Abstract
The mammalian circadian system consists of a central oscillator in the suprachiasmatic nucleus of the hypothalamus, which coordinates peripheral clocks in organs throughout the body. Although circadian clocks control the rhythmic expression of a large number of genes involved in metabolism and other aspects of circadian physiology, the consequences of genetic disruption of circadian-controlled pathways remain poorly defined. Here we report that the targeted disruption of Nocturnin (Ccrn4l) in mice, a gene that encodes a circadian deadenylase, confers resistance to diet-induced obesity. Mice lacking Nocturnin remain lean on high-fat diets, with lower body weight and reduced visceral fat. However, unlike lean lipodystrophic mouse models, these mice do not have fatty livers and do not exhibit increased activity or reduced food intake. Gene expression data suggest that Nocturnin knockout mice have deficits in lipid metabolism or uptake, in addition to changes in glucose and insulin sensitivity. Our data support a pivotal role for Nocturnin downstream of the circadian clockwork in the posttranscriptional regulation of genes necessary for nutrient uptake, metabolism, and storage.
Keywords: mRNA, clock, diabetes, posttranscriptional, lipid
Circadian clocks are present in most tissues of the body, where they control the expression of 5–10% of the tissue-specific mRNAs through both transcriptional and posttranscriptional regulation (1, 2). The widespread importance of circadian clock regulation is evident in that generalized disruption of normal clock function results in tumor formation, sleep disorders, and metabolic problems (reviewed in refs. 3 and 4). For example, mutations in the central clock genes Clock or Bmal1 result in metabolic changes found in obesity and the metabolic syndrome (5–8), and numerous genes involved in fatty acid, cholesterol, and glucose metabolism in liver are regulated in circadian or diurnal patterns (9–15), indicating that the clock plays a broad role in regulating metabolism. Nonetheless, the large number of genes, metabolic pathways, and cell/tissue types that are under general circadian control impose a major challenge in understanding the molecular details. Further advances in this area require refined understanding of the specific circadian output pathways by which the clocks regulate physiology.
For cycling mRNAs to closely reflect daily rhythmic transcriptional drive, their half-lives must be relatively short. There are several examples of rhythmic posttranscriptional regulation in which the mRNA half-life or adenylation state changes over the course of the day (16–19), but very little is known about the mechanisms responsible. A likely contributor is Nocturnin (Ccrn4l, Noc), which has been implicated in the posttranscriptional regulation of mRNA stability and/or translatability by the circadian clock (20). Noc is expressed rhythmically in many tissues, with particularly high-amplitude rhythms in liver where mRNA levels are increased 100-fold in early night (21). Noc is at a pivotal position to play a role in shaping the rhythmic pattern of gene expression either within the core molecular clockwork or in regulatory output pathways of the clock.
We report here that Noc−/− mice, produced through targeted disruption of the Noc gene, have normal circadian behavior and clock gene expression. This suggests that Noc is in an output pathway downstream of the central circadian clockwork. The importance of this output pathway in metabolic control is revealed by our finding that Noc−/− mice are resistant to diet-induced obesity and exhibit other metabolic changes. This metabolic phenotype is profoundly different from the metabolic syndrome seen in mice with general central clock disruption and suggests that Noc controls specific circadian pathways related to lipid uptake and/or utilization.
Results
Noc−/− Mice Exhibit Normal Circadian Rhythms and Clock Gene Expression.
To investigate Noc's role in clock-regulated posttranscriptional events, we generated mice with a targeted disruption of the Noc gene [supporting information (SI) Fig. 5]. Noc−/− mice lack the entire coding region of exon 3, which contains most of the protein coding sequence including the catalytic domain (20, 21). No detectable Noc protein is made in the mice, suggesting that this is a null allele. Both Noc−/− and Noc+/− mice on the C57BL/6J background (7–10 generations) used in these studies appeared grossly normal and reproduced successfully.
Noc is widely expressed in association with known elements of the molecular clockwork (21). To determine whether it is essential for the generation of circadian rhythms we measured locomotor activity in WT and Noc−/− mice maintained in running wheel cages. Differences in activity profiles were not detected in either cyclic light [light:dark (LD)] or in constant darkness (SI Fig. 6). The free-running period (τ) in the WT mice was 23.78 ± 0.03 h compared with 23.74 ± 0.02 h in the Noc−/− mice. The mutant mice entrained normally to LD cycles and also exhibited normal phase-shifts to light pulses (SI Fig. 6 and data not shown). Consistent with this finding, we also found that mRNA rhythms of several circadian clockwork genes in liver were indistinguishable in WT and Noc−/− mice (SI Fig. 6 and data not shown). Together, our data suggest that Noc is neither a part of the core circadian clockwork nor necessary for light entrainment. Instead, it is likely to provide a circadian output that affects downstream physiologic rhythms.
