Abstract
Biomphalaria glabrata is the major intermediate snail host for Schistosoma mansoni, one of the important schistosomes infecting man. Much remains to be discovered concerning specific molecules mediating the defence events in these intermediate hosts, triggered by invading schistosomes. An expressed sequence tag (EST) gene discovery strategy known as ORESTES has been employed to identify transcripts that might be involved in snail–schistosome interactions in order to examine gene expression patterns in infected B. glabrata. Over 3930 ESTs were sequenced from cDNA libraries made from both schistosome-exposed and unexposed snails using different tissue types, producing a database of 1843 non-redundant clones. The non-redundant set has been assessed for gene ontology and KEGG pathway assignments. This approach has revealed a number of signalling, antioxidant and immune-related gene homologues that, based on current understanding of molluscan and other comparative systems, might play an important role in the molluscan defence response towards infection.
Keywords: Expressed sequence tag, EST, ORESTES, Molluscan defence, Biomphalaria glabrata, Schistosoma mansoni
1. Introduction
The freshwater snail Biomphalaria glabrata is an intermediate host for Schistosoma mansoni, the digenean parasite that causes human intestinal schistosomiasis. This host–parasite relationship has become a model system for examination of snail–schistosome interactions, and as such, recent molecular work has focused on B. glabrata. Now with the continued significance of genome research, the B. glabrata genome initiative (http://biology.unm.edu/biomphalaria-genome/) aims to increase the available genetic data for this snail species, with the final goal of a complete genome sequence. Such sequence data will complement that available for the schistosome parasite from the schistosome genome/transcriptome sequencing initiatives [1–5] and for the definitive host from the human genome project [6]. In addition to genome sequencing, the generation of expressed sequence tags (ESTs), short stretches of sequence obtained from cDNA libraries [7], is valuable in a number of ways: in identifying snail homologues of genes previously described in other species; for identifying transcribed regions of the genome, useful for genome annotation and analysis; for the detection of splice variants and alternative polyadenylation gene isoforms; in the discovery of single nucleotide polymorphisms (SNPs) and finally for expression studies, such as those involving microarrays. The EST project described here was initiated with the ultimate aim of manufacturing a cDNA microarray for B. glabrata, which required a large number of sequenced cDNA clones to be available.
EST projects in other molluscs, such as oysters, have revealed a wealth of useful sequence data including signalling, antioxidant and immune-related gene homologues [8,9], demonstrating that molluscs express many of the same genes, and may therefore carry out the same processes, which have previously been described in vertebrates. A recent EST project from Lymnaea stagnalis [10] identified a number of genes that had not previously been identified in the Lophotrochozoa. Therefore initiating an EST sequencing project in B. glabrata has the potential to identify other novel molluscan genes including those that might be associated with the snail's response to infection. At the start of this project (January 2003) only 1427 B. glabrata EST sequences were available on GenBank from earlier studies [11–15]. During the course of this project several other laboratories have also developed gene discovery programmes for B. glabrata [16,17] (see also http://biology.unm.edu/biomphalaria-genome/detailing unpublished EST programmes).
Previous EST projects in B. glabrata [11,15] used traditional library construction and sequencing approaches to obtain sequence data. A complimentary EST approach called open reading frame ESTs (ORESTES) [18] has been used successfully to obtain large numbers of sequences for both human [18–20] and schistosome [4,5] transcriptome projects. The ORESTES approach preferentially targets the middle section of mRNAs [18], making it more likely coding regions will be sequenced, than in other EST methodologies where sequencing commences at the end of the cDNA, often only obtaining untranslated sequence. This alternative method has two advantages for snail ESTs; firstly, it is more likely that gene similarity to other organisms can be ascertained if coding regions are sequenced, and secondly, the data generated are likely to be complementary to, rather than redundant with, sequence data from traditional approaches. The ORESTES approach also allows the construction of a number of mini-libraries using small quantities of RNA [21], making it suitable for investigating gene expression in small amounts of tissue such as those present in B. glabrata. Producing a large number of smaller libraries also facilitates a more extensive analysis of gene expression; thus in the EST project described here, different snail strains (both resistant and susceptible to S. mansoni infection) were used and different tissue types from both parasite-exposed and unexposed material were examined. Based on gene ontology and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway assignments a number of antioxidant, signalling and immune-related gene homologues have been identified and are presented here; the potential involvement of these genes in molluscan defence is considered, particularly within the framework of comparative immunobiology.
