Abstract
Certain types of human papillomaviruses (HPVs) are closely linked to the development of human cancers. Herein, it is shown that intracellular targeting of the HPV16 E6 oncoprotein by E6-binding peptide aptamers resulted in the apoptotic elimination of HPV16-positive cancer cells, whereas HPV-negative cells were not affected. These results provide direct experimental evidence that the HPV E6 oncoprotein has antiapoptotic activity in HPV-positive tumor cells that is required for their survival. The E6-targeting molecules identified herein have implications for the development of therapeutic strategies for the treatment of HPV-associated dysplasias and cancers.
Tumor viruses play an important role in human carcinogenesis, contributing to at least 15% of the total cancer incidence (1). Certain types of human papillomaviruses (HPVs), such as HPV16 and HPV18, have been associated etiologically with a number of epithelial malignancies in humans, including cervical cancer (1). The oncogenic activity of HPVs has been linked to the expression of the viral E6 and E7 genes, both of which are invariably retained and expressed in HPV-positive cervical carcinoma cells.
The E6 protein interacts with the tumor suppressor protein p53 via the cellular ubiquitin ligase E6-AP, resulting in p53 degradation (2). In addition, E6 has been reported to bind to a number of other cellular proteins, including E6BP (3), paxillin (4), hDLG (5), IRF-3 (6), Bak (7), and E6TP1 (8). E6 has been shown to possess transforming activities in several experimental systems (9, 10) that may not necessarily depend on the inactivation of p53 (11, 12). In addition, E6 has been reported to act as a transcriptional activator (13) or repressor (14) to activate telomerase (15) and to interfere with keratinocyte differentiation (16). Furthermore, E6 can influence the apoptotic response of cells and, depending on the experimental context, has been described to exert both proapoptotic (17, 18) and antiapoptotic (19–21) effects.
The coexpression of the E6 and E7 oncogenes strongly increases their transforming potential, indicating functional cooperativity (9, 10). Because cells can react to abnormal growth stimuli, such as those exerted by the E7 oncoprotein and pRb mutations, by inducing apoptosis (19–22), it is possible that this cellular response is blocked in HPV-positive cancer cells by the antiapoptotic potential of the multifunctional E6 protein. Thus, it is one of the major open questions in HPV-associated carcinogenesis as to whether E6 indeed acts antiapoptotically in HPV-positive cancer cells. Moreover, if E6 does act in such a manner, it may well be of therapeutic benefit to block E6 specifically, while retaining the activity of E7 as a proapoptotic stimulus. A selective inhibition of E6, however, will be difficult to achieve by blocking viral transcription or targeting viral mRNAs, because the E6 and E7 genes are expressed together as polycistronic transcripts from a common promoter. Thus, to inhibit E6 specifically, it seems necessary to identify molecules that selectively target E6 on the protein level.
Peptide aptamers represent a distinct class of molecules that are selected for in vivo binding to a given target protein (23) and can block its intracellular activity selectively (24–26). They thus represent a powerful alternative to traditional approaches (e.g., knockout or mutation of a gene) for studying the phenotypic consequences of the inactivation of a target protein and should allow the analysis of the target protein in its natural cellular context, e.g., the study of E6 within cancer-derived HPV-positive tumor cells.
Materials and Methods
Peptide Aptamer Screening.
