Abstract
HPV-16 E6 and E7 genes are required to efficiently immortalize a broad spectrum of cell types including cervical keratinocytes. Therefore, the E6/E7 genes can be considered relevant targets for anti-cancer therapy. We produced several engineered hairpin (HP) ribozymes to specifically disrupt HPV-16 E6/E7 mRNA. After extensive biochemical characterization, one anti-E6 HP ribozyme (R434) was selected for in vivo testing because of its superior catalytic capabilities. When expressed in cis, R434 efficiently inhibited E6 in vitro translation. Cis-expression of the HP ribozyme with HPV-16 E6/E7 genes in normal human keratinocytes reduced the growth rate and prevented immortalization. RNA analysis by reverse transcription-PCR showed that E6/E7 transcripts were cleaved in post-transfected cells and virtually were eliminated after long term expression. Of interest, an inactive version of the HP also was able to significantly affect the immortalizing ability of E6/E7, probably through passive hybridization. The combination of passive and cleaving antisense RNA therefore is established as an effective inhibitor of HPV-16 E6/E7 immortalization.
Over one-half of invasive cervical carcinomas worldwide and many cervical carcinoma-derived cell lines contain and express DNA from human papillomavirus type 16 (HPV-16). In early stages, HPV-16 expression causes benign proliferation and efficiently immortalizes cultured human epithelial cells, including cervical keratinocytes (1–3). The HPV-16 viral, early genes E6 and E7 are required to acquire and maintain efficiently the transformed phenotype in a broad spectrum of cell types and commonly are expressed in cervical carcinoma cells. Thus, E6/E7 genes are the hallmark of cervical carcinoma (4–7). The E6 and E7 proteins bind with high affinity the p53 and Rb tumor suppressors, respectively (8, 9). The interaction of HPV-16 E6 protein with p53 results in degradation through the ubiquitin pathway, resulting in the equivalent of a mutant p53 phenotype (10–12). The association of E7 with Rb impedes the interaction of Rb with several proteins (i.e., E2F), efficiently disrupting the cell cycle (13–15). Therefore, the E6/E7 genes are ideal targets for anti-cancer therapy.
Antisense RNA and oligonucleotides have been used specifically to block translation of several genes. This effect is obtained by the hybridization of passive antisense molecules with their respective complementary mRNA to form nontranslatable double-stranded RNA molecules or DNA–RNA hybrids that promote the activity of endogenous RNase H, an enzyme that specifically digests the RNA strand of DNA–RNA hybrid molecules (16–18). In cultured tumor cells, antisense oligonucleotides have been shown to suppress effectively translation of several genes and to reverse some phenotypes (19–22). However, passive antisense therapy has the disadvantage of being active for a limited period and often causing nonspecific toxicity (23, 24). This approach has produced inhibition of E6/E7 gene expression in HPV-18-containing cell lines (e.g., C4–1, HeLa) (25). Suppression of E6/E7 has caused a significant decrease in growth rate, but the continuous addition of antisense oligonucleotide was required. Similar results were obtained when antisense E6/E7 RNA was expressed from a dexamethasone-inducible plasmid vector in stable transfected C4–1 cells (26) and in HPV-16 containing tumor cells infected with recombinant adenoviruses (27).
In the last decade, a new approach to antisense therapy has become available, consisting of small catalytic antisense RNA molecules that can hybridize and process the complementary RNA target (28). Generically known as trans-acting ribozymes, these molecules are released intact after processing the target that retains the ability to hybridize and process other target RNA molecules (i.e., multiple turnover) thus decreasing the dosage necessary for gene inhibition (29–32).
The in vitro use of engineered “hammerhead” (HH) ribozymes as antiviral and antitumor agents has been reported widely (33–35). HH ribozymes directed against E6/E7 genes have resulted in specific processing of HPV-16 and HPV-18 targets (36–39). However, only HH ribozymes directed against a HPV-18 target have been shown to inhibit growth of cultured tumor cells to some degree (39).
Hairpin (HP) ribozymes originally were derived from the 359-nt negative polarity strand satellite RNA of the tobacco ringspot virus (−sTRSV) and from other related plant satellite RNA replicons (40). The HP ribozyme represents the catalytic moiety of the satellite RNA that is responsible for site-specific cleavage and ligation reaction necessary for the minus strand replication (41–43). HP ribozymes efficiently cleave at 37°C and require minimal metal cofactors for activity (42, 44). To cleave efficiently to a specific target in vivo, recombinant HP ribozymes require specific target sequences for cleavage (GUC motif) and specific modifications in the ribozyme structure itself (44–47). HP ribozymes have been used successfully to inhibit HIV-1 expression in cultured infected lymphocytes (29, 48). However, to date, there has been no report of HP ribozymes that can process HPV-16 mRNA in vitro or in vivo.
The present results focus on the production and testing of an engineered HP ribozyme (R434) directed against the HPV-16 E6 mRNA. The ribozyme was optimized for catalytic degradation of HPV-16 E6 targets for in vivo use. HPV-16 E6/E7 gene expression and immortalization potential were inhibited significantly by the catalytic activity of the HP ribozyme expressed in cis. These results support the use of ribozymes as antiviral agents in the early stages of HPV-16 infection.
MATERIALS AND METHODS
Cell Culture and Transfection.