2. Materials and methods
2.1. Snail material
B. glabrata strains used were: resistant BS90 [22] (NHM3017) or susceptible NHM1742 or BB02 (NHM3032), the strain currently being used for the genome sequencing project (see http://biology.unm.edu/biomphalaria-genome/BB02STRAIN.html). Snails were held overnight in autoclaved snail water with 100 μg/ml ampicillin prior to killing by decapitation. The exuded haemolymph was collected, pooled and the haemocytes isolated by centrifugation at 4 °C, 10,000g for 20 min. Each snail was preserved in 800 μl RNAlater (Ambion Inc., Texas, USA) and stored at −20 °C until dissection. Haemopoietic organ, ovotestis, head/foot and brain tissue was dissected. For the exposed material, 60 snails were individually exposed to 10 S. mansoni miracidia (Belo Horizonte strain) each and 2, 4, 6, 8 and 24 h after infection, 12 of the snails were swiftly killed as above. Tissue was pooled from each time period; the extended sampling was designed to include all transcripts expressed over the first 24 h of infection.
2.2. RNA extraction
Total RNA was extracted from each dissected tissue using SV RNA extraction kit (Promega UK Ltd, Southampton, UK) according to the manufacturer's protocol. This kit includes DNAse treatment to eliminate genomic DNA contamination. Pigment from the head/foot tissue was found to block the spin columns supplied with this extraction kit, so RNA was extracted from this tissue using Trizol (Invitrogen Ltd, Paisley, UK). Briefly, 30 mg tissue was ground in 1 ml Trizol and centrifuged at 12,000g for 10 min at 4 °C. The supernatant was incubated at room temperature for 5 min then 0.2 ml chloroform added, mixed vigorously and left at room temperature for 3 min. The samples were spun at 12,000g, 4 °C for 15 min and the RNA precipitated from the supernatant using 0.5 ml propan-2-ol and centrifugation at 12,000g for 10 min at 4 °C. The pellet was washed using 75% ethanol and dissolved in 50–100 μl water. RNA extracted using Trizol was DNAse treated (Promega), according to the manufacturer's instructions prior to mRNA extraction. mRNA was extracted from the total RNA from both extraction methods using the Micro-fastTrack 2.0 mRNA extraction kit (Invitrogen) according to the manufacturer's instructions. The mRNA was eluted in 200 μl elution buffer and precipitated overnight at −70 °C using 600 μl ethanol. The mRNA was dissolved in 10 μl water and tested using specific B. glabrata actin primers [12] to check there was no DNA contamination.
2.3. cDNA synthesis and amplification
For each library, a 27 μl mastermix containing 800 U Reverse Transcriptase (MMLV-RT) (Promega), 4 μl RNAsin (Promega), 4 μl dNTPs at 2 mM, 8 μl 5× buffer (Promega) and 7 μl mRNA (70–240 ng) was prepared and 2 μl aliquoted into 12 tubes prepared with 12 different arbitrary primers (1.5 μl of 15 mM) (for primer sequences see supplementary material). The tubes were incubated at 42 °C for 1 h then heated to 70 °C for 10 min.
Amplification was carried out using Ready-to-go beads (Amersham Biosience, Amersham, UK). The 3.5 μl cDNA reactions (including primers) and 25 μl water were each added to a tube containing a single bead and amplified using the following cycling conditions: 75 °C for 5 min followed by 15 cycles at 94 °C, 52–45 °C for 1 min (touchdown PCR, dropping 0.5 °C each cycle) and 1 min 72 °C, then 26 cycles of 94 °C for 30 s, 48 °C for 1 min and 72 °C for 1 min, then 7 min at 72 °C. A negative control (no DNA) was carried out simultaneously for each primer (dissolving 2 ready-to-go beads in 50 μl water and aliquoting 3 μl into a tube containing 0.3 μl primer (at 15 mM)). Three μl of each synthesis reaction was examined by gel electrophoresis alongside the control amplification and reactions chosen for inclusion in the mini-library only if the control amplification showed no contamination and a smear without single prominent bands in the reaction profile. This ensured that a mix of products would be obtained from each library.