Yeast strain KF1 (MATa trp1-901 leu2-3,112 his3-200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ SPAL10-URA3) was generated from PJ69-4A (27) after integration at the ura3-52 locus by homologous recombination of a PCR product encompassing the SPAL10-URA3 allele from yeast strain MaV103 (28). KF1 thus contains three selectable marker genes under the transcriptional control of Gal4-binding sites: GAL2-ADE2, GAL1-HIS3, and SPO13-URA3. As bait, the complete HPV16 E6 coding sequence was fused in frame to the Gal4 DNA-binding domain into vector pPC97 (29), yielding pPC97/16E6. A yeast expression vector, pADtrx, in which the ADH1 promoter directs expression of the Escherichia coli thioredoxin A (trxA) gene fused to the Gal4 activation domain, was constructed from pRS424 (30). In addition, pADtrx contains the simian virus 40 nuclear localization signal and an influenza virus hemagglutinin epitope. A randomized peptide expression library was generated in pADtrx by cloning randomized 60-mer oligonucleotides into the unique RsrII site of trxA. Oligonucleotides contained triplets of the sequence NNK (where N = G, A, T, or C and K = G or C), which encode for all 20 amino acids but result in only one stop codon. The complexity of the peptide aptamer expression library was estimated to be in the range of 2 × 108 different members.
KF1 transformants expressing pPC97/16E6 and the peptide aptamer expression library were selected initially for growth in the absence of adenine. Subsequently, they were analyzed by replica plating for activation of the Gal4-dependent GAL2-ADE2, GAL1-HIS3, and SPO13-URA3 genes. Peptide aptamer expression vectors from clones exhibiting growth in the absence of adenine and histidine were rescued, and activation of the selectable markers was verified by rescreening. Binding specificity of HPV16 E6 binding peptide aptamers was investigated by screening for interaction with the complete hepatitis B virus (HBV) core HPV16 E7, HPV6b E6, HPV11 E6, and HPV18 E6 proteins expressed from pPC97. C2-2 was isolated from the peptide expression library by using the HBV core protein as bait (unpublished work), and CoPep was chosen arbitrarily from the peptide expression library.
In Vitro Binding Assays.
The trxA-peptide cassette from pADtrx was subcloned in frame with glutathione S-transferase (GST) into the bacterial expression vector pGEX4T3 (Amersham Pharmacia). GST-peptide-aptamer fusion proteins were synthesized in E. coli and immobilized on glutathione Sepharose beads. In vitro (wheat germ extract) translated 35S-radiolabeled HPV16 E6 protein was added, and the beads were collected by centrifugation, washed, and boiled in SDS containing loading buffer. 35S-labeled E6 that bound to the beads was detected by subsequent SDS/PAGE followed by fluorography. The positive control GST-E6AP fusion protein contains E6-AP amino acids 213–865 linked to GST and has been described (31).
Colony-Formation Assays.
The trxA-peptide cassette was subcloned into the episomal eukaryotic expression vector pCEP4 (Invitrogen) and fused 3′ to the HSV1 VP22 gene and 5′ to a 6xHis tag. Exponentially growing cells were transfected either by calcium-phosphate transfection as described (32) or by lipofection (DOTAP, Roche Molecular Biochemicals) and selected for the presence of pCEP4 by hygromycin B resistance. Colonies were fixed with formalin and stained with crystal violet.
Apoptosis Assays and Immunofluorescence Studies.
After transfection, cells were grown for ≈30 h on coverslips, and terminal deoxynucleotidyltransferase-mediated UTP end labeling (TUNEL) analysis was performed with the in situ cell death detection kit (Roche Molecular Biochemicals), followed by immunofluorescence analysis for peptide aptamer expression with a monoclonal anti-His antibody (Dianova, Hamburg, Germany; ref. 33). Total DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI, Roche Molecular Biochemicals). Apoptotic DNA strand breaks, peptide aptamer expression, and total DNA were visualized by transmission epifluorescence microscopy. For the determination of p53 contents, cells were harvested 20–24 h after transfection, and p53 was visualized by immunofluorescence with the polyclonal anti-p53 antibody FL 393 (Santa Cruz Biotechnology).