Noc−/− Mice Are Resistant to Diet-Induced Weight Gain.
Our investigation of downstream rhythms focused on the liver because Noc exhibits a high-amplitude rhythm in this tissue. Because many rhythmic mRNAs in liver encode genes involved in glucose and lipid metabolism (10, 11, 14, 22), we reasoned that Noc might play a role in their circadian regulation. In support of this hypothesis, morphological examination of the livers of the Noc−/− mice revealed that they accumulate significantly less lipid in lipid droplets than WT controls (Fig. 1A).
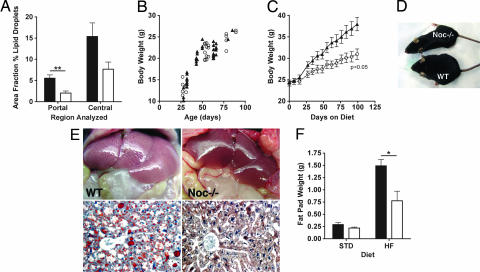
Fig. 1.
Noc−/− mice exhibit a diet-dependent lean phenotype. (A) Noc−/− mice have decreased lipid accumulation in livers. Livers were stained with ORO, and the area fraction of ORO staining was determined in liver chords surrounding portal and central veins separately by using size-calibrated images and Image J software. The values represent group means (± SEM) of sections from six mice per genotype [t test: P = 0.0035 for portal area; not significant (P = 0.06) for central area]. (B) Body weights of male WT (filled symbols) and Noc−/− (open symbols) mice from 4 to 13 weeks with ad libitum access to the standard diet were not different (WT, n = 24; Noc−/−, n = 36; analysis of covariance: F1,59 = 0.1222, P = 0.728). (C) Male WT mice (filled symbols) gained more weight than male Noc−/− mice (open symbols) when given ad libitum access to a high-fat diet (WT, n = 8; Noc−/−, n = 12) for 14 weeks beginning at 8 weeks of age (repeated-measures ANOVA: P = 0.042). Values are group means ± SEM. (D) Photograph of age-matched WT and Noc−/− mice showing body size differences after 20 weeks on the high-fat diet. (E) Noc−/− livers at 20 weeks did not accumulate excess fat on the high-fat diet. Greater accumulation of lipid in WT (Left) compared with Noc−/− (Right) livers at the macroscopic level (Upper) and in ORO-stained sections (Lower) was observed. (F) Epididymal fat pads dissected at ZT1 from Noc−/− mice (open bars) on a high-fat diet are significantly smaller than those of WT controls (filled bars). Shown are average weights (± SEM) of one epididymal fat pad from each mouse [for the standard diet, n = 5 for both genotypes (t test: P = 0.17); for the high-fat diet, n = 5 Noc−/− and n = 4 WT (t test: P = 0.024)].
The marked decrease in lipid droplets suggested alterations in lipid uptake or metabolism in Noc−/− mice; therefore, we examined weight gain in Noc−/− mice. On a standard diet (8% kcal from fat) their weights were indistinguishable from WT mice (Fig. 1B). However, Noc−/− mice fed a high-fat diet (45% kcal from fat) exhibited an obese-resistant phenotype (Fig. 1 C and D). WT mice, as expected, gained weight on this diet and became obese. However, Noc−/− mice gained less weight and were only slightly heavier than on a standard diet. This weight difference was not due to changes in general growth because the body lengths of the two genotypes were indistinguishable (data not shown). In WT mice the high-fat diet also caused a large accumulation of fat in the liver that was visible even under gross examination, whereas the Noc−/− mice had non-fatty livers (Fig. 1E). The diet-induced increase in visceral adipose tissue mass in epididymal fat pads was also less in Noc−/− mice (Fig. 1F).