2.4. Cloning and sequencing
For each library, the selected amplified cDNA samples were pooled and cloned using pGEM-T easy cloning kit (Promega) according to the manufacturer's instructions. One hundred and ninety two clones (2×96), selected at random from the cloning plates were picked into 0.5 ml LB and grown up overnight. Ten μl PCRs with M13 forward and reverse primers were carried out to check insert size and the presence of a single insert and, from these, 96 colonies were chosen for 100 μl PCRs. PCRs contained 1×NH4 reaction buffer (Bioline, London, UK), 2.5 mM MgCl2, 0.2 mM dNTP, 0.2 μM each M13 Forward and Reverse primers and 0.025 U/μl PCR Taq polymerase (Bioline, London, UK). Cycling conditions were: 94 °C for 2 min, then 35 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 1 min 30 s, then 10 min at 72 °C. Glycerol stocks for the selected colonies were stored at −80 °C.
PCR products were purified using Multiscreen PCR filter plates (Millipore, Billerica, USA) then cycle-sequenced directly using BigDye kit (Applied Biosystems, Foster City, USA) and T7 primer and run on ABI 377 or capillary sequencers. Vector, primer and poor quality sequences were removed using Sequencher 3.1.1 (GeneCodes Corp., Ann Arbor, USA).
2.5. Bioinformatics
Cluster analysis was performed in SeqTools (http://www.seqtools.dk/) using BlastN score values (cut-off value 0.5) and used to calculate percentage redundancy. For each library BlastN and BlastX [23] searches were run and any ribosomal sequences and sequences shorter then 80 bp removed. Duplicate sequences were also removed, although overlapping sequences were retained. Since each library represented different tissues, strains or infected/uninfected material, duplicate sequences between libraries were retained.
2.6. Clone nomenclature
Each clone had a unique ZB number assigned during sequencing. In addition to this the clones were also assigned a code based on strain (see Section 2.1), tissue type (B—brain, HO—haemopoietic organ, HAEM—haemocytes, HF—head/foot and OT—ovotestis), infection status (EX—parasite exposed, UN—unexposed) and plate number and position. The sequences were submitted to GenBank (accession numbers CK149151-CK149590, CK656591-CK656938, CO870183-CO870449, CV548035-CV548805, EG030731-EG030747).
2.7. Gene function
Gene ontology functions were assigned using GOblet (http://goblet.molgen.mpg.de/). KEGG pathway analysis was carried out using the KEGG automatic annotation server (KAAS) for ortholog assignment and pathway mapping (http://www.genome.jp/kegg/kaas/).
3. Results
3.1. ORESTES libraries
A total of 41 ORESTES libraries were made from five tissue types (head–foot, brain, ovotestis, haemocytes and haemopoietic organ) from the three B. glabrata strains, one resistant (NHM3017) and two susceptible (NHM1742 and BB02 (NHM3032)), using material that had either remained unexposed or had been exposed to S. mansoni (Table 1). Some of the libraries were made from the same snail strain and tissue type but were separately made with different primers, using new experimental material, so have been treated independently. Libraries were prepared from each of the two susceptible strains for each tissue, with the exception of exposed ovotestis from NHM1742 where two libraries were made. Two or three libraries were prepared per tissue and exposure type for the resistant strain, with the exception of brain tissue where material was limiting. A single plate of 96 clones was sequenced for each library and in total, 3936 clones were sequenced from 41 libraries.
Table 1.
