultures. The tumorigenicity assays were performed by Dr. Philippe Genne at ONCODESIGN. We also thank Paul Rabinow for his participation and involvement in the study as well as for reviewing the manuscript. We are grateful to Moshe Oren for his constructive criticism in discussing results. We thank Tom White and John Sninsky as well as Jacob Aten for stimulating discussion. We are indebted to Christian Rebollo for his involvement, as well as to Cécile Rouzaud and Pascale Villedieu for efficiency and help. This work was supported by the Ministère de la Recherche, the Association Française contre les Myopathies (to A.T. and R.B.A.), and the Association pour la Recherche sur le Cancer (to F.C.).
ABBREVIATIONS
- HUMSIAH
human seven in absentia homologue
- FISH
fluorescent in situ hybridization
References
- 1.Manuelidis L. Science. 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- 2.Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, et al. Cold Spring Harbor Symp Quant Biol. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- 3.Marshall W F, Dernburg A F, Harmon B, Agard D A, Sedat J W. Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchison N, Weintraub H. Cell. 1985;43:471–482. doi: 10.1016/0092-8674(85)90177-1. [DOI] [PubMed] [Google Scholar]
- 5.Puck T T, Bartholdi M, Krystosek A, Johnson R, Haag M. Somatic Cell Mol Genet. 1991;17:489–503. doi: 10.1007/BF01233173. [DOI] [PubMed] [Google Scholar]
- 6.Felsenfeld G. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence J B, Carter K C, Xing X. Cold Spring Harbor Symp Quant Biol. 1993;58:807–818. doi: 10.1101/sqb.1993.058.01.088. [DOI] [PubMed] [Google Scholar]
- 8.Krug R M. Curr Opin Cell Biol. 1993;5:944–949. doi: 10.1016/0955-0674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- 9.Hulspas R, Houtsmuller A B, Krijtenburg P J, Bauman J G J, Nanninga N. Chromosoma. 1994;103:286–292. doi: 10.1007/BF00352253. [DOI] [PubMed] [Google Scholar]
- 10.Nagele R, Freeman T, McMorrow L, Lee H. Science. 1995;270:1831–1835. doi: 10.1126/science.270.5243.1831. [DOI] [PubMed] [Google Scholar]
- 11.Csink A K, Henikoff S. Nature (London) 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- 12.Dernburg A F, Broman K W, Fung J C, Marshall W F, Philips J, Agard D A, Sedat J W. Cell. 1996;85:745–749. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 13.Marshall W F, Fung J C, Sedat J W. Curr Opin Genet Dev. 1997;7:259–263. doi: 10.1016/s0959-437x(97)80136-0. [DOI] [PubMed] [Google Scholar]
- 14.Telerman A, Tuynder M, Dupressoir T, Robaye B, Sigaux F, Shaulian E, Oren M, Rommelaere J, Amson R. Proc Natl Acad Sci USA. 1993;90:8702–8706. doi: 10.1073/pnas.90.18.8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemani M, Linares-Cruz H, Bruzzoni-Giovanelli J-P, Roperch M, Tuynder L, et al. Proc Natl Acad Sci USA. 1996;93:9039–9042. doi: 10.1073/pnas.93.17.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 17.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 18.Waga S, Hannon G J, Beach D, Stillman B. Nature (London) 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 19.Xiong Y, Hannon G, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–703. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 20.Waldman T, Lengauer C, Vogelstein B. Nature (London) 1996;381:713–716. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 21.Linares-Cruz G, Millot G, De Cremoux P, Vassy J, Olofsson B, Rigaut J P, Calvo F. Histochem J. 1995;27:15–23. doi: 10.1007/BF00164168. [DOI] [PubMed] [Google Scholar]
- 22.Amson R, Nemani J-P, Israeli L, Bougueleret L, et al. Proc Natl Acad Sci USA. 1996;93:3953–3957. doi: 10.1073/pnas.93.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberg R A. Science. 1992;254:1138–1145. doi: 10.1126/science.1659741. [DOI] [PubMed] [Google Scholar]
- 24.Hannon G J, Demetrick D, Beach D. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z Y, Perkins N D, Ohno T, Nabel E G, Nabel G J. Nat Med. 1995;1:1052–1056. doi: 10.1038/nm1095-1052. [DOI] [PubMed] [Google Scholar]
- 26.Sheikh M S, Rochefort H, Garcia M. Oncogene. 1995;11:1899–1905. [PubMed] [Google Scholar]
- 27.Wu H, Wade M, Krall L, Grisham J, Xiong Y, Van Dyke T. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- 28.Foisner R, Gerace L. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- 29.Bookstein R, Lee E, Y, To H, Young L, J, Sery T, W, Hayes R C, Friedmann T, Lee W, H. Proc Natl Acad Sci USA. 1988;85:2210–2214. doi: 10.1073/pnas.85.7.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurz A, Lampel S, Nickolenko J, Bradl J, Benner A, Zirbel R, Cremer T, Lichter P. J Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]