Abstract
In most vertebrates, a primary antibody repertoire is created through the recombination of a diverse set of Ig variable (V), diversity (D), and joining (J) gene segments. In contrast, an avian immune repertoire is generated by gene conversion of rearranged Ig genes during B cell development within the bursa of Fabricius, a lymphoid organ unique to birds. To investigate the properties of antigen-specific Igs created through the process of gene conversion, we have developed a system for the production of avian-derived mAbs. This system was used to produce multiple antibodies after a single immunization with a conserved peptide from the human cystic fibrosis transmembrane conductance regulator gene. Each antibody isolated was found to have arisen independently through a distinct series of gene conversion events. These primary antibodies displayed evidence of diversity in all of the complementarity determining regions of both heavy and light chains, and both the heavy and the light chains contributed to antigen specificity. In the light chains, diversity could be attributed to gene conversion events. The measured affinity constants of two of the antibodies were between 108 and 109 M−1, and the antibodies were functional in quantitative ELISA as well as immunohistochemical studies of cystic fibrosis transmembrane conductance regulator expression. These data demonstrate that antigen-specific antibodies produced by Ig gene conversion display both high affinity and specificity. In addition, the methods developed here provide the description of a system for the production of mAbs derived from a nonmammalian species.
The antigen-binding pocket of an antibody is formed by the juxtaposition of six polypeptide loops, three from the light chain variable region and three from the heavy chain variable region (1). Antibody specificity is primarily determined by the amino acid sequence of these loops. In humans and rodents, the first two loops, complementarity determining region (CDR) 1 and CDR 2, of both the heavy and light chains are predetermined by the sequences of germ line variable (V) gene segments. In contrast, the CDR 3 loop is produced somatically by the imprecise joining of V, diversity (D), and joining (J) segments, and diversity is further amplified by nucleotide additions and deletions that occur at the sites of recombination (reviewed in ref. 2). The primary immune repertoire produced by V(D)J recombination is capable of recognizing a wide variety of antigens. However, the initial antibodies produced in an antigenic response are usually of only moderate affinity. Subsequent somatic mutations of the rearranged Ig genes increase both the affinity and specificity of the resulting antibodies.
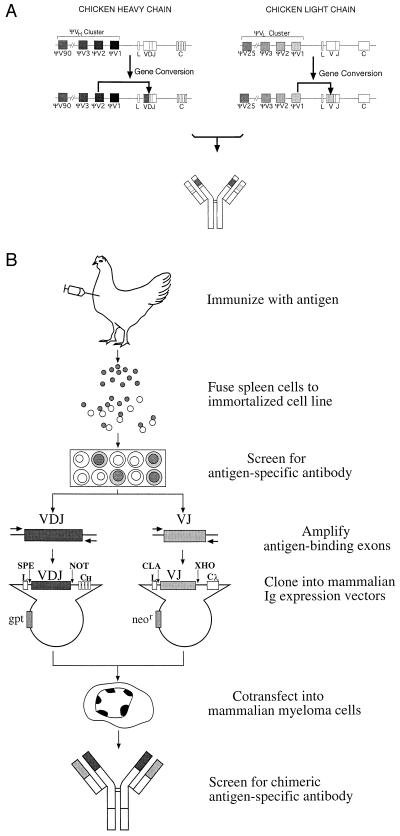
In chickens, the Ig heavy (IgH) and light chain (IgL) gene loci each contain only single V and J germ line gene segments capable of undergoing recombination (3–4). Thus, no diversity of CDR 1 or CDR 2 is created by V(D)J recombination. Furthermore, the diversity of CDR 3 is more limited than in other vertebrates because of the absence of nucleotide addition during V(D)J recombination (5). Ig recombination in the chicken serves primarily to create an expressible gene. A primary immune repertoire is created subsequently by the modification of rearranged IgH VDJ and IgL VJ exons by gene conversion (6–10). The process of gene conversion transfers sequence information encoded in pseudo-V gene segments located 5′ of the functional V gene segment to the rearranged VDJ and VJ exons (Fig. 1A). A rearranged V segment frequently undergoes 7–10 random gene conversion events during bursal B cell development. Gene conversions can result in changes in the sequence of each of the potential antigen binding loops and can alter the length of the loops through codon additions and deletions. The final sequence of the chicken antibody variable region will depend on the pseudogenes used, the extent of the pseudogene sequence transferred, and the order in which different pseudogene sequences are superimposed on each other. Thus, the unprimed antibody repertoire of each chicken arises essentially de novo and has the potential to contain diversity in all three CDRs. This mechanism for creating antibody diversity suggests that the primary immune repertoire of the chicken has the potential to be even more diverse than that of rodents or humans (11).
Figure 1.