Biomphalaria glabrata ORESTES libraries. The number of non-redundant (NR) sequences was determined after cluster and Blast analyses to remove duplicate and ribosomal sequences from within each library
| Straina | Tissueb | Parasite exposurec | No. sequences obtained | % redundancy | No. NR sequences | % Unique (and non-ribo) | Library name |
|---|---|---|---|---|---|---|---|
| 1742 | HF | EX | 93 | 36.6 | 55 | 59.1 | BgORESTES infected NHM 1742 Head/foot |
| 1742 | HF | UN | 88 | 52.3 | 35 | 39.8 | BgORESTES uninfected NHM 1742 Head/foot |
| 1742 | OT | UN | 89 | 46.1 | 36 | 40.4 | BgORESTES uninfected NHM 1742 Ovotestis |
| 3017 | OT | EX | 94 | 50 | 45 | 47.9 | BgORESTES schistosome-exposed NHM 3017 Ovotestis |
| 1742 | OT | EX | 90 | 47.8 | 51 | 56.7 | BgORESTES infected NHM 1742 Ovotestis 1 |
| 1742 | OT | EX | 91 | 58.2 | 37 | 40.7 | BgORESTES infected NHM 1742 Ovotestis 2 |
| 1742 | HAEM | EX | 91 | 53.8 | 24 | 25.3 | BgORESTES infected NHM 1742 Haemocytes |
| 1742 | HO | UN | 92 | 51.1 | 42 | 45.7 | BgORESTES uninfected NHM 1742 Haemopoietic organ |
| 1742 | HO | EX | 96 | 56.3 | 36 | 37.5 | BgORESTES infected NHM 1742 Haemopoietic organ |
| 1742 | HAEM | UN | 92 | 58.7 | 24 | 26.1 | BgORESTES uninfected NHM 1742 Haemocytes |
| 3017 | HF | EX | 94 | 30.9 | 58 | 61.7 | BgORESTES schistosome-exposed NHM 3017 Head/foot |
| 3017 | OT | UN | 94 | 40.4 | 53 | 56.4 | BgORESTES unexposed NHM 3017 Ovotestis |
| 3017 | OT | EX | 93 | 40.9 | 52 | 55.9 | BgORESTES schistosome-exposed NHM 3017 Ovotestis2 |
| 3017 | HAEM | UN | 94 | 47.7 | 35 | 37.2 | BgORESTES unexposed NHM 3017 Haemocytes |
| 3017 | HAEM | EX | 87 | 57.5 | 20 | 23.0 | BgORESTES schistosome-exposed NHM 3017 Haemocytes |
| 3017 | HO | UN | 91 | 28.6 | 56 | 61.5 | BgORESTES unexposed NHM 3017 Haemopoietic organ |
| 3017 | HO | EX | 81 | 30.9 | 47 | 58.0 | BgORESTES schistosome-exposed NHM 3017 Haemopoietic organ |
| 3017 | HF | UN | 95 | 34.7 | 52 | 54.7 | BgORESTES unexposed NHM 3017 Head/foot |
| 3017 | HF | EX | 96 | 52.1 | 34 | 35.4 | BgORESTES schistosome-exposed NHM 3017 Head/foot2 |
| 3017 | HAEM | UN | 93 | 29 | 42 | 45.2 | BgORESTES unexposed NHM 3017 Haemocytes2 |
| 3017 | HAEM | EX | 90 | 70 | 9 | 10.0 | BgORESTES schistosome-exposed NHM 3017 Haemocytes2 |
| 3017 | HO | UN | 93 | 62.4 | 16 | 17.2 | BgORESTES unexposed NHM 3017 Haemopoietic organ2 |
| 3017 | HO | EX | 96 | 71.9 | 20 | 20.8 | BgORESTES schistosome-exposed NHM 3017 Haemopoietic organ2 |
| 3017 | OT | UN | 96 | 35.4 | 57 | 59.4 | BgORESTES unexposed NHM 3017 Ovotestis2 |
| 3017 | OT | EX | 94 | 45.7 | 47 | 50.0 | BgORESTES schistosome-exposed NHM 3017 Ovotestis3 |
| 3017 | HF | UN | 94 | 43.6 | 40 | 42.6 | BgORESTES unexposed NHM 3017 Head/foot2 |
| 3017 | HF | EX | 93 | 50.5 | 39 | 41.9 | BgORESTES schistosome-exposed NHM 3017 Head/foot3 |
| 1742 | B | UN | 95 | 20 | 66 | 69.5 | BgORESTES uninfected NHM 1742 Brain |
| 1742 | B | EX | 94 | 9.6 | 82 | 87.2 | BgORESTES infected NHM 1742 Brain |
| 3017 | B | UN | 91 | 71.4 | 18 | 19.8 | BgORESTES unexposed NHM 3017 Brain |
| 3017 | B | EX | 94 | 57.4 | 32 | 34.0 | BgORESTES schistosome-unexposed NHM 3017 Brain |
| 3032 | HF | EX | 95 | 51.6 | 43 | 45.3 | BgORESTES infected NHM 3032 Head/foot |
| 3032 | OT | EX | 94 | 46.8 | 47 | 50.0 | BgORESTES infected NHM 3032 Ovotestis |
| 3032 | B | EX | 94 | 42.6 | 49 | 52.1 | BgORESTES infected NHM 3032 Brain |
| 3032 | HO | EX | 96 | 36.5 | 56 | 58.3 | BgORESTES infected NHM 3032 Haemopoietic organ |
| 3032 | HAEM | UN | 95 | 24.2 | 64 | 67.4 | BgORESTES uninfected NHM 3032 Haemocytes |
| 3032 | HAEM | EX | 96 | 33.3 | 47 | 49.0 | BgORESTES infected NHM 3032 Haemocytes |
| 3032 | HF | UN | 93 | 36.6 | 59 | 63.4 | BgORESTES uninfected NHM 3032 Head/foot |
| 3032 | OT | UN | 94 | 19.1 | 74 | 78.7 | BgORESTES uninfected NHM 3032 Ovotestis |
| 3032 | B | UN | 95 | 23.2 | 73 | 76.8 | BgORESTES uninfected NHM 3032 Brain |
| 3032 | HO | UN | 93 | 16.1 | 71 | 76.3 | BgORESTES uninfected NHM 3032 Haemopoietic organ |
| Total | 3809 | 1843 | |||||
| Mean | 43.2 | 44.9 | 48.2 |
NHM Strain: 3017—resistant snails, 1742, 3032 (BB02)—susceptible snails.
