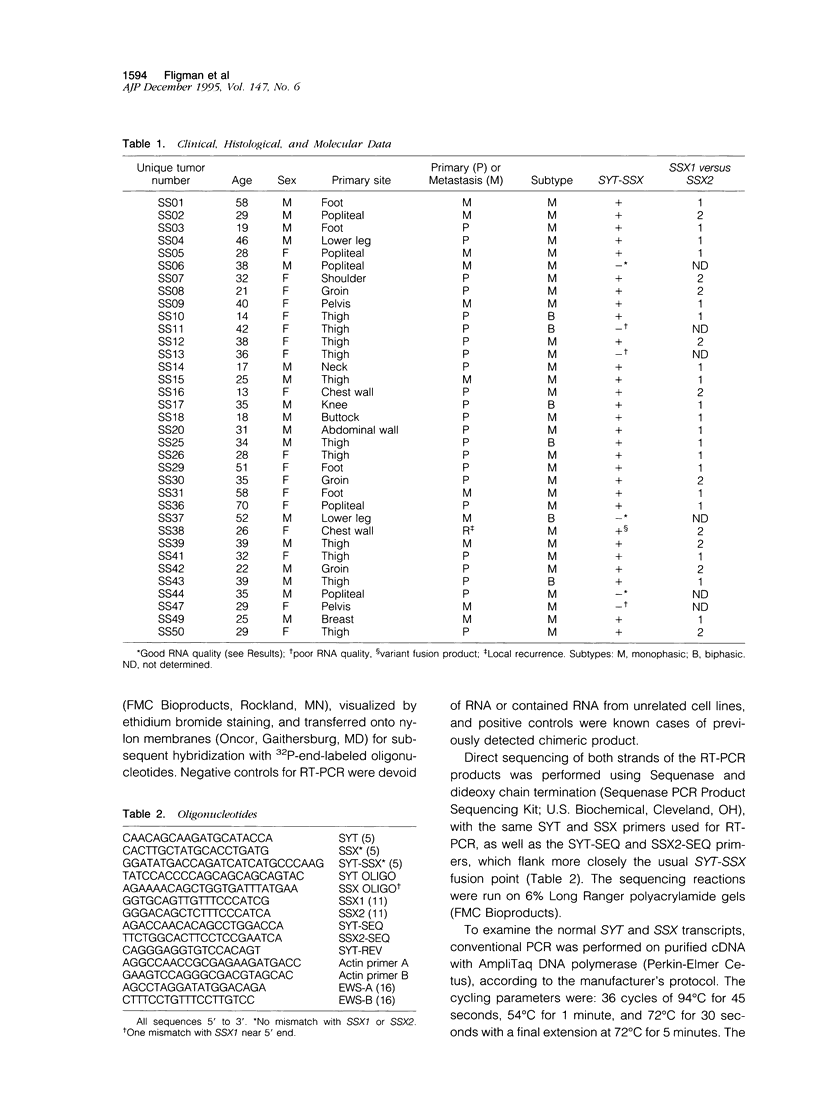
Abstract
The translocation t(X;18)(p11;q11) is seen in > 80% of synovial sarcomas (SS) with informative karyotypes. The breakpoints of the t(X;18) have been cloned and shown to involve two novel genes, SSX (at Xp11) and SYT (at 18q11), which produce a chimeric SYT-SSX transcript as a result of the translocation. Recently, SSX has been shown to be duplicated, with both copies, SSX1 and SSX2, located within distinct subregions of Xp11. We performed a reverse transcriptase polymerase chain reaction (RT-PCR) assay for both chimeric SYT-SSX transcripts in a series of 35 SS (29 monophasic, 6 biphasic) to assess its usefulness in molecular diagnosis and to evaluate the incidence of molecular variants. Of the 35 cases, 29 (83%) showed a specific SYT-SSX RT-PCR product, using a consensus primer for SSX1 and SSX2 Upon excluding three negative cases that had poor quality RNA, the proportion of positives rose to 91% (29/32). The 29 positive cases were further studied using primers specific for either SSX1 or SSX2; 19 cases were positive for SYT-SSX1 and 10 for SYT-SSX2. The relationship of histological subtype (monophasic versus biphasic) to SSX1 or SSX2 involvement was not statistically significant. In a single histologically unremarkable monophasic SS, a slightly larger SYT-SSX2 RT-PCR product was observed. Sequencing of this novel variant showed a 129-bp segment inserted between the usual SYT and SSX2 fusion points, of which 126 bp were derived from a more proximal (5') portion of SSX2 The 3 bp immediately 5' to the fusion point could not be assigned to either SYT or SSX2 and may represent an insertion-deletion or a cryptic splicing event. This fragment maintains the reading frame of the chimeric product and encodes a predicted protein larger by 43 amino acids, which nevertheless replaces the region homologous to the transcriptional repression domain Kruppel-associated box, recently recognized in the 5' portion of the SSX genes, with all but the 3' end of the SYT transcript. Thus, a diagnosis of SS may be confirmed in > 90% of cases using RT-PCR detection of the chimeric transcript resulting from the t(X;18), and the incidence of molecular variants appears low.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clark J., Rocques P. J., Crew A. J., Gill S., Shipley J., Chan A. M., Gusterson B. A., Cooper C. S. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet. 1994 Aug;7(4):502–508. doi: 10.1038/ng0894-502. [DOI] [PubMed] [Google Scholar]
- Crew A. J., Clark J., Fisher C., Gill S., Grimer R., Chand A., Shipley J., Gusterson B. A., Cooper C. S. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J. 1995 May 15;14(10):2333–2340. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. J., D'Cruz C. M., Lovell M. A., Biegel J. A., Barr F. G. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994 Jun 1;54(11):2869–2872. [PubMed] [Google Scholar]