The livers of Noc−/− mice on standard and high-fat diets were examined for changes in expression profiles of mRNAs for Pparγ, Srebp-1c, Srebp-1a, Scd1, and L-Fabp, five genes known to be involved in lipid uptake, storage, and metabolism (Fig. 2). Because Pparγ and Srebp-1c are transcription factors that control many genes in lipid-related pathways and have been reported to exhibit circadian profiles (10, 13), we collected samples at 4-h intervals around the clock to assess rhythmicity in addition to overall changes in expression. In the case of Pparγ, although average daily levels were not different between genotypes (Fig. 2A Left), the robust diurnal rhythm in WT mice on the high-fat diet is in marked contrast to the nonrhythmic and highly variable levels seen in Noc−/− mice (Fig. 2A Center and Right). Levels of Srebp-1c mRNA increased significantly in the WT mice on the high-fat diet and showed a marked rhythm, whereas the Noc−/− mice maintained low levels, similar to that seen with the standard diet (Fig. 2B).
Fig. 2.
Lipid-related genes have diet- and genotype-dependent expression profiles. Pparγ (A), Srebp-1c (B), Scd1 (C), Srebp-1a (D), and L-Fabp (E) mRNA levels were measured by using quantitative RT-PCR on total RNA isolated from livers from WT (filled bars and symbols) and Noc−/− (open bars and symbols) mice maintained on either a standard diet (STD) or a high-fat diet (HF). Samples were collected at 4-h intervals over 24 h from mice in a 12-h LD cycle. (A Left and B Left) Data pooled from all time points. (A Center and Right and B Center and Right) Data shown with respect to time of day (ZT0 is time of light onset, and ZT12 is light offset; ZT0 points are replotted as ZT24 to aid in visualization of the rhythm). There is a significant time of day effect for Pparγ in the WT mice (repeated-measures ANOVA: P = 0.022) but not for the Noc−/− mice (P = 0.95) on the high-fat diet. The time-of-day effect for Srebp-1c is not significant for either genotype (P = 0.38, WT; P = 0.65, Noc−/−) on the high-fat diet, although on this diet the WT have significantly higher levels of Srebp-1c than the Noc−/− mice (P = 0.026) (n = 2–3 mice per time point per genotype). (C–E) Only pooled data (n = 15–18 mice per genotype) from all time points are shown because these mRNAs are not rhythmic. Shown are group means ± SEM. Asterisks denote statistically significant differences between genotypes (differences between diets are not marked): ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001.
We also examined the expression of Scd1 (steroyl-CoA desaturase), Srebp-1a, and L-Fabp (liver-specific fatty acid binding protein), three nonrhythmic liver mRNAs (10, 13). Scd1 is a direct target of Srebp-1c that is involved in lipogenesis (23), Srebp-1a is a splice variant encoded by the same gene as Srebp-1c, which has roles in cholesterol synthesis in addition to lipogenesis (24), and L-Fabp is a cytosolic lipid carrier (25, 26). Scd1 and L-Fabp mRNA levels were substantially lower in Noc−/− mice than in WT on a high-fat diet; L-Fabp mRNA was also significantly lower in Noc−/− animals on a standard diet. In contrast, differences between genotypes were not seen for Srebp-1a mRNA (Fig. 2 C and D). These gene expression changes in key factors related to lipid metabolism are consistent with lack of accumulation of lipid in Noc−/− mice.
Food Intake, Activity, and Metabolic Rate of Noc−/− Mice.
The obese-resistant phenotype of the Noc−/− mice is not due to increased activity, decreased food intake, or increased metabolic rate. Overall ambulatory activity (using infrared beam breaks rather than running wheels) in Noc−/− mice was less than the WT on the standard diet and equal to the WT on the high-fat diet (Fig. 3A). Both genotypes consumed equivalent calories (Fig. 3B) and exhibited similar metabolic rates on the two diets (Fig. 3C). Furthermore, the Noc−/− mice had lower body temperatures on both diets, consistent with the idea that these animals are not generating excess heat through increased metabolism (Fig. 3D). The respiratory exchange rates were the same for both genotypes on the standard diet (Fig. 3E). On the high-fat diet the respiratory exchange rates decreased for both genotypes, consistent with an increased use of lipids as a fuel source. However, the Noc−/− mice showed a trend toward higher values when compared with WT mice, suggesting lower lipid oxidation. In conclusion, the Noc−/− mice maintain the same weight on the standard diet and remain lean on the high-fat diet, despite being less (standard diet) or equally (high-fat diet) active, eating the same amount of food, and producing less heat than the WT mice. The most likely explanation for the surprising phenotype is deficient food absorption from the intestine and/or abnormalities in lipid clearance, metabolism, or storage.
Fig. 3.
