Abstract
Centromere protein C (CENPC) is a key protein that has been localized to the inner kinetochore plate of active mammalian centromeres. Using gene targeting techniques, we have disrupted the mouse Cenpc gene and shown that the gene is essential for normal mouse embryonic development. Heterozygous mice carrying one functional copy of the gene are healthy and fertile, whereas homozygous embryos fail to thrive. In these embryos, mitotic arrest and gross morphological degeneration become apparent as early as the morula stage of development. The degenerating embryos demonstrate highly irregular cell and nuclear morphologies, including the presence of a large number of micronuclei. Mitotic chromosomes of these embryos display a scattered and often highly condensed configuration and do not segregate in an ordered fashion. These results describing the phenotype of the mutant mouse embryos indicate that CENPC has a direct role in the mitotic progression from metaphase to anaphase.
The centromere is a functional chromosomal domain that is responsible for the accurate segregation of eukaryotic chromosomes during mitotic and meiotic cell divisions. It is involved in sister chromatid cohesion and is the attachment site for spindle microtubules. Through its interaction with molecular motors, the centromere assists in the alignment of the replicated chromosomes onto the metaphase plate and the poleward movement of chromosomes during anaphase. Problems in sister chromatid separation can lead to aneuploidy, cancer, and cell death.
The centromere of the budding yeast Saccharomyces cerevisiae has been extensively characterized at the structural, biochemical, and genetic levels (1). It is made up of a 125-bp cis-acting CEN DNA unit that is known to associate with a number of proteins in forming a functional structure. Mutations in the CEN DNA and the centromere proteins have been shown to result in chromosome missegregation and mitotic arrest (2, 3). In comparison with the centromere of S. cerevisiae, the centromeres of the fission yeast Schizosaccharomyces pombe are significantly larger. They range in size from 35 to 110 kb and are made up of both repeated and unique DNA (4–6).
Mammalian centromeres, like those of Sch. pombe, are typically made up of long tracts of tandemly, repeated satellite DNA. For example, human centromeric DNA primarily consists of a 171-bp α-satellite DNA that spans several megabases on each chromosome (reviewed in ref. 7), whereas the mouse centromere is composed of a 120-bp tandem repeat known as minor satellite (8). Interestingly, a number of functional human centromeres that are devoid of normal centromeric repeats have recently been described on mitotically stable marker chromosomes (9–11).
To date, several mammalian centromere proteins have been isolated and characterized. These proteins can be subdivided into two categories, those that are present throughout the cell cycle and those that appear at specific stages of the cell cycle. Three proteins that are known to be present throughout the cell cycle include CENPA, CENPB, and CENPC (12). CENPA is a histone H3-like protein that is thought to be associated with the formation of centromere-specific chromatin (13). CENPB is a DNA-binding protein that interacts with a 17-bp CENPB box motif found on α satellite and mouse minor satellite DNA (14, 15), and it has been proposed to have a role in the specific packaging of centromeric heterochromatin (16). CENPC is a highly basic protein with DNA-binding properties and is located at the inner kinetochore plate (17, 18). All three proteins are presumed to form the kinetochore precursor onto which the transient proteins associate to form a functionally active kinetochore. The transient group of proteins includes the motor proteins CENPE and MCAK, CENPF, and the inner centromere protein, INCENP (reviewed in refs. 19 and 20).
Studies of human dicentric chromosomes (21, 22) and marker chromosomes containing neocentromeres (10, 11) have shown that CENPC is present on active but not inactive centromeres, suggesting that CENPC has an essential role in centromere function. In agreement with this idea, cells microinjected with anti-CENPC antibodies exhibit mitotic delay and formation of shortened and disrupted kinetochores (23). Transient expression studies of truncated forms of CENPC have revealed two functional regions of the protein. The first is an instability domain located at the amino terminus and is thought to be involved in the regulation of the temporal destruction of the protein at specific stages of the cell cycle. The second functional region is a DNA-binding and centromere-targeting domain located in the central portion of the protein (18, 24). Furthermore, CENPC shares a region of homology with the S. cerevisiae protein Mif2p (25, 26). Mutations in the MIF2 gene have been shown to result in defective chromosome segregation and delayed progression through mitosis (25). Recent evidence suggests that MIF2 is located at the centromere (26).
To directly investigate the role and biological significance of CENPC in mouse, we have disrupted the gene by homologous recombination. We describe here the phenotype and the consequence of such a gene disruption.
METHODS
Construction of Cenpc Targeting Construct.