Chickens create antibody diversity by a mechanism that can be exploited to generate a new type of mAb. (A) Diagram of the chicken Ig loci illustrating the process of Ig diversification by intrachromosomal gene conversion. The genomic loci of the chicken IgH and IgL chains contain only single functional V and J gene segments. Therefore, V(D)J recombination does not result in combinational diversity. Instead, Ig diversity occurs by the sequential unidirectional transfer of sequences from the upstream pseudo-V (ΨV) gene segments to the expressed V region by intrachromosomal gene conversion; a single gene conversion event at each locus is diagrammed on the second line. The antibody produced by the assembly of diversified heavy and light chains is indicated below. (B) The flow chart depicts the major steps in the process of producing chicken-derived mAbs, described in detail in the text.
Consistent with this hypothesis, chickens have been demonstrated to produce an immunologic response to a wide variety of antigens. Chicken polyclonal antibodies have been developed successfully to a variety of antigens (12–19). However, the properties of these antisera result from the combined effects of multiple independent antibodies. The antigen specificity and affinity of individual antibodies developed through the process of bursal-dependent gene conversion has not been determined. To examine these issues, we have developed a method for the generation of chimeric mAbs whose antigen binding specificity is derived from the IgL and IgH chain genes of chicken B cells.
MATERIALS AND METHODS
Immunization of the Chicken: Immortalization and Screening of Antigen-Specific Chicken B Cell Lines.
An adult female White Leghorn line SC chicken (Hyline International, Dallas Center, IA) was immunized by injecting s.c. over the rib cage 400 μg of a peptide-carrier complex emulsified in complete Freund’s adjuvant. The peptide-carrier complex was amino acids 1468–1480 at the carboxyl terminus of the human CFTR protein (20) coupled to keyhole limpet hemocyanin. The chicken was given an i.v. boost of 100 μg of antigen in saline on day 12; on day 13, the animal was killed by CO2 inhalation. Lymphocytes were extracted from the spleen by teasing and were separated from red blood cells by centrifugation on a Ficoll gradient. The cells were fused with the chicken B cell line R27H4 (21–22) (generously supplied by H. Matsuda, Hiroshima University). In brief, 1 × 108 R27H4 cells were washed, mixed with an equal number of lymphocytes, and pelleted by centrifugation. Approximately 0.5 ml of 40% polyethylene glycol was added to the dry cell pellet followed by dropwise addition of 50 ml RPMI 1640 medium. The cells were pelleted and resuspended in plating medium [50% Ham F10 nutrient supplement/50% RPMI 1640 medium/10% fetal bovine serum/10% chicken serum/10% DT40 conditioned medium (23)/1X HAT supplement (GIBCO, Gaithersburg, MD)] at 3 × 106 spleen cells/ml. The fusion mixture was plated in 24-well plates, 1 ml/well. After fusion (18–20 days), supernatants were screened for CFTR peptide-specific antibodies by ELISA. Positive wells were expanded and frozen after a limited number of passages. Before PCR, these cell lines were thawed, and genomic DNA was prepared.
Amplification and Cloning of Chicken Ig Antigen Binding Exons.
The VDJ sequences of the chicken IgH chain gene were synthesized by PCR by using the primers HCSpe (5′-GGACTAGTGTCAACGGGGGGTCTCACGGGGGGCCGGCTC) and HCNot (5′-ATAAGAATGCGGCCGCGGCAATTTTTGGGGGGGTTGAAGACT). These primers hybridize to nucleotides 101–131 in the chicken leader-V intron and nucleotides 5–35 in the sequences 3′ of the VDJ gene, respectively. The VJ sequences of the chicken IgL chain gene were synthesized by PCR by using the primers LVCla (5′-CCATCGATGACTGTGGGCACGGGGCTCTGTCCCATTGCTGC) and 839Xho (5′-CCGCTCGAGGGAAGAAAGACCGAGACGAGGTCAGCGACT). These primers hybridize to nucleotides 23–55 in the chicken light chain leader-V intron and nucleotides 5–34 in the intron 3′ of the VJ gene sequences, respectively. Approximately 100 ng of genomic DNA from each chicken B cell line, 100 pmol of each of the two heavy or light chain PCR primers, dXTP, 10x buffer, and 0.5 μl cloned pfu polymerase were assembled, and the PCR was performed at 94°C 1′30", 65°C 1′, and 72°C 4′ for 30–35 cycles. The 530-bp heavy chain and 500-bp light chain products were purified after electrophoresis in a 1% agarose gel. The heavy chain product was digested with SpeI and NotI; the light chain product was digested with ClaI and XhoI. These chicken antigen-binding exons were cloned directionally into the expression vectors at the introduced restriction sites described below. The heavy chain and light chain expression vectors each contain regulatory signals necessary for expression in a murine myeloma cell line. In addition, the heavy chain vector pEVHCγ1 (24) (kindly provided by M. Elliott, West Virginia University) contains the mouse IgH chain leader, rearranged VDJ and the IgGγ1 constant exons; the light chain vector λ2Δ2ke (25) (generously provided by M. Shulman, University of Toronto) contains the Igλ2 leader, rearranged VJ and Igλ2 constant domain exons. The modified heavy chain expression vector pEVHCγ1-SN was made by engineering two new restriction enzyme sites in pEVHCγ1: A NotI site was created in the Ig J 3′-flanking region by cloning a synthetic NotI linker into the BamHI site 197 nt from J; a SpeI site was created in the leader-V intron by two-step PCR mutagenesis. The light chain expression vector λ2Δ2ke-CX was made by engineering two new restriction enzyme sites in λ2Δ2ke. An XhoI site was created by cloning a synthetic XhoI linker into the HindIII site, 13 nt 3′ of the rearranged VJ gene segment; a ClaI site was created by cloning a synthetic ClaI linker into the PstI site in the leader-V intron. Linkers and PCR primers were synthesized by Operon Technologies (Alameda, CA) or the Howard Hughes Medical Institute Oligonucleotide Synthesis Facility at the University of Chicago.