The lean phenotype is not due to hyperactivity or reduced caloric intake in the Noc−/− mice. (A) Noc−/− (open bars) are less active than WT (filled bars) mice on a standard diet (STD), but on a high-fat diet (HF) activity measurements are similar. Mice were fed ad libitum (n = 4 per group), and total activity was measured as infrared beam crossings in an Oxymax metabolic chamber system (Columbus Instruments). Values are group means ± SEM [t test: standard diet, P = 0.013; high-fat diet, not significant (P = 0.088)]. (B) There is no difference in caloric intake between Noc−/− (open bars) and WT (filled bars) mice on either standard or high-fat diets. The same adult male mice shown in A were analyzed for total caloric intake in the Oxymax metabolic chambers. All values represent group means ± SEM. No significant difference was determined by t test (P = 0.93 for both diets). (C) There are no significant differences in metabolic parameters between Noc−/− (open bars) and WT (filled bars) adult male mice. Mice were provided ad libitum access to either standard (Left) or high-fat (Right) chow in the Oxymax metabolic chamber system. Oxygen consumption and CO2 production were measured, and resting metabolic rate (RMR) was calculated (see Materials and Methods). Values are group means ± SEM (n = 4). There was no statistical difference between groups (t test: P > 0.45). (D) Body temperature was decreased in Noc−/− mice. Six mice of each genotype were maintained on a standard diet, and temperatures were recorded throughout a 21-day period using telemetry. The mice were then switched to high-fat diets, and body temperature data were collected for an additional 21 days. Body temperature means over the 21 days were calculated for each animal. Shown are group means ± SEM [asterisks denote statistically significant differences (t tests) between groups: ∗, P < 0.05 for standard diet; ∗∗∗, P < 0.001 for high-fat diet]. (E) Respiratory exchange rates were calculated by taking the ratio of VCO2/VO2 for each animal from C. The decreased value of the respiratory exchange rate on the high-fat diet is a reflection of the increased use of lipids as an energy source in both genotypes (asterisks denote statistically significant differences between groups: ∗, P < 0.05; ∗∗, P < 0.005). The difference between genotypes is not statistically significant on either diet (standard diet, P = 0.52, high-fat diet, P = 0.24), although on the high-fat diet the Noc−/− mice have a trend toward lower lipid utilization than the WT mice.
To distinguish among these possibilities we examined circulating triglyceride, cholesterol, and free fatty acid levels. We hypothesized that lipid levels would be increased if lipid storage or insulin sensitivity were altered. However, total circulating cholesterol and triglyceride levels were the same between the two genotypes under fasting and fed conditions on both the standard and high-fat diets (SI Fig. 7). Fasting free fatty acid levels were also not statistically significantly different, but there was a trend to higher levels in the Noc−/− mice on both diets. The lack of major differences in circulating lipids in combination with reduced accumulation of lipid in liver and white adipose tissue in Noc−/− mice (Fig. 1) point toward altered uptake of lipid from the intestine.
Noc−/− Mice Exhibit Alterations in Glucose Homeostasis.
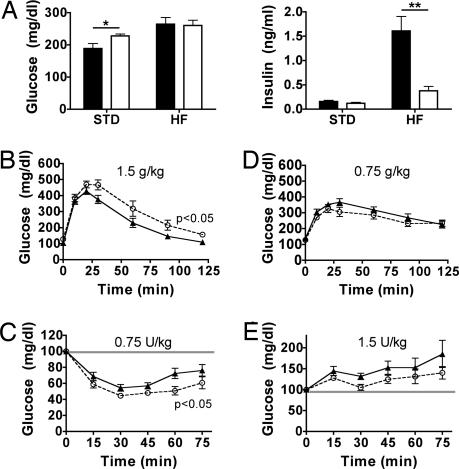
To assess whether resistance to steatosis was also associated with alterations in hepatic glucose metabolism, we also examined glucose homeostasis and insulin sensitivity. On the standard diet, circulating glucose levels were slightly increased in the Noc−/− mice (Fig. 4A Left), but there was no difference in circulating insulin levels (Fig. 4A Right). On the high-fat diet glucose levels increased relative to those on standard diet, but there was no difference between genotypes. Insulin levels also increased significantly on the high-fat diet in both genotypes, but the increase was much more dramatic in WT mice; the increase was ≈10-fold in WT compared with 3-fold in Noc−/− mice. Nonetheless, the 3-fold increase in Noc−/− mice was sufficient to maintain the same blood glucose levels as the WT mice (Fig. 4A).
Fig. 4.