Using a mouse Cenpc cDNA fragment as a probe, we isolated a clone from a mouse genomic 129/Sv phage library (Stratagene). The clone contained exons 5–11 of the mouse Cenpc gene (27). From the clone, a 6.3-kb XbaI fragment containing exons 8 and 9 was used to construct the targeting vector. A 700-bp XhoI–SalI fragment covering the junction between intron 7 and exon 8 was deleted and replaced by a 6.7-kb splice acceptor-IRES-βgeo selectable marker. This construct, when homologously recombined into the mouse Cenpc locus, causes premature protein truncation that leads to the loss of the centromere-targeting domain, resulting in the abolition of CENPC function.
Transfection and Screening for Targeted Cell Lines.
Mouse embryonic stem (ES) cell lines E14, R1, and W9.8 were used in transfection experiments to generate homologous recombinants. Cells (5 × 107) were electroporated with 40 μg of linearized construct DNA at 0.8 kV, 3 μF, and ∞ Ω (Bio-Rad Gene Pulser) and grown on STO-neoR feeder cells (28) plus 103 units/ml leukemia inhibitory factor (LIF) (Amrad-Pharmacia). After 24 hours, G418 (GIBCO/BRL) selection was applied at an active concentration of 200 μg/ml. Resistant colonies were picked 7 to 10 days later and cell lines were established. Cells were grown up in 3-cm-diameter culture dishes to confluency, and genomic DNA was extracted, digested with EcoRI (Boehringer Mannheim), electrophoresed, and blotted onto Hybond N+ (Amersham) by using standard procedures. The filters were probed with a 3′ XbaI 1.2-kb probe (see Fig. 1A).
Figure 1.
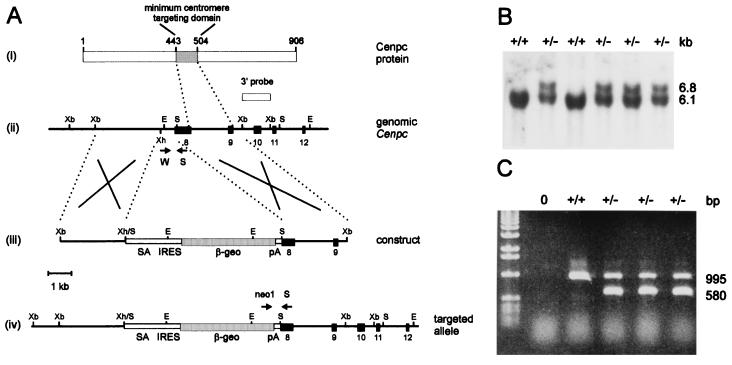
Targeted disruption of the mouse Cenpc gene. (A) Gene replacement constructs and restriction maps. (i) Mouse CENPC protein, showing the amino acid positions of the minimum centromere-targeting domain (18) and the location of this domain downstream of the gene-disruption site. (ii–iv) Restriction maps for the Cenpc locus covering exons 8 to 12 (ii), the gene replacement construct (iii), and the Cenpc locus after targeted disruption (iv). Black boxes represent exons. The selectable marker cassette contained in the targeting construct consists of a splice acceptor site (SA), a picornaviral internal ribosome-entry site (IRES), a lacZ–neomycin-resistance fusion gene (βgeo), and a simian virus 40 polyadenylation sequence (pA) (27). A 1.2-kb XbaI fragment (designated 3′ probe) located downstream of the targeted region was used in the Southern screening strategy and detected a 6.1-kb wild-type EcoRI fragment in the untargeted locus or a 6.8-kb EcoRI fragment in the targeted locus. Arrows indicate positions of primers used in mouse tail and embryo PCR. Crosses denote expected sites of homologous recombination. Abbreviations for restriction enzymes are E, EcoRI; S, SalI; Xb, XbaI; and Xh, XhoI. (B) Southern blot analysis of wild-type and correctly targeted ES cell lines. The sizes of wild-type 6.1-kb and homologous recombinant 6.8-kb bands are shown on the right. (C) PCR genotyping of mouse tail DNA or postimplantation embryos. The primer set S-W gives a 995-bp wild-type Cenpc product, whereas the neomycin-Cenpc primer set, neo1-S, gives a 580-bp targeted product.
Blastocyst Injection and Chimeric Mouse Production.
Targeted ES cell lines were injected into C57BL/6 blastocysts by standard methods (29). The injected blastocysts were then transferred into recipient pseudopregnant HSD Ola (Gpi-1bb) mice. Chimeric mice were selected by coat color and were mated with C57BL/6 mice to generate heterozygotes. Progeny from chimeric and heterozygous crosses were genotyped as described below. The heterozygous mice were crossed to obtain homozygotes.