Tissue type: HF—head/foot, OT—ovotestis, HO—haemopoietic organ, HAEM—haemocytes, B—brain.
Snails exposed (EX) or unexposed (UN) to S. mansoni miracidia.
3.2. Analysis of total ESTs and selection of non-redundant ESTs
A total of 3809 sequences were obtained (127 reactions did not work or the sequenced clones contained no insert or had mixed sequences so these were not analysed further) and were compared to the non-redundant section of GenBank. The Blast results (Table 2) showed that 28.5% of the gene fragments identified proteins on the database, including 35 previously characterized B. glabrata proteins and 127 proteins with no assigned function. Including some other non-coding gene fragment matches, less than 2% () of the sequences matched characterized B. glabrata genes or proteins in the non-redundant section of GenBank. Nearly 35% could not be assigned any function, either having no Blast matches, or having homology to a nucleotide or protein sequence on GenBank with no function described. Unfortunately, 39% of the sequences matched B. glabrata ribosomal sequences. Other workers [16] have also found a large ribosomal content in polyA selected RNA from B. glabrata, and concluded that the high A content in B. glabrata ribosomal sequences meant that polyA selection did not efficiently remove it. In the present study it was found that some 18mer primers chosen for ORESTES library construction tended to target ribosomal regions, so they were not used again. However, it was impossible to predict in advance which primers would be problematic. The ‘other’ sequences had database matches but do not necessarily code for proteins, for example retrotransposon sequences. For each library, duplicate and ribosomal sequences were removed and the % redundancy per plate ranged from 9.6% to 71.9% (Table 1).
Table 2.
Blast results summary. Breakdown of the types of sequences obtained from the B. glabrata ORESTES libraries identified with Blast searches of GenBank
| All sequences |
Non-redundant sequences |
|||
|---|---|---|---|---|
| Category | No. sequences | % sequences | No. sequences | % sequences |
| Protein | 907 | 23.8 | 739 | 40.1 |
| Mitochondrial Protein | 4 | 0.1 | 4 | 0.2 |
| Ribosomal Protein | 14 | 0.4 | 11 | 0.6 |
| Biomphalaria Protein | 35 | 0.9 | 22 | 1.2 |
| Biomphalaria fragment | 41 | 1.1 | 40 | 2.2 |
| Ribosomal | 1482 | 38.9 | 44 | 2.4 |
| Unknown (no BLAST match) | 1106 | 29.0 | 832 | 45.1 |
| Unknown EST | 84 | 2.2 | 56 | 3.0 |
| Unknown Protein | 127 | 3.3 | 88 | 4.8 |
| Other | 8 | 0.2 | 7 | 0.4 |
| Total | 3809 | 100.0 | 1843 | 100.0 |
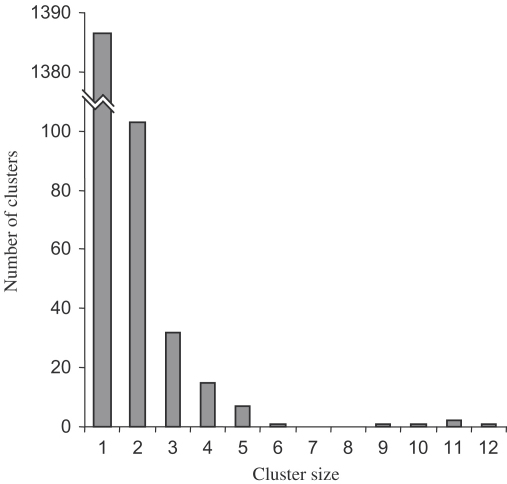
After removal of duplicate and ribosomal sequences, a total of 1843 non-redundant sequences were submitted to GenBank, ranging in size from 80 bp (shorter sequences were discarded) to 1068 bp, with a mean length of 518 bp. Cluster analysis between libraries (since the data were previously sifted to remove duplicate clones from each library) revealed 456 sequences in 163 clusters and 1387 singletons (Fig. 1). This resulted in 1550 unique sequences, with 15.9% redundancy. The most common sequence (in 12 of the libraries) was tropomyosin 2 (SwissProt accession number P43689), previously sequenced from B. glabrata [24], while two other common sequences (in 11 libraries) were a hypothetical integral membrane transporter protein (accession number XP_135742) and a sequence with no Blast match. Examining the Blast results from the 1843 non-redundant sequences (Table 2), 42% showed significant BlastX similarity to known proteins (including mitochondrial, ribosomal and Biomphalaria proteins) in the non-redundant databases, while 52.9% were of unknown function, 3.4% Biomphalaria sequences, 0.4% ‘other’ sequences (e.g. retrotransposons) and the remainder were ribosomal sequences (2.4%).