Transfection and Identification of Cell Lines Secreting Antigen-Specific Chimeric Antibodies.
Murine myeloma cells (5 × 106) P3 × 63 Ag8.653 (American Type Culture Collection) were cotransfected with 1 μg of light chain plasmid and 7.5 μg of heavy chain plasmid linearized with PvuI. Transfection was by electroporation at 960 μF and 0.3 kV (26–27). Starting 48 h posttransfection, cells were selected and grown in RPMI 1640 medium containing 10% fetal bovine serum and 1.0 mg/ml G418. Cultures were assayed for Ig production by testing tissue culture supernatants by ELISA or immunoblot. Positive cultures or single cell clones were identified and expanded. Tissue culture supernatants were harvested from stock cultures and stored at 4°C.
Antibody Purification.
Antibody was purified from tissue culture supernatants by column chromatography on Protein A [Pierce Immunopure IgG (Protein A) Purification Kit], concentrated by incubation with polyethylene glycol 8000, dialyzed into PBS, and stored at 4°C. The concentration of purified antibody was estimated by the formula OD280/1.44 = milligram of antibody per milliliter.
Immunoblot Analysis.
The CFTR antibodies were subjected to electrophoresis in native 10% or denaturing 12% polyacrylamide gels and transferred to nitrocellulose. The nitrocellulose was blocked with 5% skim milk and TBS-T (40 mM Tris, pH 7.4/100 mM NaCl/0.02% Tween-20). Antibody heavy or light chains were detected by incubating the blot with horseradish peroxidase anti-chicken IgY(IgG) (H+L) (Jackson ImmunoResearch) or horseradish peroxidase anti-mouse IgG+IgM (H+L) (Pierce) and enhanced chemiluminescence reagents (Amersham).
ELISA Analysis.
An antigen capture ELISA was used to characterize CFTR chimeric antibodies. Plates (96-well) were coated with 20 mg/ml peptide conjugate by incubating either CFTR-BSA (Research Genetics, Huntsville, AL), CFTR-keyhole limpet hemocyanin, or CSD.2–keyhole limpet hemocyanin in borate buffered saline (pH 8.8), 100 μl/well, at room temperature for at least 3 h; plates were then washed in 0.05% Triton X-100 in PBS and blocked with 1% BSA in PBS (BSA/PBS), 200 μl/well. Tissue culture supernatant or purified antibody diluted in BSA/PBS was added, 100 μl/well, and incubated at room temperature for 90–180 min. For peptide competition, a semilog dilution series from 100X to 0.001X of CFTR peptide conjugate was prepared in BSA/PBS. Each dilution either was incubated with CFTR chimeric antibody before incubation with the CFTR-coated wells or was added to test wells after CFTR chimeric antibody binding. The plates were washed and incubated with alkaline phosphatase anti-mouse IgL λ1+2 (PharMingen) or alkaline phosphatase anti-chicken IgY(IgG) (H+L) diluted 1:2000 in BSA/PBS for 90 min at room temperature. The plates were developed with 1.0 mg/ml p-nitrophenyl phosphate (Sigma 104 Phosphate Substrate Tablets) in diethanolamine/MgCl2 buffer (pH 9.8), and the OD405 was read. An isotype-specific ELISA was used to detect chimeric antibodies of undetermined specificity by using plates coated with 2 mg/ml rat anti-mouse IgG1 (PharMingen) and detected with alkaline phosphatase anti-mouse IgL λ1+2. Antibody affinity was determined by a noncompetitive enzyme immunoassay in which wells, coated with four different dilutions of CFTR–peptide/BSA conjugate, were incubated with a semilog dilution series of CFTR 1 or 2 and calculated according to published formulae (28, 29).
Immunohistochemistry.
To detect CFTR protein in mouse lung, 5-μm sections of mouse lung, formalin-fixed and embedded in paraffin, were deparaffinized in xylene and hydrated in a graded alcohol series, and the antigen was retrieved by heating in 10 mM citrate buffer (pH 6.0) in a pressure cooker. The sections were incubated with CFTR 1, CFTR 2, or a control chimeric antibody diluted in 0.1% BSA in PBS for 45 min at room temperature and then washed briefly in PBS. The presence of bound antibody was detected by incubating the slides with biotinylated anti-mouse IgG and the avidin–biotin complex reagent according to the manufacturer’s protocol (Vectastain Elite ABC Kit, Vector Laboratories). The slides were developed by the addition of 3-amino-9-ethyl carbazone (Sigma) in N-N-dimethylformamide and H2O2; the counterstain was hematoxylin.