Noc−/− mice have diet-dependent changes in glucose and insulin tolerance. (A) Circulating glucose (Left) and insulin (Right) levels in Noc−/− (open bars) and WT (filled bars) mice on standard (STD) and high-fat (HF) diets were measured from blood at ZT5 after a 5-h fast. Shown are mean levels (± SEM) from five animals from each genotype. On the standard diet, the Noc−/− mice had slightly increased glucose levels over the WT mice (t test, P = 0.048) but had no difference on the high-fat diet (t test, P = 0.88). The insulin levels on the standard diet were not significantly different between genotypes (t test, P = 0.22) but were significantly decreased in the Noc−/− mice as compared with the WT mice on the high-fat diet (t test, P < 0.005). (B) Glucose tolerance tests performed on fasted (16 h) WT (filled symbols) and Noc−/− (open symbols) male mice at ZT4 revealed impaired glucose tolerance in Noc−/− mice on a standard diet. Mice were injected i.p. with d-glucose (1.5 g/kg of body weight; Sigma), and blood glucose values were measured at 0, 10, 20, 30, 60, 90, and 120 min after glucose injection (n = 12; repeated-measures two-way ANOVA, P = 0.041). Values are group means ± SEM. (C) Insulin tolerance tests on ad libitum-fed WT (filled symbols) and Noc−/− (open symbols) male mice at ZT8 revealed greater insulin sensitivity in Noc−/− mice on a standard diet. Blood glucose values were assayed immediately before and at 15, 30, 60, and 75 min after i.p. injection of mammalian crystalline insulin (0.75 units/kg of body weight; Lilly) (n = 18 WT and n = 17 knockout; repeated measures two-way ANOVA, P = 0.029). Values are group means ± SEM. (D) Both Noc−/− and WT mice exhibit glucose intolerance when fed a high-fat diet. Glucose tolerance tests were performed as in B except that the glucose dose was decreased by half to 0.75 g/kg. Values are group means ± SEM. There was no significant difference between genotypes (repeated-measures two-way ANOVA, P = 0.055). (E) Insulin tolerance tests conducted exactly as in C except with a 2-fold-higher insulin dose (1.5 units/kg of body weight) revealed that both WT (filled symbols) and Noc−/− (open symbols) mice become insulin-resistant when maintained on a high-fat diet for 3–4 months. Values are group means ± SEM (n = 14 per group). Significant differences between genotypes were not detected by using repeated-measures ANOVA (P = 0.80). Similar results were obtained with the standard 0.75 units/kg insulin dosage (data not shown).
Insulin and glucose tolerance tests using mice on a standard diet (Fig. 4 B and C) revealed that Noc−/− mice had moderately impaired glucose tolerance and moderately increased insulin sensitivity. These observations point to a problem in responding to a glucose challenge with an appropriate increase in insulin secretion, rather than a problem in insulin action. Similar testing after a high-fat diet revealed that both WT and Noc−/− mice became increasingly intolerant to glucose and insensitive to insulin (Fig. 4 D and E). The magnitude of these effects is reflected in the fact that to obtain data comparable to the standard diet (Fig. 4 B and C) the bolus of glucose was decreased by half (Fig. 4D). In the insulin tolerance test both Noc−/− and WT mice failed to respond even after increasing the dose of injected insulin by 2-fold (Fig. 4E). This insulin insensitivity is consistent with the increased levels of circulating insulin in both genotypes (Fig. 4A). However, it should be emphasized that insulin increased to a much greater extent in WT mice. In conclusion, both WT and Noc−/− mice develop insulin resistance on the high-fat diet, but Noc−/− mice exhibit mixed insulin sensitivity and impaired glucose tolerance compared with WT mice.
Discussion
Our analysis shows that Noc−/− mice are resistant to diet-induced obesity, as reflected in lower body weight, smaller visceral fat pads, decreased fat accumulation in the liver, decreased lipogenic gene expression, and better insulin sensitivity on the high-fat diet. This suggests that Noc participates at multiple levels in energy homeostasis, including lipid and carbohydrate metabolism. Reduced accumulation of fat in liver hepatocytes is a signature feature of Noc−/− mice and is consistent with reduced expression of lipogeneic genes such as Pparγ, Srebp-1c, and L-Fabp in liver. The Noc−/− phenotype is distinct from lipodystrophic mice, a class of “lean” mice, which have defects in white adipocyte differentiation or function (27). Lipodystrophic mice lack normal adipocytes and accumulate fat in nonadipose tissues, resulting in fatty livers, elevated circulating trigylceride levels, and insulin resistance. In contrast, adipocyte size and adipose tissue mass are normal in Noc−/− mice on a standard diet and do not increase to the same extent as WT on the high-fat diet (Fig. 1 and N.D., S. Q. Duong, and C.B.G., unpublished observation). Thus, Noc−/− mice exhibit decreased lipid storage in adipose tissue but do not exhibit increased circulating triglyceride levels or hepatic steatosis.