Results
To isolate molecules that specifically bind to the HPV16 E6 oncoprotein in vivo, a peptide aptamer interaction screen was performed with a yeast strain designed to contain three Gal4-dependent selectable growth markers: GAL2-ADE2, GAL1-HIS3, and SPO13-URA3. The complete HPV16 E6 protein fused to the Gal4-binding domain was used as bait. The active loop of the E. coli trxA protein, which was fused to the Gal4 activation domain, served as a scaffold to present constrained (34) 20-mer peptides of randomized sequence as prey. Screening of ≈2 × 106 yeast clones led to the isolation of 18 different transformants exhibiting growth in the absence of adenine by virtue of the expression of the Gal4-dependent gene GAL2-ADE2. Replica platings showed that 17 of the 18 peptide aptamers also induced the Gal4-dependent GAL1-HIS3 gene, allowing growth in the absence of histidine. These aptamers were considered to be true positives, and their amino acid sequences are presented in Table 1. In addition, 8 of the 17 peptide aptamers were able to induce the Gal4-dependent SPO13-URA3 gene, allowing growth in the absence of uracil (Table 1). The SPO13 promoter contains a negative regulatory element and is activated only by relatively strong protein–protein interactions under the conditions used (28). Peptide aptamers that were arbitrarily chosen from the library (CoPep) or that bind to the HBV core protein (C2-2; unpublished results) served as negative controls for HPV16 E6 binding and were not able to induce growth under any of the selection conditions (Table 1).
Table 1.
Peptide aptamers binding to the HPV16 E6 protein
| Aptamer | Sequence | 16E* | 16E7 | HBVC | 6E6 | 11E6 | 18E6 |
|---|---|---|---|---|---|---|---|
| E61-1 | GALVHKLFSQTSGSCLVCIS | + | − | − | + | − | − |
| E61-2† | LDVLGCLVRRLGVVLVGLH | + | − | − | n.d. | − | − |
| E61-3 | CYVECGCEVLTALVNGVRVL | + | − | − | − | − | + |
| E61-5 | GVGGLCSCASCVSEDFYABV | + | − | − | n.d. | − | − |
| E61-7 | IDLLRRLGSQLHLLLVSVGG | + | − | − | n.d. | − | − |
| E61-8 | LAVLLNGYTRAIVGISFGGW | + | − | − | n.d. | − | − |
| E61-9 | LCTMCATVFRPLLVWFWSIW | + | − | − | n.d. | + | − |
| E61-10 | QLLLDLLLGSYEGMSLTSSP | + | − | − | n.d. | − | − |
| E61-11‡ | SRSNALHTLDVLLGGT | + | − | − | n.d. | − | − |
| E61-12 | GGAVYLCDAGCCFYCCGCSG | + | − | − | n.d. | − | − |
| E61-13 | CLELFDDLFLALSLLLLVGG | + | − | − | n.d. | − | − |
| E61-14‡ | PLCRTCLIESAVLIQLSRL | + | − | − | + | + | − |
| E61-15 | VFSGVYYAEFVFAASAGGTP | + | − | − | − | − | + |
| E61-16 | MAPVGAGRPCCTVCFLTARF | + | − | − | n.d. | − | − |
| E61-17 | LSMLLFAAKLPVAVLCSWQA | + | − | − | − | − | − |
| E61-19‡ | LVGRVRIGVSVFIRGGRLL | + | − | − | n.d. | − | − |
| E61-20 | LFDIFRLCAQPVLVHGHTRV | + | − | − | n.d. | − | − |
| C2-2 | IHPLSRGNFFPHVRLMGEWR | − | − | + | n.d. | − | − |
| CoPep | DRLVIVQVSLKGAAWWAATS | − | − | − | − | − | − |
HBVC, HBV core protein; n.d., not determined.
Activation of both the GAL2-ADE2 and GAL1-HIS3 genes under selection. Peptide aptamers E61-1, -2, -5, -7, -8, -13, -17, and -20 also activated the SPO13-URA3 marker.
Truncated peptide without stop codon.
Truncated peptide ending with a stop codon.