RESULTS
Production of Chicken-Derived mAbs.
The flow chart (Fig. 1B) diagrams the stages of the procedure used to produce chicken-derived mAbs. A 13-aa peptide from the human CFTR cytoplasmic domain was used as the test antigen. An adult chicken was immunized by i.m. injection with a peptide-carrier complex. Lymphocytes were isolated from the spleen at day 13 after initial immunization and fused with the chicken B cell line R27H4, a rel-transformed hybrid TK− chicken lymphoblastoid cell line. Although these hybridomas can be subcloned successfully and screened for surface antibody expression, they are not stable, and antibody expression usually is not sustained beyond 6 weeks in culture. Three hundred and sixty hybridomas were generated, and their supernatants were screened for the presence of peptide-specific antibodies by ELISA; 21 exhibited peptide-specificity.
Genomic DNA was isolated from four chicken B cell/R27H4 hybridomas and used as the template in PCRs to amplify the specific CFTR antibody heavy and light chain antigen binding exons produced by each cell line. Because the sequences 5′ of the V segment and 3′ of the J segment are identical in all chicken B cells (3–4), a specific set of PCR primers was designed that hybridize in the leader-V intron and the 3′ flanking sequences of the J segments for both IgL and IgH. These primers amplify the complete antigen binding exons and some of the adjacent flanking sequences. Each primer was designed to include a restriction enzyme site at the 5′ end so that the PCR product could be cloned directly into an expression vector.
The PCR products of the antigen binding exons of the chicken IgL and IgH chains then were cloned into expression vectors that contained the murine Igλ2 light chain gene or the murine IgGγ1 heavy chain gene. These expression plasmids had been modified to have directional cloning sites that allowed the direct replacement of the mouse VJ or VDJ by the chicken VJ or VDJ segment. Thus, the rearranged avian antigen binding exons could be cloned directionally in place of their mammalian counterparts. A productive Ig gene is recreated by in-frame RNA splicing between the mammalian leader exon and the chicken V gene segment and between the chicken J segment and the mammalian constant region exon. These chimeric chicken–mouse expression vectors then were transfected into the nonsecreting mouse myeloma cell line P3 × 63 Ag8.653, and stable transfectants were selected by growth in selective medium. Stable transfectants were subcloned and screened for antibody production by ELISA.
The Structure and Specificity of the Chimeric CFTR Antibodies.
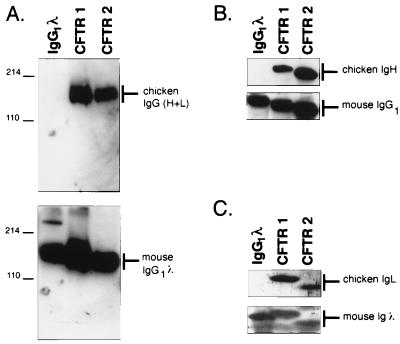
Antibodies produced by two representative transfectants were examined. Antibodies purified from cell supernatants were separated on nondenaturing and denaturing polyacrylamide gels and analyzed by immunoblotting for the presence of chimeric chicken–mouse antibodies. Each of the transfectants, CFTR 1 and CFTR 2, secretes a protein of ≈150 kDa, which reacted with antisera to both chicken and mouse Igs (Fig. 2A). When the antibody heavy and light chains were separated under denaturing conditions and probed with antisera that detected either the chicken variable regions or the appropriate mouse constant regions, the reactivity of both the heavy and light chain proteins confirmed that these chains are chimeras of chicken and murine sequences (Fig. 2 B and C).
Figure 2.
Immunoblot analysis of chimeric CFTR antibodies. (A) Purified CFTR 1 and CFTR 2 antibodies were analyzed by electrophoresis in a 10% nondenaturing polyacrylamide gel, blotted to nitrocellulose, and detected by anti-chicken IgG (H+L) (upper) and anti-mouse IgG (H+L) (lower). The marker on the right side indicates the predicted molecular component(s) of the band that are reacting with the detection reagent. Molecular mass standards are indicated in kilodaltons on the left. (B and C) The CFTR 1 and CFTR 2 antibodies, analyzed in A, were denatured in SDS/β-mercaptoethanol, separated by electrophoresis on a 12% polyacrylamide/SDS gel, blotted, and probed. (B) Region of the immunoblot containing the IgH chains: (upper) probed with anti-chicken IgG(H+L); (lower) probed with anti-mouse IgG (H+L). (C) Portion of the blots that contain the IgL chain, probed as described above.