Normal human keratinocytes (HKc) from neonatal foreskins were cultured in MCDB151-LB medium as described (49). Transfections were done by using 10 μg of total plasmid DNA with Lipofectin reagent (Life Technologies, Gaithersburg, MD) as described (50). Cells were kept in G418 (200 μg/ml) for 2 wk. Growth rate experiments were done by seeding 6-well dishes (1 × 105 cells/well) in triplicate; cells were counted after 2 wk with a Coulter Counter ZM (Coulter). For HKc immortalization experiments, selection was done with 200 μg/ml G418 for 4 days. Selected HKc were kept in MCDB151-Luria-Bertani medium and were passed continuously for 8 wk before counting.
Plasmids and Oligonucleotides.
All plasmids were constructed and sequenced by using standard procedures (51). HP ribozyme coding sequences and target sites were synthesized in an Expedite 8900 DNA synthesizer (PerSeptive Biosystems, Framingham, MA) and cloned into the pBluescript KS vector (Stratagene) for initial characterization experiments. The HPV-16 target sequence from nucleotides 413 to 446 was used for R419, R434, and R434i ribozyme kinetics experiments. The pBtV5–434 and pBtV5–434i plasmids were made by cloning double-stranded oligonucleotides with the coding sequences for R434 and R434i ribozymes into the MluI and XhoI sites of the pBluescript KS-based pBtV5 plasmid (47). The cis-acting ribozyme constructs were made as follows: the EcoRI/EcoRI fragment from p16HH-Ha (4) was removed to produce p16HH. The R434 and R434i ribozyme coding sequences were PCR-amplified from pBtV5–434 and pBtV5–434i and ligated into the StyI site from p16HH. The fragments containing the entire E6 and E7 genes (nucleotides 97–868) linked to R434 or R434i were PCR-amplified from the ligation reaction and cloned into the pCR3.1 vector by using the One-Shot TA cloning system (Invitrogen) to produce plasmids pCR16E6/E7RZ and pCR16E6/E7RZi, respectively. The pCR16HH plasmid contained the HindIII/HindIII fragment from p16HH cloned into the pCR3.1 vector. The pCR3.1lacZ plasmid expressed β-galactosidase from the pCR3.1 vector (Invitrogen).
In Vitro Transcription Assays.
Plasmids expressing HP ribozymes and target sites were linearized and cleaned through QIAquick columns (Qiagen) before use. One microgram of linear DNA template was incubated with T3 or T7 RNA polymerase and rNTPs and [α-32P]UTP (Amersham) in a 20-μl final volume as described by the manufacturer (Ambion, Austin TX). Target RNA samples were purified through a 6% polyacrylamide 7-M urea gel before use.
In Vitro Ribozyme-Mediated Cleavage of HPV-16 Substrates.
For initial evaluation, HP ribozyme digestion reactions were performed in ribozyme buffer (40 mM Tris⋅HCl, pH 7.5/12 mM MgCl2/2 mM spermidine) at 37°C with 25 nM 32P-labeled ribozyme and 50 nM 32P-labeled substrate (containing only the complementary RNA sequence) for 60 min. Complete characterization was done on modified ribozymes by incubation of 1 nM labeled ribozyme and 30 nM substrate RNA in ribozyme buffer at 37°C for 180 min (Table 1). Ribozyme expression from linear or covalently closed templates was accomplished by incubating 1 μl of the in vitro transcription reaction with 1 × 106 cpm of a 32P-labeled target RNA (HPV-16 nucleosides 413–446) in a 10-μl final volume at 37°C. The reactions were stopped by freezing the samples in dry ice until used. Samples were denatured in loading buffer (80% formamide/0.01% bromophenol blue/0.01% xylene cyanol) at 65°C for 10 min and electrophoresed through 6% polyacrylamide 7-M urea gels. After electrophoresis, gels were dried and exposed to BM-2 radiographic films (Kodak). Quantification of the bands was made with a PhosphorImager 425 (Molecular Dynamics).
Table 1.
Potential HP ribozyme target sites (HPS) in the HPV-16 E6/E7 genes
| Site name | Substrate sequence | Cleavage site | Ribozyme activity* |
|---|---|---|---|
| HPSE61 | 415-UAACU GUC AAAAGCC | 419 | ++++ |
| HPSE62 | 430-ACUGU GUC CUGAAGAA | 434 | +++ |
| HPSE63 | 487-AAGGG GUC GGUGGACC | 491 | + |
| HPSE64 | 499-GACCG GUC GAUGUAUG | 503 | + |
| HPSE65 | 510-UGUAU GUC UUGUUGCA | 514 | NT |
| HPSE7 | 675-AGAUG GUC CAGCUGGA | 679 | ND‡ |
NT, not tested.
Ribozyme activity relative to the input target RNA in 60 min at 37°C as described in Materials and Methods.
No cleavage products were detected.
Reverse Transcription–PCR (RT-PCR).