Genotyping of Mice and Southern and PCR Analyses.
DNA for Southern or PCR analysis was extracted from mouse tail by using the QIAamp Tissue Kit (Qiagen). Southern blotting and hybridization were carried out by standard methods. Mouse tail PCR was performed using a semiduplex strategy with the following primers: S, 5′-TTACCTTGAAGCAGTGCAGTG-3′; W, 5′-AACTGAGTACATGCAAGTATGG-3′; and neo1, 5′-CTTCCTCGTGCTTTACGGTATC-3′ (see Fig. 1A). PCR was performed with Taq DNA polymerase (Perkin–Elmer) with 1.5 mM MgCl2, 0.2 mM dNTPs, and 100 ng of primers in a final volume of 20 μl. The cycling conditions were 95°C for 2 min, 58°C for 1 min, and 72°C for 90 sec, over 35 cycles. The predicted PCR products for S-W (wild-type allele) and neo1-S (targeted allele) are 995 bp and 580 bp, respectively.
Genotyping of Preimplantation Embryos.
For mouse embryos up to the blastocyst stage, the limited amount of DNA template had necessitated the use of two rounds of nested PCR. Embryos were rinsed in M2 medium (Sigma) several times to remove any contaminating maternal cells, before they were taken up in 2 μl of medium and added to 23 μl of deionized H2O. Prior to PCR, the samples were denatured at 95°C for 15 min to lyse the cells and denature any proteins. First round PCR used the Cenpc primers AK, 5′-AAGATGAAGCTTCCGTCTCTC-3′; AL, 5′-TTCGTAGTCCTTTCCCATGC-3′; and the βgeo primers GF1, 5′-AGTATCGGCGGAATTCCAG-3′; GR1, 5′-GATGTTTCGCTTGGTGGTC-3′) under the following cycling conditions: first cycle 95°C for 2 min, 55°C for 3 min, and 72°C for 90 sec, and second to thirtieth cycles 95°C for 60 sec, 55°C for 60 sec, and 72°C for 90 sec. The reaction mixture included 250 ng of primers AK, AL, GF1, GR1, 1.5 mM MgCl2, 1.2 units of Taq DNA polymerase, and 0.2 mM dNTPs, in a final volume of 50 μl.
Second-round PCR involved two separate reactions using 100 ng of nested Cenpc primers: AM, 5′-CCGTCTCTCTAAAGTGTTGCAG-3′; and AN, 5′-CTTCCTCTATTGGGTGAGCC-3′; and βgeo primers, GF2, 5′-CCATTACCAGTTGGTCTGGTG-3′; and GR2, 5′-CCTCGTCCTGCAGTTCATTC-3′; 1.5 mM MgCl2, 0.5 unit of Taq DNA polymerase, and 1 μl of the first-round PCR product, in a final volume of 20 μl. The cycling conditions were as follows: first cycle 95°C for 2 min, 57°C for 60 sec, and 72°C for 90 sec, and second to thirtieth cycles 95°C for 60 sec, 57°C for 60 sec, and 72°C for 90 sec.
Embryo Staining and Determination of Mitotic Index.
Day-3.5 post coitus (pc) preimplantation embryos were placed in a droplet of M16 medium (Sigma) under mineral oil (Sigma), photographed, and kept at 37°C until fixation. These embryos were not treated with any microtubule inhibitor at any stage. Each embryo was placed into a microwell containing 0.6% trisodium citrate for 4–8 min, then taken up in a minimal volume and placed onto a glass slide. A microdrop of methanol/acetic acid (3:1) was immediately placed over the embryo, allowing it to spread and dry. After two rinses in fixative, the slides were stained in Giemsa stain, pH 6.8 (Gurr), air dried, and mounted in DPX (BDH) for analysis.
A similar procedure was followed for the chromosome analysis of 2.5-day embryos except for the addition of 0.1 mg/ml Colcemid and incubation for up to 6 hr prior to harvesting. This allowed most cells to become arrested in mitosis so that chromosome numbers per cell could be counted.
The mitotic index of the day-3.5 embryos was determined by scoring the number of mitotic events over the total number of stained nuclei (which should correspond to the total number of cells, although individual cells were not recognizable as a result of membrane rupture during the hypotonic treatment). Because severely affected embryos expressing the −/− phenotype display a wide variation in nuclear morphology, including the formation of micronuclei, a nucleus was scored to represent a cell unit if it was at least half the size of a normal-looking nucleus.