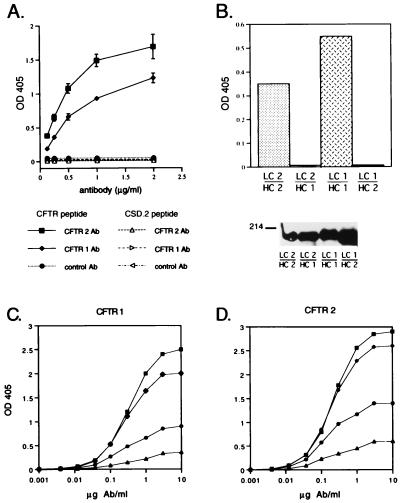
To characterize the antigen reactivity of the chimeric chicken mAbs, the CFTR antibodies and a control chimeric antibody were tested for specific binding to the CFTR peptide and to a control peptide of equivalent size, CSD.2. ELISA plates, coated with CFTR or CSD.2 peptide conjugates, were incubated with a dilution series of purified antibodies and detected with an anti-chicken Ig antiserum. The two CFTR antibodies specifically bound the CFTR peptide. In contrast, the CFTR antibodies did not bind the CSD.2 peptide, and the control chimeric chicken antibody bound neither the CFTR nor the CSD.2 peptide (Fig. 3A). As further proof of antigen specificity, the binding of the CFTR antibodies to the CFTR peptide-coated plate was competed by free CFTR peptide conjugate but not by the CSD.2 peptide (data not shown). The affinity constants of the CFTR 1 and 2 antibodies for the peptide antigen as measured by noncompetitive enzyme immunoassay were 2.3 ± 1.9 × 108 M−1 and 4.2 ± 1.7 × 108 M−1 (mean ± SD), respectively (Fig. 3 C and D).
Figure 3.
Assays demonstrating the specificity of the chicken-derived CFTR antibodies. (A) The ability of the CFTR 1 and CFTR 2 chimeric antibodies to bind the CFTR peptide and the unrelated peptide CSD.2 was assayed by ELISA (13). Filled symbols indicate the data obtained on ELISA plates coated with CFTR peptide; open symbols indicate the data obtained on the plates coated with the CSD.2 peptide. Each data point is the mean of six observations. (B) Determination of the specificity of chimeric antibodies comprised of different combinations of the heavy and light chains from the original chicken CFTR antibodies. (Upper) In an ELISA assay, wells coated with CFTR peptide were incubated with the antibodies secreted from cells transfected with each of the four combinations of the plasmids encoding CFTR 1 or CFTR 2 light chain and heavy chain. Supernatants from three independent passages of the cells were tested for binding to the CFTR peptide; the average OD405 is indicated by the bar. LC 1/HC 1, cells were transfected with the light chain of CFTR 1 and the heavy chain of CFTR 1; LC 1/HC 2, cells were transfected with the light chain of CFTR 1 and the heavy chain of CFTR 2, and so forth. (Lower) The supernatants tested in the upper panel were assayed by immunoblot to quantitate the amount of antibody present. Equal volumes of tissue culture supernatant from each transfected cell line were analyzed by electrophoresis on a 10% nondenaturing polyacrylamide gel, transferred to nitrocellulose, and probed with anti-chicken IgG(H+L). (C and D) Experimental dose–response curves of CFTR 1 and CFTR 2 antibodies at four antigen coating concentrations. The four CFTR–BSA coating concentrations were: 0.2 μg/ml (▴), 0.4 μg/ml (•), 0.8 μg/ml (♦), and 1.6 μg/ml (▪). The data points were used to calculate the functional affinity constant of each antibody according to published formulae (28, 29).
To characterize the relative contributions of the heavy and light chains to antigen specificity, transfectants were made expressing each of the four combinations of CFTR 1 and CFTR 2 heavy and light chains. All four heavy and light chain pair combinations resulted in the secretion of a 150-kDa antibody, demonstrating that heavy and light chain pairing is occurring appropriately. Only the pairings of light and heavy chains derived from the original chicken B cell clones demonstrated reactivity to the CFTR peptide (Fig. 3B). This result suggests that both the heavy and light chains in each antibody contribute to the antigen specificity of the chimeric mAbs.
Chimeric CFTR Antibodies Detect Full Length CFTR Protein.
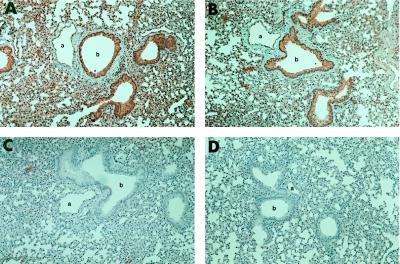
A stringent test for the specificity of an antibody is its ability to react with the intact antigen of interest. It is relatively difficult to develop antibodies that can be used for immunohistochemistry studies of low abundance, cytoplasmic antigens. CFTR 1 and CFTR 2 antibodies were used in immunohistochemical assays to localize the CFTR protein in mouse lung. Both CFTR antibodies specifically bind to epithelial cells that line the bronchioles and alveoli of the lung. In addition, the endothelial cells but not the smooth muscle cells of the pulmonary vasculature are stained specifically. In contrast, control chimeric antibodies or secondary agents do not result in specific staining (Fig. 4). A similar distribution of the CFTR gene expression has been demonstrated by RNA in situ analysis (30). Our antibody studies confirmed this pattern of expression of the CFTR gene at the protein level. In addition, these data demonstrate that, although the CFTR antibodies were raised against a peptide from human CFTR, the antibodies also bind murine CFTR, confirming that the antibodies are reactive against a conserved portion of the protein.
