Abstract
We previously produced three congenic strains carrying lupus susceptibility genes (Sle1-Sle3) from the lupus-prone NZM2410 mouse on the C57BL/6 background and characterized their component phenotypes. Sle1 mediates the loss of tolerance to nuclear antigens; Sle2 lowers the activation threshold of B cells; and Sle3 mediates a dysregulation of CD4+ T cells. We have now created a collection of bi- and tricongenic strains with these intervals and assessed the autoimmune phenotypes they elicit in various combinations. Our results indicate that Sle1 is key for the development of fatal lupus. The combination of Sle1 with Sle2, Sle3, or the BXSB-derived autoimmune accelerating gene yaa results in the development of systemic autoimmunity with variably penetrant severe glomerulonephritis culminating in kidney failure. In contrast, two locus combinations of Sle2, Sle3, and yaa failed to mediate fatal disease. These results indicate that the loss of tolerance to chromatin mediated by Sle1 is essential for disease pathogenesis and identify the pathway occupied by Sle1 as a strategic target for therapeutic intervention in systemic lupus erythematosus. The coexpression of Sle1, Sle2, and Sle3 as a B6-triple congenic results in severe systemic autoimmunity and fully penetrant, fatal glomerulonephritis. These results demonstrate the fulfillment of the genetic equivalent of Koch's postulate, where susceptibility loci in a lupus-prone strain have been identified by a genome scan, isolated and functionally characterized by congenic dissection, and finally shown to mediate full disease expression when recombined in a normal genome.
Although the effector mechanisms responsible for the clinical features of systemic lupus erythematosus (SLE) have been studied extensively (1), the primary immunological defects responsible for the initiation and progression of disease remain poorly understood. A wealth of data clearly indicates, however, that genetic predisposition is a key element in susceptibility (2, 3). Several mouse models of lupus displaying different phenotypes and genotypes have collectively contributed a great deal toward our understanding of the disease and have led to the identification of more than 20 genomic intervals associated with susceptibility to SLE or SLE-related phenotypes (4–6). In the NZB/NZW-related NZM2410 lupus-prone strain, we have specifically identified the genomic positions of three recessive loci [Sle1, Sle2, and Sle3 on chromosomes (chr) 1, 4, and 7, respectively] that are strongly associated with SLE-susceptibility (7).
Although linkage analyses have provided valuable information about the number and locations of susceptibility genes in lupus-prone strains, they have supplied little information about the nature of the component phenotypes each locus contributes, or how they interact to produce the immunopathology characteristic of the susceptible parental strains. We have adopted a congenic dissection approach to address these issues (8). The basic principle of this approach is to convert a polygenic system into a series of monogenic systems in individual congenic strains, each carrying a single susceptibility interval on a resistant genetic background. The component phenotypes expressed in these strains are amenable to genetic and functional analysis as Mendelian traits. The selection process driving the congenic derivation is purely genetic, i.e., a specific genomic segment is transferred from one strain to another (9). We have produced three congenic strains (10), B6.NZMSle1, B6.NZMSle2, and B6.NZMSle3, by moving each susceptibility interval onto the C57BL/6 (B6) background (Table 1) and used these strains to characterize the primary phenotypes associated with the Sle loci (11).
Table 1.
Genetic characteristics of the B6.NZM strains and their bi- or tricongenic combinations
| Strain* | Interval length† | Strain of origin‡ |
|---|---|---|
| B6.NZMSle1 | 37 cM | NZW |
| (B6.NZMc1) | ||
| B6.NZMSle2 | 26 cM | NZW (60%) |
| (B6.NZMc4) | NZB (40%) | |
| B6.NZMSle3 | 30 cM | NZW |
| (B6.NZMc7) | ||
| B6.NZMSle1/Sle2 | 63 cM | NZW (84%) |
| NZB (16%) | ||
| B6.NZMSle1/Sle3 | 67 cM | NZW |
| B6.NZMSle2/Sle3 | 55 cM | NZW (83%) |
| NZB (17%) | ||
| B6.NZMSle1/Yaa | 37 cM | NZW |
| +Y chr | BXSB.Yaa | |
| B6.NZMSle1/Sle2/Sle3 | 93 cM | NZW (89%) |
| NZB (11%) |
† Percent of genome based on a 1,453-cM genome that includes the 19 autosomes and the X chromosome (37).
Sle1 mediates the loss of tolerance to nuclear antigens with a high specificity for the H2A/H2B/DNA subnucleosome (12). Sle2 lowers the activation threshold of B cells, leading to polyclonal IgM Abs and expansion of the B1a cell compartment (13). Sle3 mediates a T cell dysregulation that is associated with polyclonal IgG Abs and a decrease in activation-induced cell death in CD4+ T cells (14). The Sle3 congenic interval also contains Sle5, a locus linked to anti-dsDNA Ab production (15). The relative contributions of Sle3 and Sle5 to the B6.NZMSle3 autoimmune phenotype have not been clarified yet. For simplicity in this paper, the Sle3 locus refers to the Sle3/Sle5 combination.