Fig. 1.
Histogram showing EST clusters in the non-redundant EST set, after removal of duplicates within libraries.
3.3. Cluster analysis with other B. glabrata ESTs
Sequences from the 1843 ORESTES clones were used for BlastN searches of the other 10,791 B. glabrata EST sequences available on dbEST (September 2006) including many added since the sequences presented here. Four hundred and thirty-nine of the ESTs identified B. glabrata sequences with a match greater than 1e-20. Cluster analysis of these revealed 293 clusters or unique ESTs matched sequences on dbEST. Closer examination of a subset of 1613 sequences from a B. glabrata haemocyte cDNA library [17], created in the conventional way (not from ORESTES mini-libraries) showed only 31 ORESTES sequences clustered with transcripts sequenced from that library.
3.4. Functional classification based on gene ontology assignments
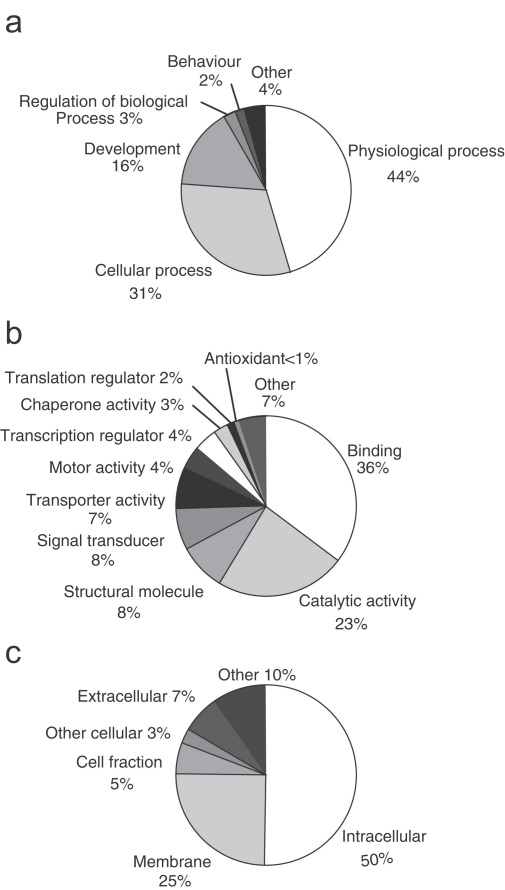
The functions of the non-redundant 1843 sequences were assessed using gene ontology, based on Blast matches with genes whose functions have been previously assessed (Fig. 2). Of the 1843 ESTs, 587 were assigned a function, in three categories, biological process, molecular function and cellular component. In the biological process categories, the largest proportion (44%) was assigned to physiological processes (Fig. 2a). The most prevalent molecular functions (Fig. 2b) were binding (36%) and catalytic activity (23%). Other molecular function assignments were signal transducers (8%) and transcription regulators (4%), and five antioxidant genes (0.4%) were also identified. Over 80% of the cellular component assignments were for genes coding for cellular proteins including 50% intracellular and 25% membrane proteins (Fig. 2c). Gene ontologies were examined to identify genes that were homologous to antioxidant molecules, signalling molecules, transcriptional regulators, immune response genes and stress response genes (Table 3), since many of these might be significant in the snail's response to parasite infection. A total of 117 homologues of genes that code for proteins involved in cell signalling or transcriptional regulation were identified; these genes were categorised as follows: signal transducers (54), cell–cell signalling (19), transcription regulator activity (28), signal transducers and transcription factor regulators (10) (Table 3). Although not the original purpose of generating these ESTs, gene ontologies were also assessed by tissue type, strain, parasite susceptibility and whether parasite exposed or unexposed, for both biological process and molecular function (see supplementary material).