Figure 4.
Immunoperoxidase detection of CFTR in mouse lung. Dilutions of purified chimeric CFTR antibodies were incubated with sections of mouse lung and detected by anti-mouse IgG. (14). (A) 1:500 dilution of CFTR 1 antibody. (B) 1:50 dilution of CFTR 2. (C) 1:10 dilution of the control antibody. (D) Biotinylated anti-mouse secondary antibody. Original magnification was ×10. a, lumen of pulmonary vessel; b, lumen of bronchiole.
Diversity of Antigen-Specific Chicken Antibodies.
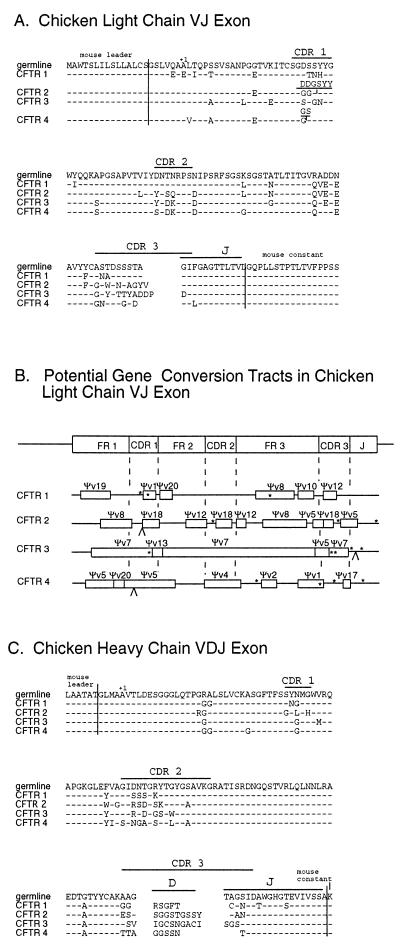
The characteristics of specific mAbs raised in a system that relies on gene conversion to generate the primary immune repertoire have not been examined previously. Therefore, the rearranged VJ and VDJ gene segments were sequenced from CFTR 1, CFTR 2, and two additional CFTR-specific chimeric chicken-mouse mAbs. The IgL and IgH genes appear to be derived independently in all four mAbs (Fig. 5). Considerable diversity in the CDR1, CDR2, and CDR3 loops of each of the IgH and IgL genes was found. None of the CDR domains contained amino acid changes common to all four antibodies. The dissimilarity in coding junctions and gene conversion tracts demonstrated that each of the four mAbs arose independently rather than through sequential alterations of a common progenitor B cell.
Figure 5.
CFTR chimeric antibody heavy and light chain sequences. (A) The amino acid sequence of the four CFTR antibody light chain VJ exons are aligned with the germ line sequence; amino acid differences are indicated. The adjacent mouse sequences are delineated. (B) Chicken antibodies are diversified by numerous gene conversion events. The location and minimal extent of gene conversion tracts are indicated by open boxes with the potential pseudogene donor indicated above the tract. Nucleotide changes that are not present in any of the pseudogenes are located with an ∗. An insertion is indicated by a ∧. (C) The amino acid sequence of the four CFTR antibody heavy chain VDJ exons are aligned with the germ line sequence; amino acid differences are indicated.
For the IgL locus, the origins of diversity could be analyzed because all pseudo-V gene segments have been sequenced. Using previously established methods for establishing candidate gene conversion events (9), a map of the potential gene conversion events that resulted in the diversification of each IgL gene segment in the CFTR antibodies was constructed (Fig. 5B). The IgL genes in the CFTR mAbs had 6.0 ± 2.4 gene conversion events. In each clone, a limited number of base pairs could not be accounted for by gene conversion. As has been noted (7), untemplated base pair changes tend to cluster at the ends of gene conversion tracks.
DISCUSSION
Based on the generation of a diverse set of specific mAbs against a short peptide with a single immunization of the chicken, our data demonstrate that the primary immune repertoire generated by gene conversion already contains an effective and diverse repertoire against peptide antigens. The immunization strategy used in these studies was designed to elicit the characteristics of the primary antibody immune response. A set of mAbs has been derived from the spleen of a single chicken after primary immunization with a peptide antigen. The resulting antibodies demonstrate high affinity for the peptide antigen. Both the IgH and IgL chains of the antibodies isolated displayed evidence of having been modified extensively by gene conversion. The specificity of the mAbs required the pairing of the heavy and light chains present in the B cells from which they were derived. This demonstrates that gene conversion of both of the heavy and light chains contributes to the potential diversity of the primary avian repertoire.