To demonstrate the E6-binding specificity of these peptide aptamers, they were analyzed for their affinity to heterologous proteins. None of the E6-binding aptamers showed detectable binding to the HPV16 E7 protein, which may be structurally related to E6 (35), to the nonrelated HBV core protein, or to the Gal4-binding domain alone, in two-hybrid assays. It should be noted, however, that 5 of the 17 peptide aptamers exhibited interactions with E6 proteins derived from other HPV types. The negative control peptide aptamers CoPep and C2-2 did not interact with any of the HPV E6 proteins (Table 1).
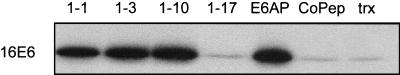
To investigate the binding to HPV16 E6 under in vitro conditions, peptide aptamers were expressed as GST-fusion proteins, and equal amounts of the fusion proteins were immobilized on glutathione Sepharose. After incubation with in vitro translated 35S-labeled HPV16 E6, bound E6 was detected by SDS/PAGE and fluorography. The 75-kDa form of the cellular E6-binding factor E6-AP, fused to GST, served as a positive control (31). GST-CoPep and GST-trxA, the latter expressing the trxA protein without a peptide insert, were included as negative controls. The level of binding considered nonspecific was the amount of HPV16 E6 protein that bound to GST-CoPep and GST-trxA. Four of six peptide aptamers tested, namely, E61-1, E61-3, E61-10, and E61-14, clearly bound to HPV16 E6, whereas no in vitro interaction was observed for E61-15 and E61-17, under the conditions used (Fig. 1 and not shown).
Figure 1.
In vitro binding of S35-labeled HPV16 E6 to GST-fused peptide aptamers E61-1, E61-3, E61-10, E61-17, and CoPep (see Table 1). E6AP, positive control, corresponding to the 75-kDa form of E6-AP fused to GST; trx, GST-trxA fusion protein.
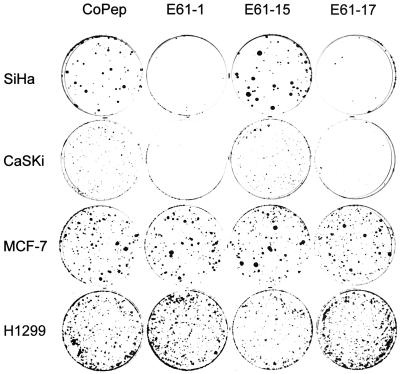
The 17 E6-binding peptide aptamers were expressed in HPV-positive and HPV-negative cells, and possible effects on cell growth were analyzed by colony-formation assays. Two of them, E61-1 and E61-17, exhibited a strong growth inhibitory effect in HPV16-positive SiHa and CaSki cervical carcinoma cells, resulting in a >90% reduction in colony numbers when compared with those of control peptide aptamers (Fig. 2). Growth inhibition was not due to unspecific toxic effects and was selective for HPV16-positive cancer cells, because the growth of HPV-negative control cells, such as MCF-7 breast cancer cells, H1299 lung carcinoma cells, C33A cervical cancer cells, and nontumorigenic HaCaT keratinocytes, was not affected (Fig. 2 and not shown). Because E61-1 and E61-17 do not interact detectably with HPV18 E6 (Table 1), the cell-type specificity and selectivity of growth inhibition is demonstrated further by the observation that their expression also did not interfere with the growth of HPV18-positive HeLa cervical carcinoma cells (not shown). Fig. 2 shows results obtained in HPV16-positive CaSki and SiHa cells and in HPV-negative MCF-7 and H1299 cells for CoPep (negative control), E61-15, which did not reduce colony formation, and the efficient growth inhibitors E61-1 and E61-17.
Figure 2.
Peptide aptamers E61-1 and E61-17 selectively block the growth of HPV16-positive cancer cells. Colony-formation assays of HPV16-positive SiHa and CaSki cells and of HPV-negative MCF-7 and H1299 cells expressing peptide aptamers E61-1, E61-15, E61-17, and CoPep, respectively.