Total RNA was obtained from cultured cells by using the RNeasy kit (Qiagen). HPV-16 E6/E7 cDNA was produced from 1 μg of total RNA using the Superscript II One Shot kit (Life Technologies). To produce differential bands for processed and nonprocessed E6/E7 mRNA, the upper primers E6U (5′-CAGCAATACAACAAACCG-3′) and E6U2 (5′-CACGTAGAAACCCAGC-3′) flanking the R434 target site (nucleotides 371–388 and 537–554, respectively) and the lower primer E7L (5′-TAGATTATGGTTTCTGAGAACA-3′) hybridizing within the E7 gene (nucleotides 862–841) were used in the following conditions: The first strand cDNA synthesis at 45°C for 30 min was followed by a denaturation step at 92°C for 2 min and 35 PCR cycles by using the denaturation step at 92°C for 1 min, the hybridization step at 45°C for 45 sec, and the polymerization step at 72°C for 1 min. This amplification produced two HPV-16 E6/E7-specific DNA fragments of 492 and 326 bp for E6U/E7L and E6U2/E7L, respectively. As a RNA integrity control, the β-actin gene was probed from 1 μg of total RNA with the oligonucleotide set 5′-TG ACGGGGTCACCCACACTGTGCCCATCTA-3′ and 5′-CTAGAAGCATTTGCGGTGGACGA TGGAGGG-3′, yielding a single 661-bp fragment, by using the PCR conditions described by the manufacturer (Stratagene). Amplified products were separated in 1.5% agarose gels and visualized with long wave UV light after ethidium bromide staining.
In Vitro Translation Assays.
HPV-16 E6/E7 proteins were produced using the T7 RNA polymerase promoter from pCR16HH, pCR16E6/E7RZ, and pCR16E6/E7RZi and using an in vitro transcription–translation system (rabbit reticulocyte TnT, Promega) and [35S]methionine (Amersham). A control expressing the beetle luciferase protein from the T7 promoter was used to monitor the reaction. Labeled proteins were separated through 15% SDS/PAGE, and gels were dried and exposed to BM-2 radiographic films (Kodak).
RESULTS
HP Ribozyme Activity Against HPV-16 E6/E7 mRNA Targets in Vitro.
Potential HP ribozyme target sites were located within the HPV-16 E6/E7 gene sequence (GeneBank accession no. K02718) by searching for 5′-GUC-3′ motifs by using the findpatterns routine of the Genetics Computer Group DNA analysis package (Genetics Computer Group, Madison, WI). Five of six potential HP ribozyme target sites were mapped to the 3′ terminus of the E6 gene (HPSE61–HPSE65), and one was mapped to the E7 gene (HPSE7) (Table 1). Synthetic HP ribozymes were derived from the satellite RNA of −sTRSV (42, 43), covalently linked to sequences complementary to their respective target sites on the HPV-16 mRNA. For initial evaluation of ribozyme activity, cleavage reactions were made by combining each 32P-labeled HP ribozyme with their complementary substrate RNA at a 1:2 M ratio. Only HP ribozymes cleaving at HPSE61 (R419) and HPSE62 (R434) significantly processed the target sequence (>60%). Ribozymes directed against HPSE63, HPSE64, and HPSE7 sites showed poor (<10%) or no cleavage activity of the target RNA and were not considered for additional use. HPSE65 was not tested because it overlaps one E6 splicing acceptor sites (nucleosides 515–526) and is located in a region with relatively high polymorphism (52).
To improve stability and catalytic ability of R419 and R434, two modifications were made in the ribozyme structure. A tetraloop modification (5′-G32GACUUCGGUCC43-3′) was introduced that resulted in a 4-base extension of helix 4 and a replacement of loop 3 of the prototype with the sequence 5′-UUCG-3′ (46) (Fig. 1). Cell-free optimization reactions for the length of helix 1 yielded active ribozymes targeted to the HPV-16 E6 sequence 5′-UAACUGUCAAAAGCC-3′ (nucleotides 415–429) for R419 and 5′-ACUGUGUCCUGAAGAA-3′ (nucleotides 430–445) for R434.
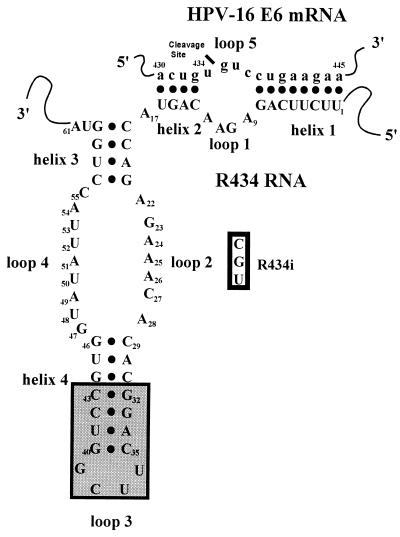
Figure 1.
HP ribozyme R434 substrate two-dimensional model. The model contains the standard four helices (helix 1–4) and five loops (loops 1–5) named as originally described for the −sTRSV hairpin ribozyme (44). Ribozyme substrate HPSE62 of the HPV-16 E6 gene is indicated by lowercase letters (a430-a445). The shaded box contains the modifications to R434 helix 4. The open box contains the specific base changes included in the inactive ribozyme R434i (A24-C, A25-G, and A26-U). •, Watson–Crick base pairs. The model is modified from the originally proposed L-shaped configuration (42, 64).
Complete biochemical characterization of the modified R419 and R434 ribozymes was accomplished by incubating ribozyme and target RNA (HPV-16 nucleotides 413–446) at a 1:30 M ratio for 180 min at 37°C. Under these conditions, >90% of the target was cleaved, showing that these two HP ribozymes must undergo multiple turnover during the duration of the reaction because there was thirtyfold excess target compared with ribozyme. From this data, it was calculated that R419 had a Km = 0.098 μM and a kcat = 0.18 min−1, and R434 had a Km = 0.021 μM and a kcat = 0.08 min−1. Of interest, the resulting catalytic efficiency (CE = kcat/Km) for R434 was twice as high (3.81 μM−1/min−1) as that of R419 (1.84 μM−1/min−1). This difference indicates that, despite being slower, R434 forms a more stable interaction with the substrate, which we considered a key feature for in vivo performance. Therefore, R434 was selected for further experimentation.