RESULTS
Targeted Disruption of the Mouse Cenpc Gene.
For the disruption of the mouse Cenpc gene in ES cells, we constructed a promoterless targeting construct that has included exons 8 and 9 of the mouse Cenpc gene (27) (Fig. 1A). In this construct, portions of intron 7 and exon 8 were deleted and replaced with a splice-acceptor–IRES–β-galactosidase–neomycin-resistance fusion marker (30). This disruption is expected to abolish any translation of the centromere-targeting region of Cenpc and render any truncated protein functionally inactive (18, 24). Following transfection of the construct into three different ES cell lines, Southern blot analysis (see Fig. 1B for typical results) revealed targeting frequencies of 11% (10 of 93 G418-resistant colonies), 74% (17 of 23), and 55% (11 of 20) for the cell lines E14, R1, and W9.8, respectively. A total of 18 cell lines carrying a targeted Cenpc allele were randomly picked from the three different cell lines and independently injected into C57BL/6 blastocysts for chimeric animal production. This resulted in the identification of three germ-line chimeras from an R1 cell line. These animals were used to generate the heterozygous (Fig. 1C) and homozygous (see below) progeny for further studies.
Disruption of Cenpc Causes Embryonic Lethality.
Heterozygous male and female mice were phenotypically normal and showed no discernible impairment of growth and fertility. Intercrossing of the heterozygous mice produced 276 progeny, 85 of which were +/+ and 191, +/−. This result clearly indicated embryonic lethality of the homozygous mutant state. To determine the stage at which embryonic death occurred, we looked at postimplantation stages of development. Embryos at days 13.5, 10.5, and 8.5 pc were dissected out for genotyping. In the 30 embryos analyzed the homozygous mutant genotype was again absent, thus suggesting that lethality had occurred at an earlier stage.
We next looked for the presence of the −/− genotype in day-3.5 preimplantation embryos. Because of the limited amount of DNA in these early embryos, a two-round nested PCR strategy was developed for this study (Fig. 2). Fifty embryos were generated from multiple +/− × +/− crosses. As shown in Table 1, 10 −/− embryos were detected. When the stages of development of the different embryos were determined, it was apparent that the −/− embryos were, as a group, morphologically less healthy and/or delayed in their development, with 80% of embryos falling into this category (that is, morula, degenerating morula, and ≤16-cell stages; Table1) compared with 12% and 28% for the +/+ and +/− groups of embryos, respectively. These results suggested that developmental problems in the −/− embryos manifested as early as 3.5 days pc. This suggestion was further addressed by a closer examination of the individual cell morphology of the embryos.
Figure 2.
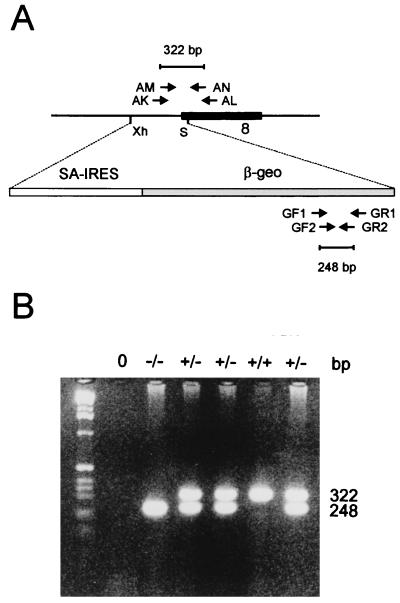
PCR analysis of preimplantation embryos from +/− × +/− crosses. (A) Nested PCR strategy for genotyping embryos up to the blastocyst stage. Cenpc primer pairs include AK–AL for first-round synthesis and AM–AN for second-round synthesis, giving a final product of 322 bp for the untargeted allele. β-geo primer pairs include GF1–GR1 for first-round synthesis and GF2–GR2 for second-round synthesis, giving a final product of 248 bp for the targeted allele. Abbreviations for restriction enzyme sites are (Xh) XhoI and (S) SalI. The black box indicates exon 8 of the mouse Cenpc gene (see Fig. 1A). (B) Nested PCR genotyping of day-3.5 embryos.
Table 1.