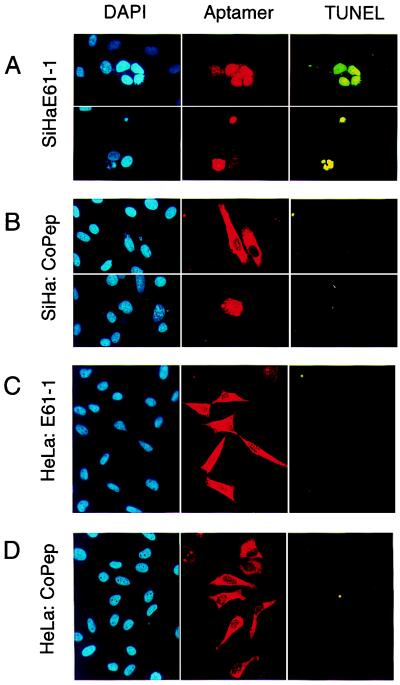
To gain insight into the mechanism of growth inhibition, E61-1-expressing cells were examined for signs of apoptosis. As shown in Fig. 3A, expression of E61-1 was linked to chromatin condensation and a positive TUNEL assay in HPV16-positive SiHa cells, indicative for apoptotic cell death (Fig. 3A). In contrast, expression of an E6-binding aptamer without growth inhibitory effect, of E61-15 (not shown), or of control aptamer CoPep (Fig. 3B) was typically not associated with apoptosis. Induction of apoptosis was specific for HPV16-positive cells and was not observed in HPV-negative MCF-7 and H1299 or in HPV18-positive HeLa cells (Fig. 3 C and D and not shown).
Figure 3.
The HPV16 E6-binding peptide aptamer E61-1 induces apoptosis selectively in HPV16-positive cells. Analysis of HPV16-positive SiHa (A and B) and HPV18-positive HeLa cells (C and D) expressing peptide aptamer E61-1 (A and C) and negative control CoPep (B and D). Total DNA was stained with DAPI; cells expressing peptide aptamers E61-1 and CoPep were detected by immunofluorescence by virtue of their His tag (Aptamer), and cells undergoing apoptosis were visualized by TUNEL analysis.
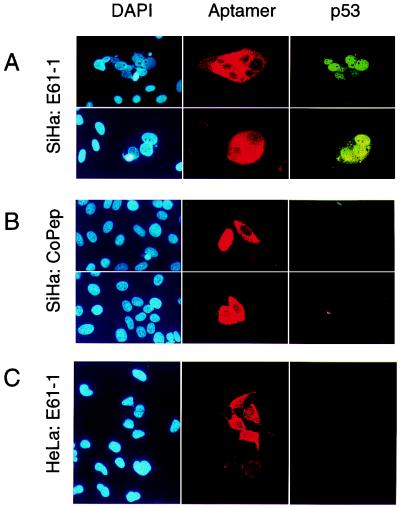
The p53 protein is considered to be an important mediator of the apoptotic response of cells to viral oncoproteins (19, 22), and inhibition of E6 could be associated with increased p53 protein levels caused by interfering with E6-mediated degradation of p53. Immunofluorescence studies showed that expression of the HPV16 E6-targeting peptide aptamer E61-1 was linked to strongly increased p53 protein levels in HPV16-positive SiHa cancer cells (Fig. 4A) when compared with the expression of control peptide CoPep (Fig. 4B) or with E6-binding peptide aptamers without growth inhibitory effects, such as E61-3 or E61-10 (not shown). Again, the effect of the HPV16 E6-targeting peptide aptamer E61-1 was specific for HPV16-positive cells, and p53 levels remained undetectable by immunofluorescence after expression of E61-1 in HPV18-positive HeLa control cells (Fig. 4C).
Figure 4.
Induction of p53 in HPV16-positive cells by growth inhibitory peptide aptamer E61-1. Analysis of HPV16-positive SiHa (A and B) and HPV18-positive HeLa cells (C) expressing peptide aptamer E61-1 (A and C) and negative control CoPep (B). Total DNA was stained with DAPI; expression of the peptide aptamers and p53 contents were monitored by immunofluorescence.