Fig. 2.
Gene ontologies. Percentage representation of gene ontology (GO) mappings for B. glabrata ESTs. (a) Biological processes, (b) molecular function and (c) cellular component. Note that individual GO categories can have multiple mappings and that the charts do not include ESTs with no blast/gene ontology matches.
Table 3.
Transcripts selected by gene ontology. Individual B. glabrata ESTs that identified antioxidant proteins, signal transducers, transcription regulators and immune or stress response proteins
| Name | GenBank accession | Gene ID (Blast result)a | Organisma | Blast scorea |
|---|---|---|---|---|
| Antioxidant (GO:0016209) | ||||
| 3032HOUN59B8 | CV548777 | Peroxidasin (mKIAA0230) [BAC65505] | M. musculus | 2E-23 |
| 1742HFEX1H9 | CK149203 | Dual oxidase 1 [Q8HZK3] | S. scrofa | 2E-31 |
| 3017OTEX4H6 | CK149399 | Putative iron dependent peroxidase [Q8XGB1] | S. typhi | 5E-15 |
| 1742OTEX5B12 | CK149417 | Putative iron dependent peroxidase [Q8XGB1] | S. typhi | 7E-11 |
| 3032HAEMEX55E1 | CV548486 | Peroxinectin [AAL05973] | P. monodon | 6E-17 |
| Signal transducers (GO: 0007165) | ||||
| 3032BEX52G8 | CV548350 | Filamin 1 [P21333] | H. sapiens | 5E-53 |
| 1742BUN46B6 | CV548064 | Adenylyl cyclase [Q9QW33] | Rattus sp. | 4E-78 |
| 3017OTUN34C11 | CO870317 | Ankyrin 2 (Brain ankyrin) [Q01484] | H. sapiens | 2E-11 |
| 3017HFEX11B10 | CK149239 | Inhibitor of apoptosis protein [Q8UWD2] | D. rerio | 4E-17 |
| 3032OTUN57G6 | CV548625 | Inhibitor of apoptosis protein [Q8UWD2] | D. rerio | 4E-11 |
| 3017BEX49B1 | CV548199 | Buccalin precursor [P20481] | A. californica | 5E-67 |
| 3032BEX52A2 | CV548323 | Buccalin precursor [P20481] | A. californica | 2E-54 |
| 3032BEX52A12 | CV548364 | Buccalin precursor [P20481] | A. californica | 2E-19 |
| 3032BUN58E5 | CV548692 | Buccalin precursor [P20481] | A. californica | 5E-46 |
| 3032BUN58B9 | CV548713 | Buccalin precursor [P20481] | A. californica | 2E-19 |
| 3032HAEMEX55A11 | CV548523 | cAMP-specific 3’,5’-cyclic phosphodiesterase [P12252] | D. melanogaster | 2E-31 |
| 3032OTUN57A5 | CV548614 | Serine/threonine-protein kinase TNNI3 K (ANK repeats) [Q7TQP6] | R. norvegicus | 1E-18 |
| 3032OTUN57B1 | CV548590 | Regulator of G-protein signalling 22 [Q9BYZ4] | H. sapiens | 9E-22 |
| 3032HOUN59G3 | CV548755 | Bent (GH07636p) [Q9V4F7] | D. melanogaster | 2E-19 |
| 3032HOUN59B1 | CV548737 | Bent (GH07636p) [Q9V4F7] | D. melanogaster | 3E-76 |
| 3032HOEX53H6 | CV548394 | Elongation factor 1 alpha [P13549] | X. laevis | 1E-48 |
| 3017BUN48A11 | CV548195 | FMRFamide neuropeptides [P19802] | L. stagnalis | 4E-45 |
| 3032BUN58F5 | CV548693 | FMRFamide neuropeptides [P19802] | L. stagnalis | 1E-34 |
| 3017OTEX17H11 | CK656694 | Myotilin (Titin immunoglobulin domain protein) [Q9UBF9] | H. sapiens | 8E-13 |