- Delattre O., Zucman J., Melot T., Garau X. S., Zucker J. M., Lenoir G. M., Ambros P. F., Sheer D., Turc-Carel C., Triche T. J. The Ewing family of tumors--a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med. 1994 Aug 4;331(5):294–299. doi: 10.1056/NEJM199408043310503. [DOI] [PubMed] [Google Scholar]
- Delattre O., Zucman J., Plougastel B., Desmaze C., Melot T., Peter M., Kovar H., Joubert I., de Jong P., Rouleau G. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992 Sep 10;359(6391):162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- Hunger S. P., Devaraj P. E., Foroni L., Secker-Walker L. M., Cleary M. L. Two types of genomic rearrangements create alternative E2A-HLF fusion proteins in t(17;19)-ALL. Blood. 1994 May 15;83(10):2970–2977. [PubMed] [Google Scholar]
- Janz M., De Leeuw B., Weghuis D. O., Werner M., Nolte M., Geurts Van Kessel A., Nordheim A., Hipskind R. A. Interphase cytogenetic analysis of distinct X-chromosomal translocation breakpoints in synovial sarcoma. J Pathol. 1995 Apr;175(4):391–396. doi: 10.1002/path.1711750405. [DOI] [PubMed] [Google Scholar]
- Jeon I. S., Davis J. N., Braun B. S., Sublett J. E., Roussel M. F., Denny C. T., Shapiro D. N. A variant Ewing's sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995 Mar 16;10(6):1229–1234. [PubMed] [Google Scholar]
- Ladanyi M., Lewis R., Garin-Chesa P., Rettig W. J., Huvos A. G., Healey J. H., Jhanwar S. C. EWS rearrangement in Ewing's sarcoma and peripheral neuroectodermal tumor. Molecular detection and correlation with cytogenetic analysis and MIC2 expression. Diagn Mol Pathol. 1993 Sep;2(3):141–146. [PubMed] [Google Scholar]
- Ladanyi M. The emerging molecular genetics of sarcoma translocations. Diagn Mol Pathol. 1995 Sep;4(3):162–173. doi: 10.1097/00019606-199509000-00003. [DOI] [PubMed] [Google Scholar]
- Lee W., Han K., Harris C. P., Shim S., Kim S., Meisner L. F. Use of FISH to detect chromosomal translocations and deletions. Analysis of chromosome rearrangement in synovial sarcoma cells from paraffin-embedded specimens. Am J Pathol. 1993 Jul;143(1):15–19. [PMC free article] [PubMed] [Google Scholar]
- Limon J., Dal Cin P., Sandberg A. A. Translocations involving the X chromosome in solid tumors: presentation of two sarcomas with t(X;18)(q13;p11). Cancer Genet Cytogenet. 1986 Sep;23(1):87–91. doi: 10.1016/0165-4608(86)90152-4. [DOI] [PubMed] [Google Scholar]
- Limon J., Mrózek K., Nedoszytko B., Babińska M., Jaśkiewicz J., Kopacz A., Zótowska A., Borowska-Lehman J. Cytogenetic findings in two synovial sarcomas. Cancer Genet Cytogenet. 1989 Apr;38(2):215–222. doi: 10.1016/0165-4608(89)90662-6. [DOI] [PubMed] [Google Scholar]
- Mandahl N., Heim S., Arheden K., Rydholm A., Willén H., Mitelman F. Multiple karyotypic rearrangements, including t(X;18)(p11;q11), in a fibrosarcoma. Cancer Genet Cytogenet. 1988 Feb;30(2):323–327. doi: 10.1016/0165-4608(88)90202-6. [DOI] [PubMed] [Google Scholar]
- Oda Y., Hashimoto H., Tsuneyoshi M., Takeshita S. Survival in synovial sarcoma. A multivariate study of prognostic factors with special emphasis on the comparison between early death and long-term survival. Am J Surg Pathol. 1993 Jan;17(1):35–44. [PubMed] [Google Scholar]