Frequency of the different genotypes in day-3.5 preimplantation embryos of +/− × +/− matings
| Developmental stage | No. of embryos (% of total) with genotype
|
||||
|---|---|---|---|---|---|
| Total | +/+ | +/− | −/− | No result | |
| Blastocyst | 30 (60%) | 15 (88%) | 13 (72%) | 2 (20%) | 0 |
| Morula | 5 (10%) | 1 (6%) | 2 (11%) | 2 (20%) | 0 |
| Degenerate morula | 8 (16%) | 1 (6%) | 3 (17%) | 3 (30%) | 1 |
| ≤16 cells | 7 (14%) | 0 | 0 | 3 (30%) | 4 |
| Total | 50 | 17 | 18 | 10 | 5 |
Aberrant Mitosis and Micronuclei Formation in Day-3.5 Embryos.
One-hundred and fifty-five embryos at day 3.5 pc were collected from +/− × +/− matings and subdivided into the different developmental stages by their morphology (Table 2). As controls, 64 embryos at day 3.5 pc were obtained from either +/+ × +/+ or +/+ × +/− matings. Comparison of the average litter sizes in the experimental and control groups indicated these to be 8.2 and 8.0, respectively, suggesting that fertilization was normal in both groups. Overall, the experimental group showed only 56.8% of embryos reaching the blastocyst stage compared with the control group value of 75%, indicating that the experimental group has a higher proportion of embryos that were developmentally delayed and/or unhealthy compared with the control group. Such a difference was consistent with the results shown in Table 1, in that it could be best explained by the developmental problems associated with the −/− embryos generated in the +/− × +/− matings.
Table 2.
Phenotypes of day 3.5 preimplantation embryos
| Developmental stage | +/− × +/−
|
+/+ × +/+ or +/− × +/+
|
||
|---|---|---|---|---|
| No. of embryos (% of total) | No. of embryos with micronuclei (% of total) | No. of embryos (% of total) | No. of embryos with micronuclei | |
| Blastocyst | 88 (56.8%) | 4 (10.3%) | 48 (75%) | 0 |
| Morula | 38 (24.5%) | 13 (33.3%) | 8 (12.5%) | 0 |
| Degenerate morula | 26 (16.8%) | 22 (56.4%) | 0 | 0 |
| ≤16 cells | 2 (1.3%) | 0 | 3 (4.5%) | 0 |
| Dead | 1 (0.6%) | 0 | 5 (8%) | 0 |
| Total no. of embryos | 155 | 39 | 64 | |
| No. of litters | 19 | 8 | ||
| Avg. no. of embryos/litter | 8.2 | 8.0 | ||
To further investigate the embryos, they were fixed onto slides and stained with Giemsa stain. The results indicated that a substantial number of embryos in the experimental group showed irregular-sized nuclei and a high level of micronuclei (Fig. 3C–F). In total, 39 of the 155 embryos (or 25.2%) were shown to have this phenotype—a value that closely approximated the 25% expected frequency for the −/− genotype in +/− × +/− crosses. In contrast, none of the 64 control embryos showed any sign of micronuclei formation. When the relative number of embryos with irregular nuclei and micronucleated cells was ascertained in terms of developmental stage, a grossly disproportionate number (89.7%) was shown to be associated with embryos that demonstrated developmental delay or embryonic degeneration (Table 2). This result provided evidence that expression of the abnormal nuclear and micronuclear phenotype was directly responsible for the observed developmental problems in the −/− progeny.
Figure 3.
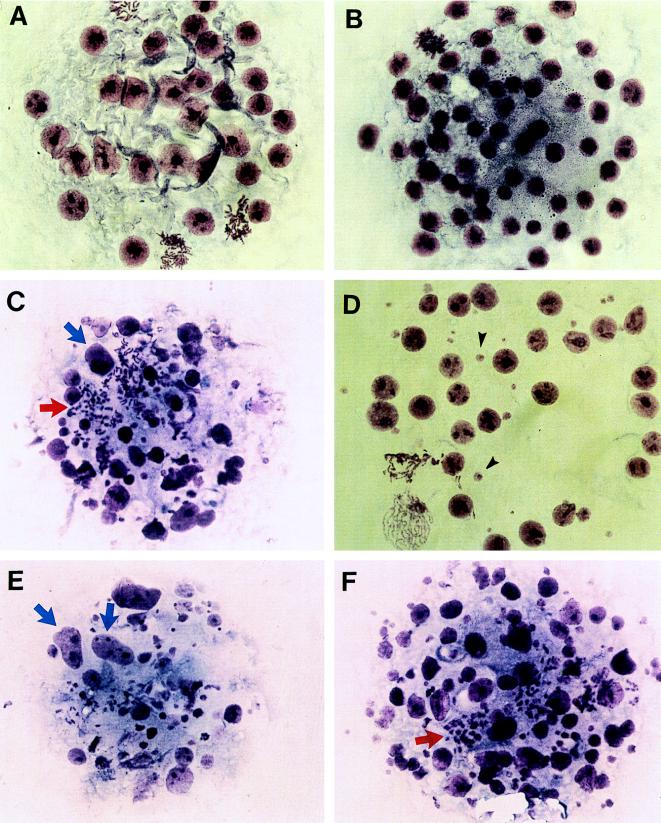
Morphological analysis of Giemsa-stained day-3.5 embryos from +/− × +/− crosses. (A and B) Normal embryos, showing regular, round interphase nuclei and a number of cells undergoing mitosis. (C–F) Putative Cenpc homozygous mutant embryos, showing oversized interphase nuclei (blue arrows), scattered and highly condensed metaphase chromosomes (red arrows), and an abundance of micronuclei (black arrows). (×200.)