Discussion
The results of this study show that it is possible to eliminate virus-positive cancer cells by molecules specifically targeting a viral oncogene product. In addition, the apoptotic elimination of HPV16-positive cancer cells by E6-targeting peptide aptamers, as observed in this study, provides direct experimental evidence that E6 acts as an antiapoptotic factor within HPV-transformed tumor cells. This activity of E6 seems to be required for the survival of HPV-positive cancer cells and provides a molecular explanation for the apparent selection pressure to maintain expression of the E6 gene in HPV-positive tumors and cell lines derived therefrom.
The interactions between HPV16 E6 and the peptide aptamers isolated in this study were specific, in that binding to heterologous proteins was not observed. However 5 of 17 of the aptamers showed some binding to E6 proteins of other HPV types, indicating the existence of structurally conserved regions between these closely related (36) proteins. Interestingly, 6 of 17 peptide aptamers contained Cys-residues spaced as C-X-X-C (E61-1, -5, -9, -12, -14, and -16), raising the possibility that they interact with E6, which itself contains four C-X-X-C sequences, via chelating zinc. Furthermore, although not exactly matching the consensus sequences proposed for the interaction domains of some cellular E6 binding factors (37, 38), several of the peptides isolated in this study were characterized by the presence of hydrophobic residues in similar motifs (e.g., D-I-L-G-related sequences, such as D-V-L-G in E61-2). In addition, however, HPV16 E6 was also bound by peptides that did not have obvious sequence homologies to known natural binding partners, such as the potent growth inhibitor E61-1.
In view of the anticipated intracellular inhibition of a given target protein, it is also probably advantageous that the screening procedure used herein already preselects for molecules that are stable under in vivo conditions and also may allow the detection of interactions not readily accessible in vitro. Indeed, one of the peptide aptamers, E61-17, which did not exhibit detectable in vitro binding to E6 under standard conditions, specifically interacted with E6 in vivo, both in yeast and in a mammalian version of the two-hybrid assay (not shown), and exhibited profound biological effects selectively in HPV16 E6-expressing cells. This divergence in binding behavior could be due, for example, to inadequate folding of the contact regions between E6 and the respective aptamer under in vitro conditions and is reminiscent of the lack of correlation between in vitro and in vivo binding observed for certain intracellular antibodies (39).
It is worth noting that 15 of 17 of the E6-binding peptides did not inhibit the growth of HPV16-positive cancer cells, although inhibitory and noninhibitory peptides were expressed at comparable levels (not shown). This result indicates that only a subset of the peptide aptamers binding to a target protein under screening conditions can inhibit its intracellular function. Lack of E6 inhibition by some aptamers could, for example, be due to binding affinities that are lower than those of cellular partners interacting with the same or a similar region of E6. In these cases, affinity maturation (40) of the peptides by randomized mutagenesis may result in molecules with increased binding affinities and convert some of the aptamers into dominant in vivo inhibitors. An alternative possibility is that some of the peptides bind to regions of E6 that are not involved in mediating the effects of E6 on cellular growth.
As noncellular factors, viral proteins are attractive targets for therapeutic intervention, because they should allow a specific attack on virus-positive cells. To our knowledge, the present work is the first study to show that it is indeed feasible to eliminate virus-positive cancer cells by specifically targeting an antiapoptotic viral gene product. Induction of apoptosis, rather than growth inhibition, would be particularly desirable for therapeutic agents, because such agents may not require continuous application. It is noteworthy that in previous attempts to inhibit HPV oncogene expression, e.g., by antisense constructs or ribozymes directed against the polycistronic E6/E7 mRNA, (partial) growth inhibition of HPV-positive cells was reported, rather than induction of apoptosis (41–46). This result could be due to the concomitant reduction of proapoptotic E7 levels, which has been observed even for antisense constructs directed selectively against the E6 portion of the polycistronic E6/E7 transcripts (43, 46).