Other classes of lean mice with normal adipocytes but depleted lipid stores have been described. In general, those mice have normal, non-fatty livers and normal triglyceride levels, similar to the Noc−/− mice (27). Decreased lipid storage can result from such factors as increased metabolic rates in peripheral tissues or decreased energy availability, appetite, or intestinal absorption. These effects may be due to local changes in one or more specific tissues or to systemic changes caused by altered CNS regulation. In comparison to these models, we show that Noc−/− mice eat the same, exhibit equal or less activity, and have similar whole-body energy expenditure as WT mice, suggesting that a more general lipid uptake mechanism may be altered.
The Noc−/− mice show similarity to one lean mouse model, the L-Fabp-deficient mouse (28). L-Fabp−/− mice are resistant to weight gain and fat accumulation in the liver and also develop changes in glucose homeostasis on a high-fat diet. L-Fabp is expressed mainly in the liver and the upper gastrointestinal tract, and its down-regulation in Noc−/− livers (Fig. 2E) on both standard and high-fat diets is likely to account for at least some features of the phenotype such as reduced lipid storage in the liver.
Likewise, the altered Pparγ and Srebp-1c expression levels in the Noc−/− mice are likely to contribute to the protection from hepatic steatosis on the high-fat diet. Pparγ is a principal regulator of adipogenesis in adipocytes but is also expressed in a circadian pattern in liver, and Srebp-1c activates transcription of several lipogenic genes. Furthermore, overexpression of either Paprγ or Srebp-1c in liver results in a hepatic steatosis (29, 30). The reduced expression of these factors in Noc−/− mice on the high-fat diet is consistent with their lack of hepatic steatosis.
Noc as a Clock Output in Metabolic Regulation.
The striking phenotype of the Noc−/− mice, along with data that place Noc in an output pathway of the circadian clock, illustrate that nutrient uptake, metabolism, and/or storage are controlled by the circadian clock. Several recent studies have demonstrated a link between the circadian clock and metabolism. For example, Clock and Bmal1, two core genes in the circadian clock mechanism, have been shown to be important for normal glucose homeostasis (6), and Bmal1 contributes to the regulation of adipogenesis (8). Furthermore, Clock mutant mice (on a C57BL/6J background) exhibit an obese phenotype associated with features of the metabolic syndrome (5). The clearly distinct phenotype of the Noc−/− mice compared with the Clock mutant mice is consistent with the idea that Noc is outside of the clock and that its loss affects only a specific subset of clock output pathways. The circadian system likely controls metabolic processes at numerous levels including transcriptional and posttranslational control (15). It therefore seems reasonable that loss of circadian regulation of mRNA half-life could have effects different from the more universal loss of circadian timing, as occurs in the Clock mutant mice, with a defective central clock mechanism.
Noc as a Deadenylase and Circadian mRNA Decay.
Because Noc has been shown to exhibit deadenylase activity, an attractive hypothesis is that Noc targets specific mRNAs for degradation. There are five currently known deadenylases in mammalian cells, and Noc is unique among them in its circadian regulation (10, 20, 21) and in its response to acute stimuli such as serum shock in cultured cells (31). Because Noc is highly expressed in the early evening in a number of different tissues, its general function could be to down-regulate mRNAs in the night as cells transition to a new metabolic state. To date, however, identification of specific targets has proven elusive. In our analysis we have identified several genes with altered expression in Noc−/− mice on a high-fat diet. Although these gene expression changes provide support for the resistance of the Noc−/− animals to the high-fat diet, these genes are unlikely to be direct targets of Noc deadenylase activity, as one would expect such targets to be increased in the Noc−/− mice. More likely, changes in the expression of these genes are secondary to the dysregulation of the primary targets of Noc.
In addition to providing a means for understanding how the circadian clock controls specific output rhythms, the Noc−/− mice provide a new tool for understanding the interplay between the circadian clock and the regulation of body weight and glucose and lipid homeostasis in response to different diets. This is an issue of much importance because the prevalence of obesity in affluent western cultures has increased dramatically over the past several decades and is rising to the forefront of health problems facing our population. Also, irregular and hectic lifestyles that are prevalent in the contemporary world may disrupt the circadian clock and change the susceptibility to high-fat diets. The Noc−/− mice should provide a valuable tool for studying the relationships of these two pathways.
Materials and Methods
Targeted Disruption of Noc.