The mAbs we isolated each appear to be derived from separate progenitors. Thus, Ig gene conversion has resulted in the development of B cells with convergent specificity. Thus, it appears that the primary repertoire created during B cell development in the bursa of Fabricius is sufficiently diverse to contain multiple independent antibodies with overlapping specificity. The repertoire of the progeny of the individual bursal progenitors can overlap despite undergoing widely divergent patterns of Ig modification through gene conversion. These data demonstrate that the use of gene conversion for the generation of Ig diversity provides avian species with a robust system for the production of a primary antibody repertoire.
Although the antigen reactive B cells we obtained did not display evidence of having diversified from a common progenitor, clonal dominance may still arise during secondary immune responses. Repetitive immunization may result in further Ig modification by either somatic mutation or gene conversion during avian B cell selection in germinal centers (30–31). Such clonal selection could lead to affinity maturation during secondary immune responses and antibodies of even higher affinities than those we have observed during primary immune responses.
The data also demonstrate that chickens can be a useful species for the production of mAbs against conserved mammalian proteins. This method has several advantages over existing techniques. The probability of generating a strong mAb response to a highly conserved mammalian protein is increased by generating mAbs in a relatively evolutionary distant species, such as the chicken. Polyclonal chicken antibodies often have high background cross-reactivity; chicken-derived mAbs should eliminate this problem. Furthermore, the chicken has approximately one hundredfold more B cells than a mouse and therefore has a larger initial cell repertoire from which to select reactive B cells after in vivo priming (33). By raising antibodies in chickens, the power of the in vivo immune system is used to select B cells producing high affinity antibodies. This advantage is important because phage display technology has been reported to be limited by the inability to recover pairs of heavy and light chains that make the highest affinity antibodies (34–35). Therefore, chicken mAb technology offers an alternative strategy to the use of phage display libraries or existing monoclonal techniques.
Acknowledgments
We thank past and current members of the Thompson lab for advice and encouragement. In addition, Louise Carlson, David Wang, and Carol Sampson provided careful technical support. H. Matsuo, M. Elliott, M. Shulman, J. Pena, M. Drumm, and C. Duckett were generous in providing the chicken hybridoma cell line, the heavy and light chain expression plasmids, the mouse lung sections, and the CFTR or CSD.2 peptides, respectively. This work was supported in part by grants from the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: CDR, complementarity determining region; V, variable; D, diversity; J, joining; IgH, Ig heavy; IgL, Ig light; CFTR, cystic fibrosis transmembrane conductance regulator; CSD.2, peptide encompassing amino acids 251–267 of the dILP protein.
References
- 1.Searle S J, Pedersen J T, Henry A H, Webster D M, Rees A R. In: Antibody Engineering. 2nd ed. Borrebaeck C A K, editor. New York: Oxford Univ. Press; 1995. pp. 3–51. [Google Scholar]
- 2.Alt F W, Blackwell T K, Yancopoulos G D. Science. 1987;238:1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- 3.Reynaud C-A, Anquez V, Dahan A, Weill J-C. Cell. 1985;40:283–291. doi: 10.1016/0092-8674(85)90142-4. [DOI] [PubMed] [Google Scholar]
- 4.Reynaud C-A, Dahan A, Anquez V, Weill J-C. Cell. 1989;59:171–183. doi: 10.1016/0092-8674(89)90879-9. [DOI] [PubMed] [Google Scholar]
- 5.McCormack W T, Tjoelker L W, Carlson L M, Petryniak B, Barth C F, Humphries E H, Thompson C B. Cell. 1989;56:785–791. doi: 10.1016/0092-8674(89)90683-1. [DOI] [PubMed] [Google Scholar]
- 6.Thompson C B, Neiman P E. Cell. 1987;48:369–378. doi: 10.1016/0092-8674(87)90188-7. [DOI] [PubMed] [Google Scholar]