The small size (20 amino acids) and the preferred conformation of the constrained peptides, displayed in the context of a trxA platform of known structure, should help in their structural elucidation (34). Besides representing useful tools for future structure/function analyses of E6, the peptides identified in this study should also provide a basis for the design of pharmacologically active small molecules that specifically target HPV-positive dysplasias and cancers.
Acknowledgments
We thank Dr. H. zur Hausen for continuous support, Drs. P. James, D. Nathans, H. Oie, M. Vidal, and H. Zentgraf for biological materials, Dr. R. Assheuer for subcloning some of the bacterial GST-aptamer expression vectors, Dr. N. Whitaker for critical reading of the manuscript, and Mrs. U. Ackermann for photographic work. These studies were supported by grants from the Wilhelm-Sander-Stiftung and the Deutsche Krebshilfe.
Abbreviations
- HPV
human papillomavirus
- GST
glutathione S-transferase
- HBV
hepatitis B virus
- TUNEL
terminal deoxynucleotidyltransferase-mediated UTP end labeling
- DAPI
4′,6-diamidino-2-phenylindole
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110538897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110538897
References
- 1.zur Hausen H. Science. 1991;254:1167–1172. doi: 10.1126/science.1659743. [DOI] [PubMed] [Google Scholar]
- 2.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 3.Chen J J, Reid C E, Band V, Androphy E J. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 4.Tong X, Howley P M. Proc Natl Acad Sci USA. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S S, Weiss R S, Javier R T. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronco L V, Karpova A Y, Vidal M, Howley P M. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas M, Banks L. Oncogene. 1998;17:2943–2954. doi: 10.1038/sj.onc.1202223. [DOI] [PubMed] [Google Scholar]
- 8.Gao Q, Srinivasan S, Boyer S N, Wazer D E, Band V. Mol Cell Biol. 1999;19:733–744. doi: 10.1128/mcb.19.1.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawley-Nelson P, Vousden K H, Hubbert N L, Lowy D R, Schiller J T. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Münger K, Phelps W C, Bubb V, Howley P M, Schlegel R. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Band V, Dalal S, Delmolino L, Androphy E J. EMBO J. 1993;12:1847–1852. doi: 10.1002/j.1460-2075.1993.tb05833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pim D, Storey A, Thomas M, Massimi P, Banks L. Oncogene. 1994;9:1869–1876. [PubMed] [Google Scholar]
- 13.Desaintes C, Hallez S, van Alphen P, Burny A. J Virol. 1992;66:325–333. doi: 10.1128/jvi.66.1.325-333.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etscheid B G, Foster S A, Galloway D A. Virology. 1994;205:583–585. doi: 10.1006/viro.1994.1684. [DOI] [PubMed] [Google Scholar]
- 15.Klingelhutz A J, Foster S A, McDougall J K. Nature (London) 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 16.Sherman L, Schlegel R. J Virol. 1996;70:3269–3279. doi: 10.1128/jvi.70.5.3269-3279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu C, Meikrantz W, Schlegel R, Sager R. Proc Natl Acad Sci USA. 1995;92:7829–7833. doi: 10.1073/pnas.92.17.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahl A F, Donaldson K L, Fairchild C, Lee F Y, Foster S A, Demers G W, Galloway D A. Nat Med. 1996;2:72–79. doi: 10.1038/nm0196-72. [DOI] [PubMed] [Google Scholar]