The Noc (Ccrn4l) gene was disrupted by homologous recombination in mouse D3 embryonic stem cells, and mouse chimeras were produced by blastocyst injection in the Medical College of Wisconsin Transgenic Facility (see SI Methods for detailed strategy). Antibodies to a mouse Noc peptide (amino acids 125–150) were produced in guinea pigs by Covance Research Products (Madison, WI). Mice used in these experiments are either siblings from heterozygous matings or age-matched mice from homozygous matings bred in our facility between 3 and 6 months of age.
Quantitation of Clock Gene Expression by Real-Time PCR.
Mice raised in a 12:12 h LD cycle were transferred to constant darkness for 24 h and were killed by cervical dislocation under CO2 anesthesia. RNA from freshly frozen liver was extracted by using the RNeasy Mini Kit (Qiagen). cDNA was produced from 250 ng of total RNA by using the iScript cDNA Synthesis Kit, diluted 1:3, and 1 μl was used in 25-μl PCRs using iQ SYBR Green Supermix in a MyiQ thermal cycler and detection system (Bio-Rad). Relative mRNA abundance was calculated and normalized to the levels of GAPDH. The highest value from among WT samples was set to 100, and all other values were normalized to that. Primers are included in SI Table 1.
Quantitation of Lipid-Related Gene Expression by Real-Time PCR.
Liver tissue was collected from mice killed at 4-h intervals over 24 h in a 12-h LD cycle. RNAs were extracted from tissues (three mice per genotype per time point) by using TRIzol reagent (Invitrogen), and cDNA was synthesized with the SuperScript II system (Invitrogen) with random hexamers according to the manufacturer's instructions. The real-time PCRs were performed with a Bio-Rad iCycler using iQ SYBR Green Supermix (Bio-Rad) as described for the clock genes above. Relative mRNA levels were calculated by normalization to β2-microglobulin mRNA (see SI Table 1 for primers).
Oil Red O (ORO) Staining and Quantification.
Lipid accumulation was assessed by ORO staining of 10-μm frozen sections of livers fixed in phosphate-buffered 4% paraformaldehyde. ORO staining was quantified by using digital images taken adjacent to either portal or central veins. Color images were acquired by using a CoolSnap Color camera, saved as TIFF files, and analyzed by using Image J software. An overlay consisting of a 100-μm2 grid was generated over each image, and the area fraction, defined as (points over ORO)/(points over image) × 100, was determined for multiple images from five to seven different animals for each experimental group. Values are presented as area fraction percentage.
Feeding and Weight Collection.
Mice were maintained on a 12-h LD cycle on a standard diet containing 8% kcal from fat (8604 rodent chow; Harlan Teklad) or a high-fat diet containing 45% kcal from fat (D12451; Research Diets, New Brunswick, NJ). Weights were determined gravimetrically.
Metabolic Cage Measurements.
Mice were maintained on a 14 h:10-h LD cycle in a pathogen-free animal facility. Food intake, ambulatory activity, oxygen consumption, and CO2 production were simultaneously determined for four mice per experiment in an Oxymax metabolic chamber system (Columbus Instruments, Columbus, OH). Adult male mice (8–14 weeks old when on a standard diet and 14–19 weeks old when on a high-fat diet) were placed in a chamber, and every 15 min one reading per mouse was taken over 72 h. The mice were allowed to adjust to the cages during the first 24 h, and thus only the last 48 h of each experimental run was used for data analysis (n = 4 for both genotypes). Resting metabolic rate was determined by measuring VO2 for each animal during a 150-min period of inactivity.
Measurement of Body Temperature.
Temperature transponders (G2 E-mitter; Mini Mitter, Bend, OR) were implanted i.p. in six WT and six Noc−/− mice. Mice were individually housed in 12-h LD cycles, and data were collected every 10 min by using ER-4000 Energizer/Receivers and VitalView software.
Glucose and Insulin Tolerance Tests and Serum Insulin Concentrations.
For the glucose tolerance tests male mice were fasted for 16 h before an i.p. injection of d-glucose (1.5 or 0.75 g/kg of body weight; Sigma, St. Louis, MO) at 4 h after light onset (ZT4). Blood glucose was measured from tail blood by using a OneTouch Ultra glucometer (Lifescan, Milpitas, CA). For the insulin tolerance test male mice were fed ad libitum and injected i.p. at ZT8 with mammalian crystalline insulin (0.75 or 1.5 units/kg of body weight; Lilly, Indianapolis, IN). Serum insulin was determined by using an insulin RIA (Linco, St. Charles, MO).