The mitotic indices for the micronucleated and control group of embryos were determined. This involved counting the number of mitotic spreads over the total number of cells for each embryo. When the results for all the embryos within each group were pooled, an average mitotic index of 6.9% was obtained for the micronucleated embryos, whereas the value was 3.6% for their normal littermates, suggesting that some mitotic delay or arrest has occurred in the affected embryos. Furthermore, analysis of the mitotic stages of these embryos has indicated that metaphase chromosomes were not correctly aligning onto the metaphase plate and then not proceeding through an ordered anaphase, in contrast to the control group in which a significant proportion of cells showed normal progression through these stages. As shown in Fig. 3, the chromosomes of the affected embryos displayed a scattered configuration, with a substantial proportion of these chromosomes showing a highly condensed morphology.
To look for evidence of chromosome missegregation and aneuploidy at an earlier developmental stage, we have analyzed 2.5-day-old preimplantation embryos. Fifty-two embryos were obtained from +/− × +/− matings, and 32 embryos from +/+ × +/+ matings. The embryos were treated similarly to the 3.5-day embryos, except for a 3- to 6 hr-incubation in Colcemid to arrest mitosis to determine the chromosome number. The results indicated that 78.9% of embryos from the experimental group were at the expected eight- (or occasionally slightly more) cell stage compared with 78.1% in the control group, suggesting that embryonic development was not delayed at this stage. No micronuclei were observed in any of the embryos. When the number of chromosomes of the individual cells were counted, no aneuploidy was detected. These results therefore established that mitosis and embryonic development proceeded normally during the first three cell division cycles.
DISCUSSION
In a previous study, Tomkiel et al. (23) microinjected anti-human CENPC antibodies into cultured HeLa cells and demonstrated a transient arrest of cell division at metaphase. The arrested cells exhibited poor kinetochore structure and defective microtubule binding. The use of such a strategy, whilst offering useful preliminary insights into possible roles of centromere proteins, has a number of limitations. For example, the CENPC–antibody–antigen complex may interfere with other centromere or chromosomal proteins ,obscuring the real phenotype, or the anti-CENPC antibody may cross-react with other nuclear proteins to create a more complex effect. The question of whether the antibody used can completely inhibit the function of CENPC is also difficult to resolve completely. The production of specific null mutations of the Cenpc gene by homologous recombination in transgenic mice circumvents these uncertainties and provides a useful model system for the understanding of the functions of this protein. We report here the production of such a model for Cenpc, and describe the phenotype of this targeted transgenic mouse mutant.
Both human and mouse CENPC genes have previously been shown to be single-copy genes (17, 31). Our results demonstrate that disruption of this gene has no apparent effect on the growth and fertility of the heterozygous mice, suggesting that one functional copy of this gene is sufficient for full centromere activity. However, disruption of both the Cenpc alleles results in embryo lethality, as evident from our failure to detect the −/− genotype in any liveborns and in embryos at days 13.5, 10.5, and 8.5 pc. Clues to the observed lethality have come from direct examination of preimplantation embryos. When 3.5-day old embryos from +/− × +/− crosses were analyzed, approximately 25% of embryos displayed poor nuclear morphology and an abundance of micronuclei, which corresponds well with the expected frequency for the homozygous genotype. As a group, these embryos show slight to severe developmental delay or morphological degeneration. Although we have not defined the exact stage of embryonic death, the severity of the phenotype (Fig. 3) suggests that the embryos are not likely to implant.