These Sle loci are not sufficient individually to mediate fatal lupus nephritis on a C57BL/6 background. Genetic modeling of the inheritance of lupus susceptibility in our original testcross of B6 and NZM2410 indicated that disease liability increases with the number of susceptibility loci and suggested that significant penetrance of disease would require two or more loci (7). Consistent with this prediction, we recently reported that Sle1 and Sle3 in combination as a B6-bicongenic strain (designated B6.NZMSle1/Sle3) develop severe humoral autoimmunity with moderately penetrant fatal glomerulonephritis (GN) (16). In this paper, we report a complete analysis of all possible combinations of Sle1, Sle2, and Sle3. We also examined the phenotypes produced by the combination of each Sle locus with the autoimmune accelerator gene Yaa, which has been shown to amplify systemic autoimmunity on the appropriate background (17). Taken together, the various polycongenic combinations that we have produced identify Sle1 as a key locus required for the initiation of the autoimmune pathogenic process in this model system.
Materials and Methods
Mice.
B6, B6.Yaa, NZW, and NZM2410 were originally obtained from The Jackson Laboratory and subsequently bred in our colony. The genetic characteristics of the polycongenic strains used in this study are summarized in Table 1. For clarity, we will identify each strain with the specific combination of Sle susceptibility genes they contain, rather than the nomenclature that we have used in the past with specific congenic intervals (B6.ZMc1, -c4, -c7). B6.NZMSle1/Sle2 and B6.NZMSle2/Sle3 were produced by intercrossing the two monocongenic strains and selecting for homozygosity along both congenic intervals with simple-sequence repeat markers at the termini of the congenic intervals, plus one internal marker corresponding to the peak linkage values, as indicated previously (10). B6.NZMSle1/Sle2/Sle3 was produced by intercrossing B6.NZMSle1/Sle2 and B6.NZMSle1/Sle3, and selecting for homozygosity at the chr 4 and 7 intervals, as described for the bicongenics. The Sle/Yaa combinations were obtained by intercrossing each monocongenic strain to B6.Yaa and selecting male progeny that were homozygous for the Sle congenic interval. The genetic characteristics of the resulting strains are shown in Table 1. Mice were up to 1 yr of age, and animals with proteinuria >300 mg/dl and blood urea nitrogen >50 mg/dl (measured with Albustix; Ames, Elkhart, IN) were killed. All mice used in this study were raised concurrently in conventional housing at the University of Florida Department of Animal Resources. An equal number of males and females were used, except for the B6.NZM.Yaa strains, where only males carry the Y-linked Yaa locus.
Histology.
Kidneys were fixed and stained with hematoxylin, eosin, and period acid-Schiff. Mice were scored as GN-positive when ≥25% of their glomeruli showed a lesion on multiple sections. In addition, glomeruli were assessed for segmental and/or global mesangial, hyaline, or proliferative lesions. When more than one type of lesion was present, the sample was scored with the dominant type of lesion. The cumulative mortality at 12 m was significantly correlated with the penetrance of proliferative lesions (r2 = 0.83, ANOVA; P = 6 × 10−4), whereas there was no correlation between either mesangial lesions or hyaline deposits and mortality. Moreover, proteinuria was only observed in animals with proliferative GN, defining this type of lesion as severe GN.
Serology.
ELISA detection of IgG Ab directed at chromatin and its subcomponents, including dsDNA, was performed as previously described (11, 12). Test serum samples were assayed at 1:100 dilution. For interplate comparisons, serial dilutions of a NZM2410 serum were included on each plate to construct a standard curve. The OD value for a 1:100 dilution was assigned a value of 100 units. Sera were identified as positive if their OD value exceeded 2 SD above the values obtained for 10 randomly chosen B6 (aged 12 mo) sera tested on the same plate. The glomerular binding assay was performed as previously described (18). Briefly, ELISA plates were coated with sonicated whole rat glomerular extracts. Tests sera (from 7-mo-old B6.NZMSle1.Yaa and B6.NZMSle1/Sle2/Sle3 mice, 9–12 mo for the other strains) were added in serial dilutions starting at 1:100, and bound IgG Ab were revealed by using peroxidase-coupled anti-mouse IgG and peroxidase substrate TMB (3,3′,5,5′-tetramethylbenzidine). A MRLlpr sample was used on each plate to standardize the assay.
Flow Cytometry.