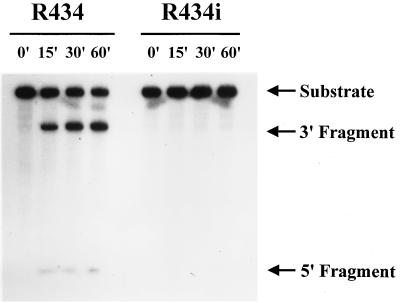
Nucleotide changes were introduced in the catalytic domain (loop 2) of R434 (A24 to C, A25 to G, and A26 to U) to produce an inactive ribozyme, R434i (Fig. 1). These changes have been reported previously to abolish HP ribozyme activity (45, 53). A comparison of the in vitro activities of R434 and R434i showed that the introduced mutations abolished the catalytic features of R434 (Fig. 2). Because theoretically R434i hybridizes to the same target site to the same extent as R434, it could have the effect of an antisense oligonucleotide. Thus, R434i is an appropriate control for the in vivo catalytic activities of R434.
Figure 2.
Ribozymes R434 and R434i biochemical analysis. The R434 and R434i ribozymes were produced by in vitro transcription from linear or covalently closed circular DNA templates with the conditions described in Materials and Methods. Ribozymes were incubated with the 32P-labeled HPSE62 substrate in a 1:2 M ratio at 37°C for up to 60 min. The substrate RNA and processed fragments are indicated.
Cis-Expressed Ribozymes Inhibit HPV-16 E6/E7 in Vitro Translation.
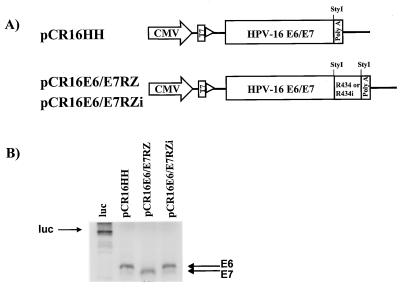
Because the biochemical characterization was performed on short synthetic targets, it was imperative to test ribozyme performance against a full length target RNA with secondary structures similar to those present in vivo. The cleavage of HPV-16 E6/E7 mRNA by R434 should result in the inhibition of E6 translation. To test the ability of R434 to produce this effect, the ribozyme coding sequence was cloned downstream from the HPV-16 E6/E7 genes. The constructs that were generated contained the HPV-16 E6/E7 genes alone (pCR16HH) or were linked at the 3′ end to the ribozymes R434 (pCR16E6/E7RZ) and R434i (pCR16E6/E7RZi), all under control of the CMV promoter/enhancer (Fig. 3A). The protein products from these plasmids were analyzed by in vitro translation by using T7 RNA polymerase and rabbit reticulocyte lysates (Fig. 3B). Labeled E6 and E7 proteins were produced from pCR16HH and pCR16E6/E7RZi, but only E7 was produced from pCR16E6/E7RZ. This result confirmed the potential of R434 as a translational inhibitor for HPV-16 E6. Under the controlled conditions of in vitro translation (i.e., presence of ribonuclease inhibitors), the E7 mRNA remained intact and produced a protein. Because, in the absence of complete E6 mRNA, more E7 transcripts would be available and an increase in the E7 protein synthesis also would be expected. With in vivo conditions, the E7 mRNA would be susceptible to exonuclease attack because of the lack of a cap structure present on the 5′ end of the polycistronic E6/E7 message.
Figure 3.
Cis-expression of R434 ribozyme inhibits HPV-16E6/E7 in vitro translation. (A) Map of HPV-16E6/E7 cis-expression constructs with R434 and R434i ribozymes. PCR-amplified fragments containing the entire HPV-16 E6/E7 genes (nucleotides 97–868) linked to R434 (pCR16E6/E7RZ) or R434i (pCR16E6/E7RZi) ribozymes were cloned in the pCR3.1 vector. The pCR16HH plasmid contains only the HPV-16E6/E7 genes. The relative positions of the StyI sites used for cloning the ribozymes and the vector poly(A) signal are shown. (B) The protein products produced by plasmids pCR16HH, pCR16E6/E7RZ, and pCR16E6/E7RZi were examined by in vitro translation reactions using T7 RNA polymerase and rabbit reticulocyte lysates in the presence of [35S]methionine. ←, the position of E6 and E7 proteins. Luc, luciferase protein reaction control.
Catalytic Activity of Cis-Acting HP Ribozymes Against HPV-16 E6/E7 mRNA in Vivo.