Based on the observed morphological and nuclear phenotype, the following model of mitotic and developmental events for the putative −/− embryos may be formulated. Because mitosis and the development of the 2.5-day embryos are normal, it can be assumed that the egg cytoplasm provides sufficient CENPC mRNA and/or protein for centromere function for up to three mitotic cell division cycles. After this period, the −/− embryos begin to misdivide, at first giving rise to predominantly normal-looking nuclei, with some micronuclei beginning to develop (Fig. 3D), However, without further replenishment of the maternal pool of CENPC, centromere function becomes progressively deficient, leading to increasing mitotic disarray and ultimately cell death.
Two observations suggest that mitotic disarray has its origin at the metaphase stage. First, the higher mitotic index for the affected group of embryos indicates that mitosis is delayed or arrested. Such a delay may be the result of a metaphase checkpoint that monitors a signal-generating mechanism at the kinetochore to delay anaphase onset until metaphase is completed, with all the chromosomes attaining bipolar attachment to microtubules and aligning properly at the spindle midzone (32). Our failure to detect any evidence for chromosomal congression at the metaphase plate in the 3.5-day affected embryos lends direct support to this possibility. Second, the mitotic chromosomes of the more severely affected embryos display a scattered configuration and are often more condensed than chromosomes in the control embryos. These features, together with the observed failure of the chromosomes to congress at the spindle midzone, are similar to those of mitotic spreads from control embryos that have been treated with a microtubule inhibitor such as Colcemid.
Despite our failure to detect any proper congression of metaphase chromosomes and progression of the cells through anaphase, the metaphase arrest phenotype appears to be eventually lifted to allow the cells to proceed further along mitosis. These post-metaphase steps presumably involve highly disordered chromosomal segregation, as evident from the extensive number of micronuclei that are detected in the affected embryos; these micronuclei structures, which encapsulate missegregating or lagging chromosomes, are formed during nuclear membrane reformation at telophase, prior to cell cleavage. The finding of some nuclei that are significantly larger (occasionally doubling) in size compared with normal nuclei (e.g., Fig. 3 C and E) further suggests that in some cells, most or all replicated chromosomes may have failed to segregate, leading to a nuclear membrane being reformed around a presumed near-tetraploid genome.
The above observations provide in vivo evidence that CENPC is essential for proper mitotic cell division. Absence of this protein may prevent the ability of the other components of the centromere to form a mature kinetochore. This in turn could affect spindle morphology, or the function of kinetochores in segregating chromosomes. The severe phenotype of the −/− embryos indicates that Cenpc is functionally nonredundant; recently, a number of other proteins that are involved in mitosis, including the S. cerevisiae centromere motor protein Kar3p, have been reported to show functional redundancy (33–35). On the basis of the severity of the observed phenotype in our mouse mutant, it may be extrapolated that null mutations of CENPC in humans will similarly lead to very early embryonic degeneration and spontaneous loss.
Earlier studies have shown that the phenotypes of some mouse mutants can be altered by genetic modifiers when crossed onto different mouse strains, as seen in the Egfr −/− and the Min/+ mice (36–38). At present, it is not known whether any genetic modifiers exist for the Cenpc locus that may elicit a mitotic or meiotic phenotype in our heterozygous mice or partially correct the severe phenotype in the homozygous null mutants. We are currently determining this by breeding the targeted Cenpc allele onto mouse strains of different genetic background. In further studies, it would also be important to use targeted gene disruption in mice to determine the phenotype of null mutations for other known centromere proteins. Two of these proteins, CENPB and CENPE, are particularly interesting for this approach, because indirect evidence suggests that the former may be dispensable for function (9–11, 14, 39) whereas the latter, which is a motor protein (40), may be functionally redundant, as is the case for Kar3p (33–35). Judging from the amount of useful information that has come from the study of chromosome segregation mutants identified in S. cerevisiae and Sch. pombe, it would be feasible that the creation of a collection of mouse mutants would assist in the understanding of mitosis and meiosis in higher eukaryotes.
Acknowledgments
We thank Dr. Peter Mountford for the splice-acceptor-IRES-βgeo selection marker, Dr. Steve Delaney for the gift of R1 cells, and Dr. Leanda Wilton for technical advice on mouse embryos. We acknowledge Sophie Gazeas, Robin Breslin, and Rachel O’Dowd for excellent technical assistance. This work was supported by the National Health and Medical Research Council of Australia. K.H.A.C. is a Principal Research Fellow of the Council.