Splenocytes were depleted of red blood cells with 0.83% NH4Cl, and single cell suspensions were prepared. FACS analysis was performed as previously described (12, 13). All primary Abs. [CD4 (RM4–5), CD45R/B220 (RA3–6B2), CD69 (H1.2F3), and CD86/B7.2 (GL1)] were purchased from PharMingen and used at pretitrated dilutions. Briefly, cells were first blocked on ice with staining medium (PBS/5% horse serum/0.05% sodium azide) containing 10% rabbit serum. Cells were then stained with optimal amounts of FITC and biotin-conjugated primary Abs diluted in staining medium for 30 min. After two washes, biotin-conjugated Abs were revealed by using streptavidin-Quantum red (Sigma). Cell staining was analyzed with a FACScan (Becton Dickinson Immunocytometry Systems). Dead cells were excluded based on scatter characteristics, and 10,000 events were acquired per sample.
Statistical Analysis.
Because most of the phenotypic data were not distributed normally, we used, unless specified, the one-tailed Wilcoxon rank sum test. Comparisons were reported as significant when P < 0.05.
Results
We produced and characterized the immunologic and autoimmune phenotypes expressed in a collection of bi- and tricongenic strains carrying various combinations of Sle1, Sle2, and Sle3 from NZM2410 and the yaa autoimmune accelerator gene from BXSB on a B6 background.
Impact of Epistasis Between Sle Loci on Lupus Nephritis.
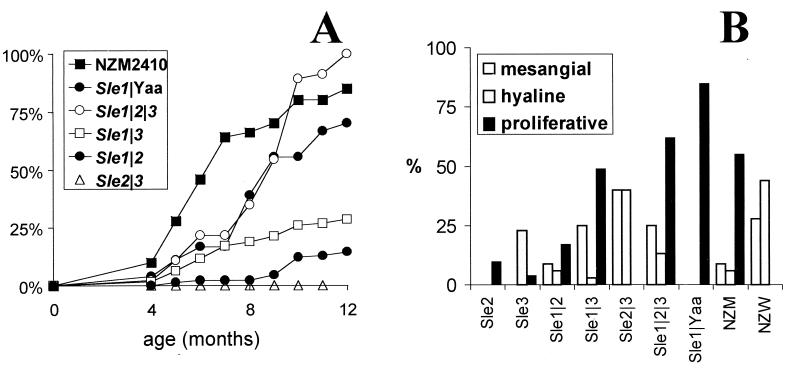
The three Sle bicongenic combinations resulted in different cumulative death rates (Fig. 1A). The Sle1/Sle3 combination resulted in the highest mortality (44% in females and 16% in males, starting at about 24 wk of age), followed by Sle1/Sle2 (18% in females and 10% in males, starting at about 36 wk of age). The Sle2/Sle3 combination did not induce any mortality by 1 yr of age, with now over 50 healthy mice maintained to that age. With both Sle1/Sle3 and Sle1/Sle2, the mortality was higher in females than in males, although the difference was only significant for Sle1/Sle3 (χ2 = 8.13, P < 4 × 10−3). From the Sle/Yaa combinations, only Sle1/Yaa resulted in significant mortality (72% by 1 yr of age). Combinations of Sle2 or Sle3 with Yaa were, as for Sle2/Sle3, unsuccessful in generating a phenotype different from the single Sle2 or Sle3 locus (data not shown).
Figure 1.
Clinical lupus nephritis is reconstituted in B6-Sle1/Sle2/Sle3. (A) Cumulative mortality in the bicongenic and tricongenic strains, as compared with NZM2410. Not shown on the figure are the parental monocongenic strains and B6-Sle2/Yaa and B6-Sle3/Yaa, which did not suffer any mortality during the first year of life. A total of 30–90 mice were used per strain. (B) GN penetrance according to the predominant type of lesion—mesangial, hyaline, or proliferative. Only lesions affecting at least 25% of the glomeruli were included. Ten (for monocongenic strains) to 35 mice per strain were used. Only NZW females were used because the penetrance of GN in NZW males at 1 yr of age is extremely low (20). B6-Sle1 does not show any severe glomerular lesion at 12 mo of age (11).
The Sle1/Sle2/Sle3 combination resulted in 100% mortality in 20 females and 25 males aged up to 12 mo. The clinical presentation of disease in these mice was identical to that of NZM2410, with abrupt kidney failure resulting in heavy proteinuria and elevated blood urea nitrogen, anasarca, and death within days. Despite a delayed onset in B6.NZMSle1/Sle2/Sle3 compared with NZM2410 mice, the overall mortality was higher in the triple congenics than in the parental strain. We have recently shown (19) that NZW and NZM2410 carry a strong suppressive allele, Sles1z, at a locus on chr 17. B6.NZMSle1/Sle2/Sle3 mice, therefore, not only combine the three NZM2410-derived susceptibility loci, but also the B6-derived Sles1b permissive allele. A complete disease penetrance was predicted in our threshold model for such a combination (7).