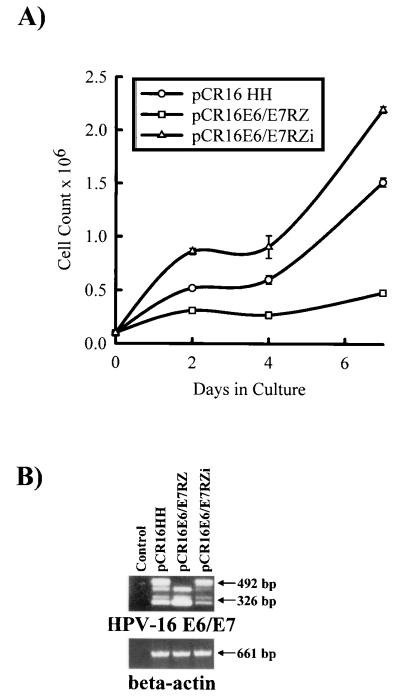
To first assess the functionality of R434 in vivo, pCR16HH, pCR16E6/E7RZ, and pCR16E6/E7RZi were transfected into HKc and kept under G418 selection (200 μg/ml) for 2 wk before examination for growth inhibition. The expression of pCR16E6/E7RZ caused a significant delay in the growth rate of transfected HKc compared with HKc transfected with pCR16HH or pCR16E6/E7RZi (Fig. 4A). To confirm that this effect was caused by ribozyme cleavage, a qualitative RT-PCR test was developed to examine the integrity of the E6/E7 mRNA. Upper primers (E6U and E6U2) were designed to hybridize to the flanking sides of the cleavage site, and a single lower anchor primer was designed to hybridize to the E7 gene (E7L). Two specific bands of 492 and 326 bp were formed in intact E6/E7 transcripts, and only the 326-bp band was formed in cleaved transcripts. Sample RNA isolated from transfected HKc showed that E6/E7 transcripts from pCR16E6/E7RZ were cleaved whereas transcripts from pCR16HH and pCR16E6/E7RZi were not (Fig. 4B). Controls without reverse transcriptase showed no amplified products, demonstrating that there was no contamination by vector nor genomic DNA. The additional bands reflected the production of incomplete E6/E7 transcripts present in the unstable transfected cell population. The β-actin control showed the integrity of the sample mRNA. These results suggest strongly that the observed effect in growth rate is a direct consequence of the inhibition of E6/E7 expression.
Figure 4.
Short term growth inhibition of HPV-16 E6/E7-transfected HKc by cis-expressed R434. (A) HKc were transfected with the pCR16HH, pCR16E6/E7RZ, or pCR16E6/E7RZi constructs and were kept in G418 (200 μg/ml) for 2 wk, and 105 cells were seeded for counting in 6-well dishes. The graphics are the mean of three experiments. (B) RT-PCR analysis of RNA from HKc transfected with HPV-16 E6/E7 (pCR16HH) and cis-expressed active and inactive ribozymes (pCR16E6/E7RZ and pCR16E6/E7RZi, respectively). Total RNA (1 μg) was subjected to a coupled RT-PCR reaction with primers specific to both sides of the ribozyme cleavage site as described in Material and Methods. A contamination control without reverse transcriptase (control) was included. Separate RT-PCR reactions were performed by using the same RNA sample with primers specific to the human β-actin gene to show RNA integrity. The HPV-16 E6/E7-amplified products (492 and 326 bp) were separated through agarose gel electrophoresis and visualized with ethidium bromide staining. The arrows indicate the position and size of the amplified products.
Cis-Expressed Ribozymes Inhibit HPV-16 E6/E7 Immortalization of HKc.
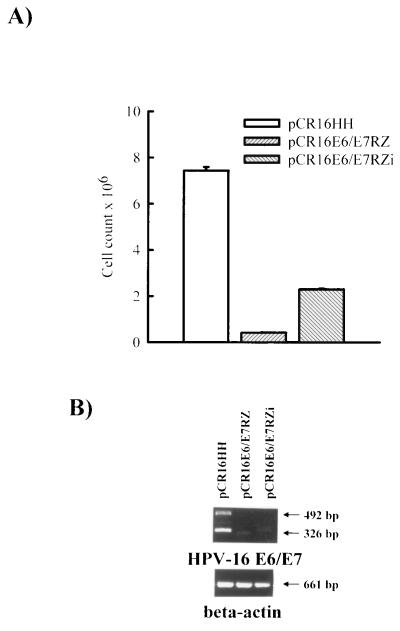
For long term ribozyme expression experiments, the plasmids pCR16HH, pCR16E6/E7RZ, and pCR16E6/E7RZi were transfected in HKc and selected with G418 (200 μg/ml) for 4 days to eliminate nontransfected cells. Selected cells were kept growing and continuously were passed for 8 wk by which time the control HKc transfected with the pCR3.1lacZ control plasmid (expressing the bacterial β-galactosidase gene) had senesced. As expected, transfection of the pCR16HH construct (which contains and expresses the E6/E7 genes) efficiently immortalized HKc as indicated by the number of surviving cells (Fig. 5A). In contrast, pCR16E6/E7RZ expression resulted in poor cell survival (<10%) relative to pCR16HH. This effect partially can be attributed to the passive antisense abilities of R434 because transfection of pCR16E6/E7RZi (which expresses a nonactive ribozyme) resulted in a significant decrease in cell survival (40%) (Fig. 5A). Surprisingly, the RT-PCR assays showed that both pCR16E6/E7RZ and pCR16E6/E7RZi but not p16HH-transfected cells lacked E6/E7 transcripts (Fig. 5B). The absence of E6/E7 transcripts in the active ribozyme-containing construct can be explained by the long term ablation of full length E6/E7 and E6*I/E7 transcripts, resulting in the senescence of the transfected HKc. No extra amplified bands were featured in these samples as the cell population became more homogeneous after 8 wk of continuous passage. The fact that the inactive ribozyme produced a reduction in the E6/E7 transcripts in the long term suggested that both the cleavage activity and passive antisense effect of R434 are associated to E6/E7 inhibition. Additional results using antisense oligodeoxynucleotides demonstrate that passive hybridization within the region covered by R434 and R434i can direct RNase H activity on the E6/E7 mRNA (L.M.A.-S. and J.A.D., unpublished work). Taken together, these results suggest that HPV-16 E6/E7 expression can be inhibited effectively by both passive and cleaving antisense RNA.