ABBREVIATIONS
- CENP
centromere protein
- ES
embryonic stem
- pc
post coitus
- IRES
internal ribosome-entry site
References
- 1.Clarke L, Carbon J. Annu Rev Genet. 1985;19:29–55. doi: 10.1146/annurev.ge.19.120185.000333. [DOI] [PubMed] [Google Scholar]
- 2.Doheny K F, Sorger P K, Hyman A A, Tugendreich S, Spencer F, Hieter P. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lechner J, Carbon J. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 4.Clarke L, Amstutz H, Fishel B, Carbon J. Proc Natl Acad Sci USA. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. EMBO J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahnenberger K M, Carbon J, Clarke L. Mol Cell Biol. 1991;11:2206–2215. doi: 10.1128/mcb.11.4.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalitsis P, Choo K H A. In: The Centromere. Choo K H A, editor. Oxford: Oxford Univ. Press; 1997. pp. 97–142. [Google Scholar]
- 8.Wong A K, Rattner J B. Nucleic Acids Res. 1988;16:11645–11661. doi: 10.1093/nar/16.24.11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voullaire L E, Slater H R, Petrovic V, Choo K H A. Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 10.du Sart D, Cancilla M R, Earle E, Mao J I, Saffery R, Tainton K M, Kalitsis P, Martyn J, Barry A E, Choo K H A. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 11.Depinet T W, Zackowski J L, Earnshaw W C, Kaffe S, Sekhon G S, Stallard R, Sullivan B A, Vance G H, Van Dyke D L, Willard H F, Zinn A B, Schwartz S. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- 12.Earnshaw W C, Rothfield N. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 13.Palmer D K, O’Day K, Trong H L, Charbonneau H, Margolis R L. Proc Natl Acad Sci USA. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broccoli D, Miller O J, Miller D A. Cytogenet Cell Genet. 1990;54:182–186. doi: 10.1159/000132989. [DOI] [PubMed] [Google Scholar]
- 16.Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. J Cell Biol. 1992;116:585–596. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saitoh H, Tomkiel J, Cooke C A, Ratrie H, Maurer M, Rothfield N F, Earnshaw W C. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 18.Yang C H, Tomkiel J, Saitoh H, Johnson D H, Earnshaw W C. Mol Cell Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choo K H A, editor. The Centromere. Oxford: Oxford Univ. Press; 1997. [Google Scholar]
- 20.Pluta A F, Mackay A M, Ainsztein A M, Goldberg I G, Earnshaw W C. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- 21.Earnshaw W C, Ratrie H, Stetten G. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- 22.Page S L, Earnshaw W C, Choo K H A, Shaffer L G. Hum Mol Genet. 1995;4:289–294. doi: 10.1093/hmg/4.2.289. [DOI] [PubMed] [Google Scholar]
- 23.Tomkiel J, Cooke C A, Saitoh H, Bernat R L, Earnshaw W C. J Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanini L, McKeon F. Mol Biol Cell. 1995;6:1049–1059. doi: 10.1091/mbc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown M T, Goetsch L, Hartwell L H. J Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meluh P B, Koshland D. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalitsis, P., MacDonald, A. C., Newson, A. J., Hudson, D. F. & Choo, K. H. A. (1998) Genomics, in press. [DOI] [PubMed]
- 28.Robertson E J. In: Teratocarcinomas and Embryonic Stem Cells: A Practical Approach. Robertson E J, editor. Oxford: IRL Press; 1987. pp. 71–112. [Google Scholar]
- 29.Bradley A. In: Teratocarcinomas and Embryonic Stem Cells: A Practical Approach. Robertson E J, editor. Oxford: IRL Press; 1987. pp. 113–152. [Google Scholar]
- 30.Mountford P, Zevnik B, Duwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Proc Natl Acad Sci USA. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKay S, Thomson E, Cooke H. Genomics. 1994;22:36–40. doi: 10.1006/geno.1994.1342. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Nicklas R B. Nature (London) 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 33.Meluh P, Rose M. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- 34.Saunders W, Hoyt M. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- 35.Goldstein L S. J Cell Biol. 1993;120:1–3. doi: 10.1083/jcb.120.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Threadgill D W, Dlugosz A A, Hansen L A, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris R C, Barnard J A, Yuspa S H, Coffey R J, Magnuson T. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 37.Sibilia M, Wagner E F. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- 38.Moser A R, Dove W F, Roth K A, Gordon J I. J Cell Biol. 1992;116:1517–1526. doi: 10.1083/jcb.116.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Earnshaw W C, Bernat R L, Cooke C A, Rothfield N F. Cold Spring Harbor Symp Quant Biol. 1991;56:675–685. doi: 10.1101/sqb.1991.056.01.076. [DOI] [PubMed] [Google Scholar]
- 40.Yen T J, Li G, Schaar B T, Szilak I, Cleveland D W. Nature (London) 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]