Because nephritis is the major cause of death in the NZM2410 model, we examined the kidney pathology associated with the Sle locus combinations (Fig. 1B). The bicongenic combinations of Sle1 with either Sle2 or Sle3 resulted in proliferative lesions, although the penetrance was significantly reduced. The combination of Sle1 with either Yaa or the combination of Sle2 and Sle3 was sufficient to restore the high levels of proliferative lesions associated with high mortality in NZM2410. These results are consistent with the observed mortality in these strains. B6.NZMSle2/Sle3 kidney histology was uniquely characterized by the presence of numerous hyaline deposits and no proliferative lesions. Interestingly, predominantly hyaline deposit pathology is not observed in either B6.NZMSle2 or B6.NZMSle3, indicating that this pathology is mediated by epistatic interactions between genes in these two intervals. It is intriguing that this type of pathology is indistinguishable from that observed in aged NZW females (20). The results show that the three Sle intervals contain all of the necessary genetic material to promote fully penetrant lupus nephritis on a B6 background.
Impact of Epistasis Between Sle Loci on Autoantibody Production.
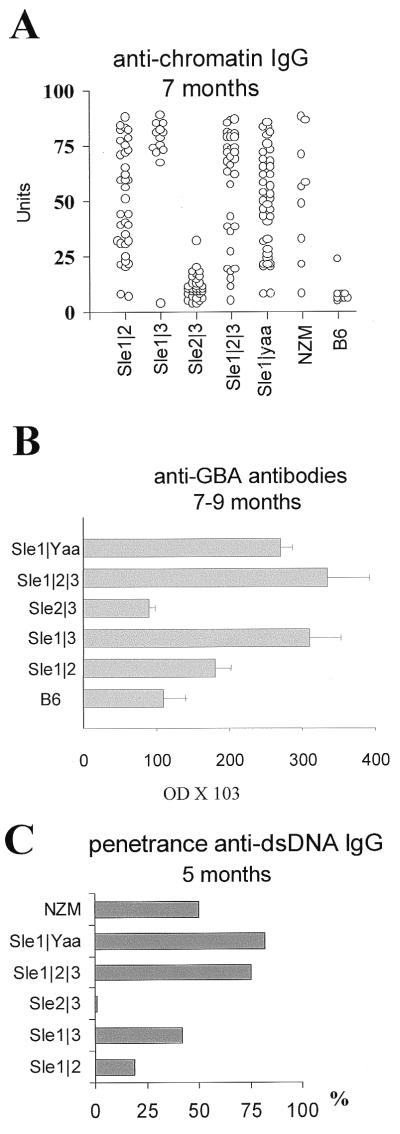
We have shown that the Sle1/Sle3 combination results in antigenic determinant spreading from a bias toward the H2A/H2B-DNA subnucleosome component to a broad spectrum recognition of nuclear components, accompanied by the production of large amounts of nephrophatic Abs binding to glomerular basement membrane components (16). Surprisingly, there was no significant difference between B6.NZMSle1/Sle3, -Sle1/Sle2/Sle3, -Sle1/Yaa, and NZM2410 anti-chromatin Ab levels at 5 and 7 mo of age (Fig. 2A), when these strains show significant differences in lupus nephritis mortality. There was even a trend toward lower levels in the triple congenics and NZM2410, probably because of renal dysfunction and fluid accumulation. B6.NZMSle1/Sle2 anti-chromatin Ab levels were significantly lower than that of B6.NZMSle1/Sle3 (P < 0.01). Finally, B6.NZMSle2/Sle3 levels were not significantly different from that of Sle3 alone. Similar results were obtained at 5, 7, and 9–12 mo of age, showing that the highly penetrant nephritis in B6.NZMSle1/Sle2/Sle3 or -Sle1/Yaa mice did not correlate with a higher production of anti-nuclear Ab production.
Figure 2.
Reconstitution of SLE serological defects with polycongenic combinations. (A) Anti-chromatin IgG Ab assayed in 7-mo-old sera. Each symbol represents a different mouse. (B) Glomerular-binding autoantibodies assayed in sera from 9–12 mo for the bicongenic and B6 mice, or 7–9 mo old for the triple congenic and B6-Sle1/Yaa mice; mean + SE, 10–15 mice per strain. (C) Penetrance of dsDNA IgG Ab production in 5-mo-old mice, 15–20 mice per strain.
The pattern of epitope spreading across chromatin was also similar between the nephropathic strains, although the significant gender difference that we have shown in Sle1/Sle3 combination (16) does not exist with Sle1/Sle2/Sle3 (data not shown). The level of Abs binding to glomeruli in B6.NZMSle1/Sle2/Sle3 or -Sle1/Yaa mice at peak disease incidence was similar to that of B6.NZMSle1/Sle3 (Fig. 2B). The generation of pathogenic nephropathic Abs in triple congenics is therefore predominantly or exclusively the result of Sle1/Sle3 interactions.