Figure 5.
Cis-expression of R434 ribozyme inhibits immortalization by HPV-16 E6/E7 mRNA in vivo. (A) HKc were transfected with the pCR16HH, pCR16E6/E7RZ, or pCR16E6/E7RZi constructs and selected with G418 (200 μg/ml) for 4 days. Transfected cells were counted after 8 wk of continuous growth. The results are the mean of three experiments. (B) Total RNA was isolated from transfected HKc 8 wk after transfection, and coupled RT-PCR reactions for HPV-16 E6/E7 and β-actin genes were performed as described in the legend of Fig. 4B.
DISCUSSION
The present results demonstrate that HP ribozymes are potentially effective antiviral agents. Because papillomaviruses covalently are closed circular, double-stranded DNA viruses, their genomes are less sensitive to ribozymes than are some other viral systems (i.e., HIV). In tumors, the HPV-16 DNA is found predominantly integrated to the cellular genome, lacking the regulatory viral-encoded E2 protein (54). Because papillomaviruses are completely dependent on cellular transcription factors for transcriptional regulation, they can be considered homologous to a nonessential cellular gene. During ribozyme therapy, the target gene translation can be inhibited but the DNA will remain intact and transcribing as long as the host cell is alive. Therefore, anti-HPV-16 ribozymes should maintain activity for long intervals until the host cell terminally differentiates or senesces.
In cervical tumors, the E6/E7 genes disrupt the function of cellular genes that are essential for the control of proliferation and cell division. However, because E6 and E7 expression does not necessarily result in malignant transformation, the elimination of these proteins would tend to decrease cell growth rather than cell death. Moreover, in tumor cells containing E6/E7, multiple changes will occur in the cellular genome. Therefore, anti-HPV-16 ribozymes, when used as potential antitumor agents, must be accompanied by additional treatments and/or enhanced with ribozymes directed against cellular genes. In contrast, because of the critical roles of E6 and E7 proteins in the early stages of infection, ribozymes feasibly may be used as antiviral agents in low grade, HPV-16-positive, precancerous lesions. Therefore, ribozymes that target E6 (i.e., R434) are effective mainly in preventing the interaction of E6 with p53 which is reported to be responsible for chromosomal instability and a high mutation rate in infected cells (55–57).
The use of ribozymes against HPV-16 targets has for the most part been limited to their in vitro activity. In contrast, many ribozymes targeting different cellular genes and other viruses have proven effective as in vivo antiviral and antitumor agents. Most of these ribozymes are HH-derived. Ribozyme therapy requires three fundamental steps: (i) expression, (ii) hybridization with the target (colocalization and site accessibility), and (iii) cleavage of the target. Because none of these steps can be extrapolated from in vitro assays, experimentation within the cellular environment and against full length transcripts are the most relevant approaches for evaluating ribozymes as therapeutic agents.
The comparison of well documented studies of HP and HH ribozymes as in vivo anti-HIV agents does not reveal a clear difference between them (47, 54, 58, 59). Moreover, the in vitro catalytic features of each ribozyme type do not necessarily correlate with its observed in vivo performance (58–60). Most of the 5′-GUC-3′ sites within the HPV-16 E6/E7 region produced HP ribozymes of limited efficiency, suggesting that the sites are not accessible or that more target site limitations exist than previously observed. When expressed in cis, the specific cleavage of the E6/E7 mRNA by R434 resulted in the growth inhibition of transfected cells. This caused the long term abrogation of HKc immortalization by E6/E7, showing the effectiveness of R434 ribozyme as an antiviral agent.
The cis-expression implies colocalization of ribozyme and target within the cell and a 1:1 ribozyme-to-substrate molar ratio. Additionally, in our experimental model, the cis-expressed ribozyme is attached to the E6/E7 mRNA, and its structural integrity is therefore protected from ribonuclease attack. Initial experiments with trans-acting R419 and R434 resulted only in passive antisense-mediated growth inhibition. This is because the integrity of the trans-expressed ribozyme structure was compromised probably by 5′ exonucleolytic degradation of the ribozyme, resulting in inhibition of the catalytic function but not hybridization ability (data not shown). Therefore, the design of future trans-acting HP ribozymes must consider exonucleolytic protection in the 5′ side toward improvement of cleavage efficiency. Additional inhibitory features can be added to ribozymes to enhance antiviral function. For example, an enhanced antiviral activity has been observed in ribozymes targeted against HIV with decoy sequences for viral-encoded transcriptional factors in addition to the therapeutic ribozyme itself (61, 62). Expression of multiple independent ribozymes would allow the targeting of different regions and produce higher turnover ratios (63). The present experiments lead to the expectation that trans-acting HP ribozymes directed against E6/E7 targets, when provided with adequate exonuclease protection, will become a highly effective and specific therapy against HPV-16 infection.
Acknowledgments
We thank Dr. Karen Vousden (Advanced BioScience Laboratories Basic Research Program, National Cancer Institute-Frederick Cancer Research and Development Center) for the p16HH-Ha plasmid.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HP, hairpin; HH, hammerhead; HPV-16, human papillomavirus type 16; HKc, human normal keratinocytes; RT, reverse transcription.