- 7.Reynaud C-A, Anquez V, Grimal H, Weill J-C. Cell. 1987;48:379–388. doi: 10.1016/0092-8674(87)90189-9. [DOI] [PubMed] [Google Scholar]
- 8.Carlson L M, McCormack W T, Postema C E, Humphries E H, Thompson C B. Genes Dev. 1990;4:536–547. doi: 10.1101/gad.4.4.536. [DOI] [PubMed] [Google Scholar]
- 9.McCormack W T, Thompson C B. Genes Dev. 1990;4:548–558. doi: 10.1101/gad.4.4.548. [DOI] [PubMed] [Google Scholar]
- 10.McCormack W T, Hurley E A, Thompson C B. Mol Cell Biol. 1993;13:821–830. doi: 10.1128/mcb.13.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson C B. Trends Genet. 1992;8:416–422. doi: 10.1016/0168-9525(92)90324-w. [DOI] [PubMed] [Google Scholar]
- 12.Fertel R, Yetiv J Z, Coleman M A, Schwarz R D, Greenwald J E, Bianchine J R. Biochem Biophys Res Commun. 1981;102:1028–1033. doi: 10.1016/0006-291x(81)91641-7. [DOI] [PubMed] [Google Scholar]
- 13.Carroll S B, Stollar B D. J Biol Chem. 1983;258:24–26. [PubMed] [Google Scholar]
- 14.Horton J J, Holden C A, Ward P J, MacDonald D M, Sanderson A R. J Invest Derm. 1984;85:96–99. doi: 10.1111/1523-1747.ep12274979. [DOI] [PubMed] [Google Scholar]
- 15.Song C-S, Yu J-H, Bai D H, Hester P Y, Kim K-H. J Immunol. 1985;135:3354–3359. [PubMed] [Google Scholar]
- 16.Bauwens R M, Kint J A, Devos M P, Van Brussel K A, De Leenheer A P. Clin Chim Acta. 1987;170:37–44. doi: 10.1016/0009-8981(87)90381-0. [DOI] [PubMed] [Google Scholar]
- 17.Gassmann M, Thömmes P, Weiser T, Hübscher U. FASEB J. 1990;4:2528–2532. doi: 10.1096/fasebj.4.8.1970792. [DOI] [PubMed] [Google Scholar]
- 18.Lee K, Ametani A, Shimizu M, Hatta H, Yamamoto T, Kaminogawa S. Agric Biol Chem. 1991;55:2141–2143. [PubMed] [Google Scholar]
- 19.Woolley J A, Landon J. J Immunol Methods. 1995;178:253–265. doi: 10.1016/0022-1759(94)00263-v. [DOI] [PubMed] [Google Scholar]
- 20.Riordan J R, Rommens J M, Kerem B-S, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J-L, Drumm M L, Iannuzzi M C, Collins F S, Tsui L-C. Science. 1989;245:1066–1072. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 21.Nishinaka S, Matsuda H, Murata M. Int Arch Allergy Immunol. 1989;89:416–419. doi: 10.1159/000234985. [DOI] [PubMed] [Google Scholar]
- 22.Nishinaka S, Suzuki T, Matsuda H, Murata M. J Immunol Methods. 1991;139:217–222. doi: 10.1016/0022-1759(91)90191-h. [DOI] [PubMed] [Google Scholar]
- 23.Baba T W, Giroir B P, Humphries E H. Virology. 1985;144:139–151. doi: 10.1016/0042-6822(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 24.Simon T, Rajewsky K. Nucleic Acids Res. 1988;16:354. doi: 10.1093/nar/16.1.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alonso A, Chang L A, Murialdo H. Mol Immunol. 1990;27:115–127. doi: 10.1016/0161-5890(90)90106-a. [DOI] [PubMed] [Google Scholar]
- 26.Morrison S L, Johnson M J, Herzenberg L A, Oi V T. Proc Natl Acad Sci USA. 1984;81:6851–6855. doi: 10.1073/pnas.81.21.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coloma M J, Hastings A, Wims L A, Morrison S L. J Immunol Methods. 1992;152:89–104. doi: 10.1016/0022-1759(92)90092-8. [DOI] [PubMed] [Google Scholar]
- 28.Beatty J D, Beatty B G, Vlahos W G. J Immunol Methods. 1987;100:173–179. doi: 10.1016/0022-1759(87)90187-6. [DOI] [PubMed] [Google Scholar]
- 29.Loomans E E M G, Roelen A J M, Van Damme H S, Bloemers H P J, Gribnau T C J, Schielen W J G. J Immunol Methods. 1995;184:207–217. doi: 10.1016/0022-1759(95)00089-s. [DOI] [PubMed] [Google Scholar]
- 30.Whitsett J A, Dey C R, Stripp B R, Wikenheiser K A, Clark J C, Wert S E, Gregory R J, Smith A E, Cohn J A, Wilson J M, Engelhardt J. Nat Genet. 1992;2:13–20. doi: 10.1038/ng0992-13. [DOI] [PubMed] [Google Scholar]
- 31.Parvari R, Ziv E, Lantner F, Heller D, Schecter I. Proc Natl Acad Sci USA. 1990;87:3072–3076. doi: 10.1073/pnas.87.8.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arakawa H, Furusawa S, Ekino S, Yamagishi H. EMBO J. 1996;15:2540–2546. [PMC free article] [PubMed] [Google Scholar]
- 33.Pink J R L. Immunol Rev. 1986;91:115–128. doi: 10.1111/j.1600-065x.1986.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 34.Winter G, Milstein C. Nature (London) 1991;349:293–299. doi: 10.1038/349293a0. [DOI] [PubMed] [Google Scholar]
- 35.Gherardi E, Milstein C. Nature (London) 1993;357:201–202. doi: 10.1038/357201a0. [DOI] [PubMed] [Google Scholar]