However, the addition of Sle2 to that combination, or of Yaa to Sle1 alone, resulted in a significantly earlier onset of the production of anti-nuclear Abs (anti-chromatin and anti-H2A/H2B/DNA Abs at 3 mo of age, P < 0.01). This, in turn, resulted in an accelerated production of pathogenic Abs, as illustrated by the significantly higher penetrance of anti-dsDNA Abs in 5-mo-old B6.NZMSle1/Sle2/Sle3 or -Sle1/Yaa mice compared with -Sle1/Sle3 (Fig. 2C; χ2 test, P < 0.03). The presence of Abs directed against dsDNA or glomerular components has been shown to best correlate to renal damage (21). This suggests that the increased pathogenicity in these strains results from a more efficient (i.e., affecting more mice) early production of nephropathic Abs. In addition, in B6.NZMSle1/Sle2/Sle3, the highly penetrant kidney lesions produced by the Sle2/Sle3 combination, although nonpathogenic by themselves, may amplify the effects of nephropathic Abs.
Impact of Epistasis Between Sle Loci on Lymphocyte Activation.
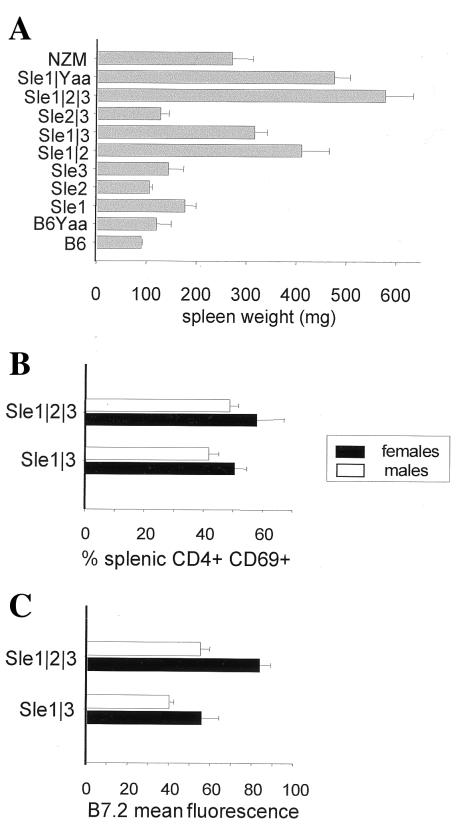
NZM2410 is characterized by splenomegaly (7), which indicates an increased number of splenocytes and hyperactivation of the immune system (16). We have previously described (16) that Sle1 is associated with a moderate but significant increase in spleen size over that of B6, and the Sle1/Sle3 combination results in a larger spleen weight increase to a level similar to that of NZM2410 (Fig. 3A). The Sle2/Sle3, Sle2/Yaa, or Sle3/Yaa combinations did not result in any significant increase in spleen size compared with the parental strains B6.NZMSle2 and B6.NZMSle3. Spleen sizes of B6.NZMSle1/Sle2 or -Sle1/Sle3 mice were comparable to that of NZM2410. The Sle1/Yaa or triple congenic combinations resulted in spleen sizes that were significantly higher than that of NZM2410, or any of the bicongenic strains (P < 0.001). This result is even more striking considering that, because of their higher mortality, the data were collected on average in younger mice in the triple congenic than in the bicongenic strains. The combination of Sle1 with Sle2, Sle3, or Yaa is therefore sufficient to restore the NZM2410 splenomegaly, and coexpression of the three loci resulted in an even greater spleen size. These results illustrate the impact of combining Sle1 with Sle2, Sle3, or Yaa, in comparison to any combination of the latter loci. This supports the notion that Sle1 disrupts an immunologic function that is synergetic with any of several alleles that amplify immune responsiveness to mediate severe autoimmune responses.
Figure 3.
Reconstitution of lymphocyte activation with polycongenic combinations. (A) Spleen weights at sacrifice, 15–25 mice per strain, means and SE. (B) Percent CD4+ splenocytes expressing CD69 and (C) mean fluorescence intensity of B7.2 on B220+ splenocytes in B6-Sle1/Sle3 and B6-Sle1/Sle2/Sle3 B6-Sle1/Sle2/Sle3; six males (□)and six females (▪) in each strain; means and SE.