- 19.Pan H, Griep A E. Genes Dev. 1994;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- 20.Puthenveettil J A, Frederickson S M, Reznikoff C A. Oncogene. 1996;13:1123–1131. [PubMed] [Google Scholar]
- 21.Stöppler H, Conrad-Stöppler M, Johnson E, Simbulan-Rosenthal C M, Smulsen M E, Rosenthal D S, Schlegel R. Oncogene. 1998;17:1207–1214. doi: 10.1038/sj.onc.1202053. [DOI] [PubMed] [Google Scholar]
- 22.Bates S, Philips A C, Clark P A, Stott F, Peters G, Ludwig R L, Vousden K H. Nature (London) 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 23.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Nature (London) 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 24.Cohen B A, Colas P, Brent R. Proc Natl Acad Sci USA. 1998;95:14272–14277. doi: 10.1073/pnas.95.24.14272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolonin M G, Finlay R L. Proc Natl Acad Sci USA. 1998;95:14266–14271. doi: 10.1073/pnas.95.24.14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norman T C, Smith D L, Sorger P K, Drees B L, O'Rourke S M, Hughes T R, Roberts C J, Friend S H, Fields S, Murray A W. Science. 1999;285:591–595. doi: 10.1126/science.285.5427.591. [DOI] [PubMed] [Google Scholar]
- 27.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vidal M, Brachmann R K, Fattaey A, Harlow E, Boeke J D. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chevray P M, Nathans D. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 31.Huibregtse J M, Scheffner M, Howley P M. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butz K, Shahabeddin C, Geisen C, Spitkovsky D, Ullmann A, Hoppe-Seyler F. Oncogene. 1995;10:927–936. [PubMed] [Google Scholar]
- 33.Zentgraf H, Frey M, Schwinn S, Tessmer C, Willemann B, Samstag Y, Velhagen I. Nucleic Acids Res. 1995;23:3347–3348. doi: 10.1093/nar/23.16.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ladner R C. Trends Biotechnol. 1995;13:426–430. doi: 10.1016/S0167-7799(00)88997-0. [DOI] [PubMed] [Google Scholar]
- 35.Münger K, Phelps W C. Biochim Biophys Acta. 1993;1155:111–123. doi: 10.1016/0304-419x(93)90025-8. [DOI] [PubMed] [Google Scholar]
- 36.Huibregtse J M, Beaudenon S L. Sem Cancer Biol. 1996;7:317–326. doi: 10.1006/scbi.1996.0041. [DOI] [PubMed] [Google Scholar]
- 37.Chen J J, Hongy Y, Rustamzadeh E, Baleja J D, Androphy E J. J Biol Chem. 1998;273:13537–13544. doi: 10.1074/jbc.273.22.13537. [DOI] [PubMed] [Google Scholar]
- 38.Elston R C, Naphtine S, Doorbar J. J Gen Virol. 1998;79:371–374. doi: 10.1099/0022-1317-79-2-371. [DOI] [PubMed] [Google Scholar]
- 39.Visintin M, Tse E, Axelson H, Rabbits T H, Cattaneo A. Proc Natl Acad Sci USA. 1999;96:11723–11728. doi: 10.1073/pnas.96.21.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katz B A. Annu Rev Biophys Biomol Struct. 1997;26:27–45. doi: 10.1146/annurev.biophys.26.1.27. [DOI] [PubMed] [Google Scholar]
- 41.von Knebel Doeberitz M, Oltersdorf T, Schwarz E, Gissmann L. Cancer Res. 1988;48:3780–3786. [PubMed] [Google Scholar]
- 42.Steele C, Sacks P G, Adler-Strothz K, Shillitoe J. Cancer Res. 1992;52:4706–4711. [PubMed] [Google Scholar]
- 43.Tan T M, Ting R C. Cancer Res. 1995;55:4599–4605. [PubMed] [Google Scholar]
- 44.He Y, Huang L. Cancer Res. 1997;57:3993–3999. [PubMed] [Google Scholar]
- 45.Alvarez-Salas L M, Cullinan A E, Siwkowski A, Hampel A, DiPaolo A. Proc Natl Acad Sci USA. 1998;95:1189–1194. doi: 10.1073/pnas.95.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venturini F, Braspenning J, Homann M, Gissmann L, Sczakiel G. Nucleic Acids Res. 1999;27:1585–1592. doi: 10.1093/nar/27.7.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]