- Panagopoulos I., Mandahl N., Ron D., Höglund M., Nilbert M., Mertens F., Mitelman F., Aman P. Characterization of the CHOP breakpoints and fusion transcripts in myxoid liposarcomas with the 12;16 translocation. Cancer Res. 1994 Dec 15;54(24):6500–6503. [PubMed] [Google Scholar]
- Shipley J. M., Clark J., Crew A. J., Birdsall S., Rocques P. J., Gill S., Chelly J., Monaco A. P., Abe S., Gusterson B. A. The t(X;18)(p11.2;q11.2) translocation found in human synovial sarcomas involves two distinct loci on the X chromosome. Oncogene. 1994 May;9(5):1447–1453. [PubMed] [Google Scholar]
- Sorensen P. H., Lessnick S. L., Lopez-Terrada D., Liu X. F., Triche T. J., Denny C. T. A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet. 1994 Feb;6(2):146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- Sreekantaiah C., Ladanyi M., Rodriguez E., Chaganti R. S. Chromosomal aberrations in soft tissue tumors. Relevance to diagnosis, classification, and molecular mechanisms. Am J Pathol. 1994 Jun;144(6):1121–1134. [PMC free article] [PubMed] [Google Scholar]
- Turc-Carel C., Dal Cin P., Limon J., Li F., Sandberg A. A. Translocation X;18 in synovial sarcoma. Cancer Genet Cytogenet. 1986 Sep;23(1):93–93. doi: 10.1016/0165-4608(86)90153-6. [DOI] [PubMed] [Google Scholar]
- Turc-Carel C., Dal Cin P., Limon J., Rao U., Li F. P., Corson J. M., Zimmerman R., Parry D. M., Cowan J. M., Sandberg A. A. Involvement of chromosome X in primary cytogenetic change in human neoplasia: nonrandom translocation in synovial sarcoma. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1981–1985. doi: 10.1073/pnas.84.7.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucman J., Melot T., Desmaze C., Ghysdael J., Plougastel B., Peter M., Zucker J. M., Triche T. J., Sheer D., Turc-Carel C. Combinatorial generation of variable fusion proteins in the Ewing family of tumours. EMBO J. 1993 Dec;12(12):4481–4487. doi: 10.1002/j.1460-2075.1993.tb06137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw B., Balemans M., Olde Weghuis D., Geurts van Kessel A. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum Mol Genet. 1995 Jun;4(6):1097–1099. doi: 10.1093/hmg/4.6.1097. [DOI] [PubMed] [Google Scholar]
- de Leeuw B., Balemans M., Weghuis D. O., Seruca R., Janz M., Geraghty M. T., Gilgenkrantz S., Ropers H. H., Geurts van Kessel A. Molecular cloning of the synovial sarcoma-specific translocation (X;18)(p11.2;q11.2) breakpoint. Hum Mol Genet. 1994 May;3(5):745–749. doi: 10.1093/hmg/3.5.745. [DOI] [PubMed] [Google Scholar]
- de Leeuw B., Suijkerbuijk R. F., Olde Weghuis D., Meloni A. M., Stenman G., Kindblom L. G., Balemans M., van den Berg E., Molenaar W. M., Sandberg A. A. Distinct Xp11.2 breakpoint regions in synovial sarcoma revealed by metaphase and interphase FISH: relationship to histologic subtypes. Cancer Genet Cytogenet. 1994 Apr;73(2):89–94. doi: 10.1016/0165-4608(94)90191-0. [DOI] [PubMed] [Google Scholar]