Expression of the costimulatory molecule B7-2 in B cells and the early activation marker CD69 in T cells is elevated in NZM2410, and to a lesser extent, in the B6.NZM strains. The higher contribution to B7-2 increased expression is associated with Sle2, and CD69 expression is contributed predominantly by Sle3, consistent with these two loci affecting mainly B and T cells, respectively (refs. 12 and 13, and this study). We compared the expression of these two activation markers in B220+ and CD4+ splenocytes, respectively, between 5-mo-old B6.NMZSle1/Sle3 and -Sle1/Sle2/Sle3 mice (Fig. 3B). In both cases, females express higher levels of activation markers, with an accentuated difference for B7-2 (P < 0.01). CD69 expression was not different between the two strains, and similar expression levels were obtained at older ages (data not shown). We have shown that the coexpression of Sle1 and Sle3 resulted in CD69+CD4+ levels similar to that of NZM2410 (16). The coexpression of the three loci did not result in an increased CD69 expression, indicating that the Sle1/Sle3 interaction was sufficient to account for the NZM2410 level of T cell activation. However, B7-2 expression in splenic B cells was significantly higher in B6.NZMcSle1/Sle2/Sle3 than in -Sle1/Sle3, in both males (P = 0.005) and females (P = 0.02). Similar results were obtained in 7- and 9-mo-old mice (data not shown). Consequently, a significant difference in B7-2 expression in the splenic B cells of these two strains is detectable at an early age and persists throughout their lifespan. An elevated expression of the B7-2 costimulatory molecule is not sufficient by itself to confer pathogenicity, as illustrated in B6-Sle2 and NZW mice (data not shown), which suffer minimal lupus nephritis (Fig. 1B). The generation of pathogenic Abs through the Sle1/Sle3 combination is still necessary for full disease expression. It is possible that B7-2 expression merely reflects B cell activation and does not have any specific contribution per se to SLE pathogenesis. B7-1 expression levels, however, do not correlate with either autoimmune phenotypes or pathogenicity in our model (data not shown).
Discussion
The development of fatal lupus nephritis in mice carrying specific combinations of lupus susceptibility genes on the B6 background provides fulfillment of the genetic equivalent of Koch's postulate. Sle1–3 have been individually identified as the major NZM2410 SLE-susceptibility loci (7), their immunophenotypes have been characterized (11–13), and here we show that their combination can fully reconstitute fatal lupus nephritis on the B6 genetic background. Although other susceptibility loci exist (15), the genetic and functional characterization of Sle1–3 will provide all of the necessary information for the full development of SLE immunopathology from a normal mouse. The B6.NZMSle1/Sle2/Sle3 strain therefore constitutes a promising animal model of systemic autoimmunity, where functional and genetic studies can be performed in a very significantly reduced level of complexity.
Although our results indicate that as few as two congenic intervals from NZM2410 are sufficient to mediate fatal disease when incorporated into the B6 genome, we anticipate that the genetic basis for this pathology is still quite complex within this simplified system. We have already shown that the Sle3 congenic interval contains at least two loci, Sle3 and Sle5 (15). We also have evidence that Sle1 and Sle2 each correspond to several SLE-susceptibility loci with overlapping, but distinct immune phenotypes (L.M., unpublished observations). One of the upcoming challenges will be to analyze the interactions between these subloci and to dissect out which of them are responsible for the immunopathology that is described in this paper as the results of interactions between whole susceptibility clusters.
This study also supports the multistep process that we proposed for lupus pathogenesis (8), where Sle1 plays the initiating role of breaking tolerance to chromatin. The addition of Sle3 results in the diversification of the humoral immune response, leading to the recognition of pathogenic determinants and a significant increase in T cell activation (16). Finally, the combination of Sle2 to Sle1/Sle3 amplifies and accelerates the pathogenic process by means of a mechanism that has yet to be elucidated. Our results, however, indicate that the significant difference in pathogenicity associated with the addition of Sle2 correlates with an early increase of B7-2 expression. It has been shown in several human and murine systems that costimulation through the CD28/B7-2 (CD86) pathway can drive Th2 lineage commitment (22–24). Th2 cytokine production has been shown to be more dependent on the B7 molecules than that of Th1 (25), and CD86 provides the initial signal to induce naive T cell to become IL-4 producers, whereas CD80 provides a more neutral differentiation signal (26). In lupus patients, CD86 expression is significantly increased on PBLs (27), and expression of CD80 and CD86 is regulated distinctly (28). In murine SLE, anti-B7-2 Ab treatments (29, 30), or genetic elimination of B7-2 with B7-2null alleles (30), have resulted in decreased anti-dsDNA production, and reduced kidney pathology, although complete immunosuppression through blockage of both B7-1 and B7-2 was necessary to obtain disease-free animals. In light of these results, it is tempting to speculate that a major contribution of Sle2 to lupus immunopathogenesis is a preferential Th2 commitment through B7-2 overexpression. The impact of the addition of Sle2 might also result from nonimmune factors affecting kidney susceptibility to end-organ damage, as suggested by the increase in kidney pathology detected in B6.NZMSle2/Sle3.