References
- 1.DiPaolo J A, Popescu N C, Alvarez L, Woodworth C D. Crit Rev Oncog. 1993;4:337–360. [PubMed] [Google Scholar]
- 2.Zur Hausen H, de Villiers E M. Annu Rev Microbiol. 1994;48:427–447. doi: 10.1146/annurev.mi.48.100194.002235. [DOI] [PubMed] [Google Scholar]
- 3.Schiffman M H. J Natl Cancer Inst. 1995;87:1345–1347. doi: 10.1093/jnci/87.18.1345. [DOI] [PubMed] [Google Scholar]
- 4.Hawley-Nelson P, Vousden K H, Hubbert N L, Lowy D R, Schiller J T. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phelps W C, Yee C L, Munger K, Howley P M. Cell. 1988;53:539–547. doi: 10.1016/0092-8674(88)90570-3. [DOI] [PubMed] [Google Scholar]
- 6.Viallet J, Liu C, Emond J, Tsao M S. Exp Cell Res. 1994;212:36–41. doi: 10.1006/excr.1994.1115. [DOI] [PubMed] [Google Scholar]
- 7.Yokoyama M, Tsutsumi K, Pater A, Pater M M. Obstet Gynecol. 1994;83:197–204. [PubMed] [Google Scholar]
- 8.Dyson N, Howley P M, Munger K, Harlow E. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 9.Werness B A, Levine A J, Howley P M. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 10.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 11.Huibregtse J M, Scheffner M, Howley P M. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbert N L, Sedman S A, Schiller J T. J Virol. 1992;66:6237–6241. doi: 10.1128/jvi.66.10.6237-6241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zerfass K, Schulze A, Spitkovsky D, Friedman V, Henglein B, Jansen-Durr P. J Virol. 1995;69:6389–6399. doi: 10.1128/jvi.69.10.6389-6399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu E W, Clemens K E, Heck D V, Munger K. J Virol. 1993;67:2402–2407. doi: 10.1128/jvi.67.4.2402-2407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walder J. Genes Dev. 1988;2:502–504. doi: 10.1101/gad.2.5.502. [DOI] [PubMed] [Google Scholar]
- 17.Cotten M, Schaffner G, Birnstiel M L. Mol Cell Biol. 1989;9:4479–4487. doi: 10.1128/mcb.9.10.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen J S. Antisense Res Dev. 1991;1:191–193. doi: 10.1089/ard.1991.1.191. [DOI] [PubMed] [Google Scholar]
- 19.Calabretta B. Cancer Res. 1991;51:4505–4510. [PubMed] [Google Scholar]
- 20.Persaud S J, Jones P M. J Mol Endocrinol. 1994;12:127–130. doi: 10.1677/jme.0.0120127. [DOI] [PubMed] [Google Scholar]
- 21.Anazodo M I, Wainberg M A, Friesen A D, Wright J A. J Virol. 1995;69:1794–1801. doi: 10.1128/jvi.69.3.1794-1801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichols G L. Antisense Res Dev. 1995;5:67–69. doi: 10.1089/ard.1995.5.67. [DOI] [PubMed] [Google Scholar]
- 23.Henry S P, Zuckerman J E, Rojko J, Hall W C, Harman R J, Kitchen D, Crooke S T. Anti-Cancer Drug Des. 1997;12:1–14. [PubMed] [Google Scholar]
- 24.Henry S P, Grillone L R, Orr J L, Bruner R H, Kornbrust D J. Toxicology. 1997;116:77–88. doi: 10.1016/s0300-483x(96)03532-9. [DOI] [PubMed] [Google Scholar]
- 25.Steele C, Cowsert L M, Shillitoe E J. Cancer Res. 1993;53:2330–2337. [PubMed] [Google Scholar]
- 26.von Knebel Doeberitz M, Gissmann L. Haematol Bluttransfus. 1987;31:377–379. doi: 10.1007/978-3-642-72624-8_80. [DOI] [PubMed] [Google Scholar]
- 27.Hamada K, Sakaue M, Alemany R, Zhang W W, Horio Y, Roth J A, Mitchell M F. Gynecol Oncol. 1996;63:219–227. doi: 10.1006/gyno.1996.0310. [DOI] [PubMed] [Google Scholar]
- 28.Cech T R. J Am Med Assoc. 1988;260:3030–3034. [PubMed] [Google Scholar]
- 29.Ojwang J O, Hampel A, Looney D J, Wong-Staal F, Rappaport J. Proc Natl Acad Sci USA. 1992;89:10802–10806. doi: 10.1073/pnas.89.22.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koizumi M, Kamiya H, Ohtsuka E. Biol Pharm Bull. 1993;16:879–883. doi: 10.1248/bpb.16.879. [DOI] [PubMed] [Google Scholar]
- 31.Efrat S, Leiser M, Wu Y J, Fusco-DeMane D, Emran O A, Surana M, Jetton T L, Magnuson M A, Weir G, Fleischer N. Proc Natl Acad Sci USA. 1994;91:2051–2055. doi: 10.1073/pnas.91.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cech T R. Gene. 1993;135:33–36. doi: 10.1016/0378-1119(93)90046-6. [DOI] [PubMed] [Google Scholar]
- 33.Kashani-Sabet M, Scanlon K J. Cancer Gene Ther. 1995;2:213–223. [PubMed] [Google Scholar]
- 34.Kiehntopf M, Esquivel E L, Brach M A, Herrmann F. Lancet. 1995;345:1027–1031. doi: 10.1016/s0140-6736(95)90762-9. [DOI] [PubMed] [Google Scholar]