Measurement of Blood Lipids.
Blood lipid assays were performed on serum isolated from whole blood from animals on various feeding regimens as described in the figure legends. Cholesterol and triglycerides were measured by using a VetTest Chemistry Analyzer (IDEXX, Westbrook, ME). Free fatty acid levels were determined by an enzymatic colorimetric endpoint test (NEFA C test; WAKO Chemicals, Cape Charles, VA).
Supplementary Material
Acknowledgments
We thank Yunxia Wang and Minhong Zhuang for help in generating the Noc−/− mice and the members of the C.B.G. and J.C.B. laboratories for many useful discussions of our data. We are grateful to the Diabetes and Endocrinology Research Center at the University of Virginia for help in performing the metabolic cage studies and for insulin and free fatty acid measurements (supported by National Institutes of Health Grant DK063609). This work was supported by National Institutes of Health Grants EY11489 (to C.B.G.), GM076626 (to C.B.G.), and EY02414 (to J.C.B.); Cell and Molecular Biology Predoctoral Training Grant 2T32 GM008136-21 from the National Institutes of Health (to N.D.); and Medical College of Wisconsin Research Program Development Funds (to J.C.B.).
Abbreviations
- LD
light:dark
- ORO
Oil Red O
- ZTn
n hours after light onset.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 9553.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702448104/DC1.
References
- 1.Lowrey PL, Takahashi JS. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Hastings MH, Reddy AB, Maywood ES. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 4.Fu L, Lee CC. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 5.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oishi K, Atsumi G, Sugiyama S, Kodomari I, Kasamatsu M, Machida K, Ishida N. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 8.Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Proc Natl Acad Sci USA. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kita Y, Shiozawa M, Jin W, Majewski RR, Besharse JC, Greene AS, Jacob HJ. Pharmacogenetics. 2002;12:55–65. doi: 10.1097/00008571-200201000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 11.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 12.Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, Ohkura N, Azama T, Mesaki M, Yukimasa S, et al. J Biol Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 13.Brewer M, Lange D, Baler R, Anzulovich A. J Biol Rhythms. 2005;20:195–205. doi: 10.1177/0748730405275952. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 15.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, et al. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 16.Lidder P, Gutierrez RA, Salome PA, McClung CR, Green PJ. Plant Physiol. 2005;138:2374–2385. doi: 10.1104/pp.105.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.So WV, Rosbash M. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson BG, Frim DM, Schwartz WJ, Majzoub JA. Science. 1988;241:342–344. doi: 10.1126/science.3388044. [DOI] [PubMed] [Google Scholar]
- 19.Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L. Plant J. 2003;33:361–371. doi: 10.1046/j.1365-313x.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- 20.Baggs JE, Green CB. Curr Biol. 2003;13:189–198. doi: 10.1016/s0960-9822(03)00014-9. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, Green CB, Besharse JC. BMC Dev Biol. 2001;1:9. doi: 10.1186/1471-213X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storch K, Lipan O, Leykin I, Viswanathan N, Davis F, Wong W, Weitz C. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 23.Sampath H, Ntambi JM. Curr Opin Clin Nutr Metab Care. 2006;9:84–88. doi: 10.1097/01.mco.0000214564.59815.af. [DOI] [PubMed] [Google Scholar]
- 24.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Biochem J. 2005;391:549–560. doi: 10.1042/BJ20050296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Am J Physiol. 2006;290:G36–G48. doi: 10.1152/ajpgi.00510.2004. [DOI] [PubMed] [Google Scholar]
- 27.Reitman ML. Annu Rev Nutr. 2002;22:459–482. doi: 10.1146/annurev.nutr.22.010402.102849. [DOI] [PubMed] [Google Scholar]
- 28.Newberry EP, Xie Y, Kennedy SM, Luo J, Davidson NO. Hepatology. 2006;44:1191–1205. doi: 10.1002/hep.21369. [DOI] [PubMed] [Google Scholar]
- 29.Horton JD, Goldstein JL, Brown MS. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uno K, Katagiri H, Yamada T, Ishigaki Y, Ogihara T, Imai J, Hasegawa Y, Gao J, Kaneko K, Iwasaki H, et al. Science. 2006;312:1656–1659. doi: 10.1126/science.1126010. [DOI] [PubMed] [Google Scholar]
- 31.Garbarino-Pico E, Niu S, Rollag MD, Strayer CA, Besharse JC, Green CB. RNA. 2007;13:745–755. doi: 10.1261/rna.286507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.