Interestingly, the Yaa locus combined with Sle1 also resulted in very severe immunopathology. Yaa expression is very dependent of background genes: whereas B6.Yaa mice are essentially normal, Yaa expressed on an NZW background (the strain of origin of Sle1), either in NZW.Yaa congenics or in (NZW × B6.Yaa)F1 leads to a similar level of pathogenicity to that of BXSB (31). A recent linkage analysis of SLE susceptibility has identified a BXSB locus, Bxs3, located in the Sle1 region (32). Our results functionally confirm a strong epistasis between Yaa and this region of chr 1. The mode of action of Yaa has not yet been clearly defined. It has been shown, however, that Yaa increases antibody production, not only to self antigens, but also to T cell-dependent foreign antigens (33). This indicates that Yaa is a type II gene in our model of SLE pathogenesis (8), amplifying the autoantibody production induced by genes provided by selected background strains such as BXSB and NZW. Bone marrow chimera experiments have clearly established that Yaa is expressed in B cells (34), and it has been postulated that Yaa increases the expression of a B cell adhesion molecule promoting low-avidity Th–B cell interaction (35). We have shown that Sle1 is also expressed in B cells (36) and T cells (E. S. Sobel and L.M., unpublished observations). By dissecting out the functional relationships between Sle1 and Yaa, our model opens new avenues to understand the role of the elusive Yaa locus in the autoimmune process.
The breakdown of tolerance to chromatin accomplished by Sle1 stands out as a necessary step in this model. Without this locus, neither the Sle2/Yaa, Sle3/Yaa nor the Sle2/Sle3 combinations result in any increased autoimmunity over the phenotypes of Sle2 or Sle3 expressed alone. We have recently shown the existence of epistatic interactions between Sle1 and NZW-derived negative modifiers, Sles1–4 (19). Moreover, the NZW allele of the most potent of these loci, H2-linked Sles1, is sufficient to completely abrogate the pathogenic process initiated by Sle1. The protective effects of the NZW-derived Sles alleles and the permissive effects of the B6-derived Sles alleles were again revealed in the immunopathology resulting from the Sle loci combinations, as compared with NZM2410. Taken together, these results identify Sle1 as a strategic locus in SLE pathogenesis and put a high priority on the identification of the corresponding genes and their functional characterization as a potential window of intervention to manipulate the disease process.
Acknowledgments
We thank Jocelyn Tulsian for expert technical assistance with the management of the mouse colony. This work was supported by National Institutes of Health Grants RO1 AR42563 and PO1 AI39824 (to E.K.W.).
Abbreviations
- SLE
systemic lupus erythematosus
- chr
chromosome
- B6
C57BL/6
- GN
glomerulonephritis
References
- 1.Kotzin B L, O'Dell J R. In: Samter's Immunologic Diseases. Frank M M, Austen K F, Claman H N, Unanue E R, editors. Brown, Boston: Little; 1995. pp. 667–698. [Google Scholar]
- 2.Harley J B, Moser K L, Gaffney P M, Behrens T W. Curr Opin Immunol. 1998;10:690–696. doi: 10.1016/s0952-7915(98)80090-3. [DOI] [PubMed] [Google Scholar]
- 3.Gulko P S, Winchester R J. In: Lupus: Molecular and cellular pathogenesis. Kammer G M, Tsokos G C, editors. Totowa, NJ: Humana; 1999. pp. 101–123. [Google Scholar]
- 4.Theofilopoulos A N. In: Systemic Lupus Erythematosus. Lahita R G, editor. New York: Churchill Livingstone; 1993. pp. 121–194. [Google Scholar]
- 5.Vyse T J, Kotzin B L. Annu Rev Immunol. 1998;16:261–292. doi: 10.1146/annurev.immunol.16.1.261. [DOI] [PubMed] [Google Scholar]
- 6.Morel L, Wakeland E K. Curr Opin Immunol. 1998;10:718–725. doi: 10.1016/s0952-7915(98)80094-0. [DOI] [PubMed] [Google Scholar]
- 7.Morel L, Rudofsky U H, Longmate J A, Schiffenbauer J, Wakeland E K. Immunity. 1994;1:219–229. [PubMed] [Google Scholar]
- 8.Wakeland E K, Morel L, Mohan C, Yui M. J Clin Immunol. 1997;17:272–281. doi: 10.1023/a:1027370514198. [DOI] [PubMed] [Google Scholar]
- 9.Wakeland E K, Morel L, Achey K, Yui M, Longmate J. Immunol Today. 1997;18:473–477. doi: 10.1016/s0167-5699(97)01126-2. [DOI] [PubMed] [Google Scholar]