- 35.Thompson J D, Macejak D, Couture L, Stinchcomb D T. Nat Med. 1995;1:277–278. doi: 10.1038/nm0395-277. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Kamath P, Zhang S, Weil M M, Shillitoe E J. Cancer Gene Ther. 1995;2:263–271. [PubMed] [Google Scholar]
- 37.Shillitoe E J, Kamath P, Chen Z. Cancer Gene Ther. 1994;1:193–204. [PubMed] [Google Scholar]
- 38.He Y K, Lu C D, Qi G R. FEBS Lett. 1993;322:21–24. doi: 10.1016/0014-5793(93)81102-6. [DOI] [PubMed] [Google Scholar]
- 39.Chen Z, Kamath P, Zhang S, St. John L, Adler-Storthz K, Shillitoe E J. Cancer Gene Ther. 1996;3:18–23. [PubMed] [Google Scholar]
- 40.Prody G A, Bakos J T, Buzayan J M, Schneider I R, Bruening G. Science. 1986;321:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 41.Feldstein P A, Buzayan J M, Bruening G. Gene. 1989;82:53–61. doi: 10.1016/0378-1119(89)90029-2. [DOI] [PubMed] [Google Scholar]
- 42.Hampel A, Tritz R. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- 43.Haseloff J, Gerlach W L. Gene. 1989;82:43–52. doi: 10.1016/0378-1119(89)90028-0. [DOI] [PubMed] [Google Scholar]
- 44.Hampel A, Tritz R, Hicks M, Cruz P. Nucleic Acids Res. 1990;18:299–304. doi: 10.1093/nar/18.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berzal-Herranz A, Joseph S, Chowrira B M, Butcher S E, Burke J M. EMBO J. 1993;12:2567–2573. doi: 10.1002/j.1460-2075.1993.tb05912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson P, Monforte J, Tritz R, Nesbitt S, Hearst J, Hampel A. Nucleic Acids Res. 1994;22:1096–1100. doi: 10.1093/nar/22.6.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu M, Poeschla E, Yamada O, DeGrandis P, Leavitt M C, Heusch M, Yees J K, Wong-Staal F, Hampel A. Virology. 1995;206:381–386. doi: 10.1016/s0042-6822(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 48.Yu M, Ojwang J, Yamada O, Hampel A, Rapapport J, Looney D, Wong-Staal F. Proc Natl Acad Sci USA. 1993;90:6340–6344. doi: 10.1073/pnas.90.13.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pirisi L, Yasumoto S, Feller M, Doniger J, DiPaolo J A. J Virol. 1987;61:1061–1066. doi: 10.1128/jvi.61.4.1061-1066.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarez-Salas L M, Velazquez A, Lopez-Bayghen E, Woodworth C D, Garrido E, Gariglio P, DiPaolo J A. Cancer Lett. 1995;91:85–92. doi: 10.1016/0304-3835(95)03721-8. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 52.Alvarez-Salas L M, Wilczynski S P, Burger R A, Monk B J, DiPaolo J A. Int J Oncol. 1995;7:261–266. doi: 10.3892/ijo.7.2.261. [DOI] [PubMed] [Google Scholar]
- 53.Siwkowski A, Shippy R, Hampel A. Biochemistry. 1997;36:3930–3940. doi: 10.1021/bi9628735. [DOI] [PubMed] [Google Scholar]
- 54.Zhou C, Bahner I, Rossi J J, Kohn D B. Antisense Nucleic Acid Drug Dev. 1996;6:17–24. doi: 10.1089/oli.1.1996.6.17. [DOI] [PubMed] [Google Scholar]
- 55.Slebos R J, Kessis T D, Chen A W, Han S M, Hedrick L, Cho K R. Virology. 1995;208:111–120. doi: 10.1006/viro.1995.1134. [DOI] [PubMed] [Google Scholar]
- 56.Yan Y, Ouellette M M, Shay J W, Wright W E. Mol Biol Cell. 1996;7:975–983. doi: 10.1091/mbc.7.6.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X L, Han S, Baluda M A, Park N H. Oncogene. 1997;14:2347–2353. doi: 10.1038/sj.onc.1201078. [DOI] [PubMed] [Google Scholar]
- 58.Rossi J J, Elkins D, Taylor N, Zaia J, Sullivan S, Deshler J O. Antisense Res Dev. 1991;1:285–288. [PubMed] [Google Scholar]
- 59.Rossi J J, Sarver N. Adv Exp Med Biol. 1992;312:95–109. doi: 10.1007/978-1-4615-3462-4_9. [DOI] [PubMed] [Google Scholar]
- 60.Crisell P, Thompson S, James W. Nucleic Acids Res. 1993;21:5251–5255. doi: 10.1093/nar/21.22.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuyama N, Ohkawa J, Koguma T, Shirai M, Taira K. Nucleic Acids Res. 1994;22:5060–5067. doi: 10.1093/nar/22.23.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamada O, Kraus G, Luznik L, Yu M, Wong-Staal F. J Virol. 1996;70:1596–1601. doi: 10.1128/jvi.70.3.1596-1601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohkawa, J., Yuyama, N., Takebe, Y., Nisikawa, S., Homann, M., Sczakiel, G. & Taira, K. (1993) Nucleic Acids Symp. Ser. 121–122. [PubMed]
- 64.Altschuler M, Tritz R, Hampel A. Gene. 1992;122:85–90. doi: 10.1016/0378-1119(92)90035-n. [DOI] [PubMed] [Google Scholar]