- 10.Morel L, Yu Y, Blenman K R, Caldwell R A, Wakeland E K. Mamm Genome. 1996;7:335–339. doi: 10.1007/s003359900098. [DOI] [PubMed] [Google Scholar]
- 11.Morel L, Mohan C, Croker B P, Tian X-H, Wakeland E K. J Immunol. 1997;158:6019–6028. [PubMed] [Google Scholar]
- 12.Mohan C, Alas E, Morel L, Yang P, Wakeland E K. J Clin Invest. 1998;101:1362–1372. doi: 10.1172/JCI728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohan C, Morel L, Yang P, Wakeland E K. J Immunol. 1997;159:454–465. [PubMed] [Google Scholar]
- 14.Mohan C, Yu Y, Morel L, Yang P, Wakeland E K. J Immunol. 1999;162:6492–6502. [PubMed] [Google Scholar]
- 15.Morel L, Mohan C, Yu Y, Rudofsky U H, Schiffenbauer J, Longmate J A, Wakeland E K. Mamm Genome. 1999;10:176–181. doi: 10.1007/s003359900964. [DOI] [PubMed] [Google Scholar]
- 16.Mohan C, Morel L, Yang P, Wanatabe H, Croker B P, Gilkeson G S, Wakeland E K. J Clin Invest. 1999;103:1685–1695. doi: 10.1172/JCI5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izui S, Merino R, Fossati L, Iwamoto M. Int Rev Immunol. 1995;11:211–230. doi: 10.3109/08830189409061728. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein K A, Valerio R D, Lefkowith J B. J Immunol. 1995;154:2424–2433. [PubMed] [Google Scholar]
- 19.Morel L, Tian X-H, Croker B P, Wakeland E K. Immunity. 1999;11:131–139. doi: 10.1016/s1074-7613(00)80088-6. [DOI] [PubMed] [Google Scholar]
- 20.Kelley V E, Winkelstein A. Clin Immunol Immunopathol. 1980;16:142–150. doi: 10.1016/0090-1229(80)90198-1. [DOI] [PubMed] [Google Scholar]
- 21.Gilkeson G S. In: Lupus: Molecular and Cellular Pathogenesis. Kammer G M, Tsokos G C, editors. Totowa, NJ.: Humana; 1999. pp. 448–470. [Google Scholar]
- 22.Freeman G J, Boussiotis V A, Anumanthan A, Bernstein G M, Ke X Y, Rennert P D, Gray G S, Gribben J G, Nadler L M. Immunity. 1995;2:523–532. doi: 10.1016/1074-7613(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 23.Kuchroo V J, Pradhu Das M, Brown J A, Ranger A M, Zamvil S S, Sobel R A, Weiner H L, Nabavi N, Glimcher L H. Cell. 1995;80:707–718. doi: 10.1016/0092-8674(95)90349-6. [DOI] [PubMed] [Google Scholar]
- 24.Larche M, Till S J, Haselden B M, North J, Barkans J, Corrigan C J, Kay A B, Robinson D S. J Immunol. 1998;161:6375–6382. [PubMed] [Google Scholar]
- 25.Schweitzer A N, Sharpe A H. J Immunol. 1998;161:2762–2771. [PubMed] [Google Scholar]
- 26.De Becker G, Moulin V, Tielemans F, De Mattia F, Urbain J, Leo O, Moser M. Eur J Immunol. 1998;28:3161–3171. doi: 10.1002/(SICI)1521-4141(199810)28:10<3161::AID-IMMU3161>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 27.Folzenlogen D, Hofer M F, Leung D Y M, Freed J H, Newell M K. Clin Immunol Immunopathol. 1997;83:199–204. doi: 10.1006/clin.1997.4353. [DOI] [PubMed] [Google Scholar]
- 28.Liu M-F, Li J-S, Weng T-H, Lei H-Y. Scand J Immunol. 1999;49:82–87. doi: 10.1046/j.1365-3083.1999.00452.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima A, Azuma M, Kodera S, Nuriya S, Terashi A, Abe M, Hirose S, Shirai T, Yagita H, Okumura K. Eur J Immunol. 1995;25:3060–3069. doi: 10.1002/eji.1830251112. [DOI] [PubMed] [Google Scholar]
- 30.Liang B, Gee R J, Kashgarian M J, Sharpe A H, Mamula M J. J Immunol. 1999;163:2322–2329. [PubMed] [Google Scholar]
- 31.Izui S, Higaki M, Morrow D, Merino R. Eur J Immunol. 1988;18:911–917. doi: 10.1002/eji.1830180612. [DOI] [PubMed] [Google Scholar]
- 32.Hogarth M B, Slingsby J H, Allen P J, Thompson E M, Chandler P, Davies K A, Simpson E, Morley B J, Walport M J. J Immunol. 1998;161:2753–2761. [PubMed] [Google Scholar]
- 33.Fossati L, Iwamoto M, Merino R, Izui S. Eur J Immunol. 1995;25:166–173. doi: 10.1002/eji.1830250128. [DOI] [PubMed] [Google Scholar]
- 34.Merino R, Fossati L, Lacour M, Izui S. J Exp Med. 1991;174:1023–1029. doi: 10.1084/jem.174.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izui S, Merino R, Fossati L, Iwamoto M. Int Rev Immunol. 1994;11:211–230. doi: 10.3109/08830189409061728. [DOI] [PubMed] [Google Scholar]
- 36.Sobel E S, Mohan C, Morel L, Schiffenbauer J, Wakeland E K. J Immunol. 1999;162:2415–2421. [PubMed] [Google Scholar]
- 37.Silver L M. Mouse Genetics. London: Oxford Univ. Press; 1995. [Google Scholar]