| 3017HAEMUN28C9 | CO870195 | Protein kinase C inhibitor 1 (14-3-3-like) [P35214] | H. sapiens | 3E-52 |
| 3032HFEX50F2 | CV548237 | Hemolectin [Q9U5D0] | D. melanogaster | 2E-20 |
| 3032HFUN56E2 | CV548539 | Hemolectin [Q9U5D0] | D. melanogaster | 2E-20 |
| 3032HFUN56B8 | CV548566 | Hemolectin [Q9U5D0] | D. melanogaster | 4E-28 |
| 3017BEX49E3 | CV548205 | Molluscan insulin-related peptide 3 [P80090] | L. stagnalis | 6E-13 |
| 3032HFUN56E6 | CV548560 | Multiple EGF-like-domain protein 3 [O75095] | H. sapiens | 1E-30 |
| 3032HFUN56F9 | CV548575 | Multiple EGF-like-domain protein 3 [O75095] | H. sapiens | 8E-24 |
| 3032HOUN59C2 | CV548745 | Nidogen 1 [P10493] | M. musculus | 2E-23 |
| 3017HAEMEX19E1 | CK656743 | Feline leukemia virus subgroup C receptor [Q9N1F2] | F. catus | 2E-28 |
| 3017HOUN20E11 | CK656783 | PERQ amino acid rich, with GYF domain 1 [Q99MR1] | M. musculus | 1E-23 |
| 3032HFEX50E11 | CV548269 | PERQ amino acid rich, with GYF domain 1 [Q99MR1] | M. musculus | 1E-20 |
| 3017HFUN44E6 | CO870392 | Polycystic kidney disease protein 2 [Q7Z2B5] | S. purpuratus | 9E-45 |
| 3032HOUN59B11 | CV548796 | Polydom protein precusor [Q9ES77] | M. musculus | 3E-12 |
| 3017HAEMUN28F10 | CO870213 | Polyserase 1B protein [Q7Z410] | H. sapiens | 3E-22 |
| 3017HAEMEX29B4 | CO870226 | Polyserase 1B protein [Q7Z410] | H. sapiens | 1E-22 |
| 3032BEX52E7 | CV548346 | Serine/threonine protein phosphatase 2A [P11493] | S. scrofa | 4E-63 |
| 3032OTUN57F6 | CV548624 | pRb-interacting protein RbBP-36 [Q8IZZ0] | H. sapiens | 1E-20 |
| 3032BEX52H9 | CV548355 | Tyrosine phosphatase IA-2beta [Q9Y4I9] | H. sapiens | 6E-57 |
| 3032BUN58D5 | CV548691 | Tyrosine phosphatase IA-2beta [Q9Y4I9] | H. sapiens | 2E-46 |
| 3032OTUN57B12 | CV548658 | Muscle M-line assembly protein UNC-89 [O01761] | C. elegans | 2E-12 |
| 3032HOUN59F4 | CV548760 | Muscle M-line assembly protein UNC-89 [O01761] | C. elegans | 8E-25 |
| 3032HOEX53B12 | CV548417 | Muscle M-line assembly protein UNC-89 [O01761] | C. elegans | 2E-20 |
| 1742HOUN8B5 | CK149518 | RAS related protein Rab21 [Q9UL25] | H. sapiens | 3E-29 |
| 3017HFEX11H12 | CK149287 | RAS related protein Rab21 [Q9UL25] | H. sapiens | 1E-29 |
| 1742HFEX1G2 | CK149192 | GTP-binding nuclear protein Ran [P79735] | D. rerio | 2E-80 |
| 3017OTUN34B9 | CO870310 | Serine/threonine protein kinase SSTK [Q9BXA6] | H. sapiens | 3E-18 |
| 3032BUN58F11 | CV548727 | E3 ubiquitin-protein ligase HECTD1 [Q9ULT8] | H. sapiens | 6E-66 |
| 3032HOEX53G8 | CV548401 | SNF4/AMP-activated protein kinase gamma subunit [O96613] | D. melanogaster | 2E-29 |
| 3017OTEX17B2 | CK656650 | Transportin-SR [Q9Y540] | H. sapiens | 9E-58 |
| 1742HFEX1H2 | CK149197 | Twitchin [Q7YT99] | M. galloprovincialis | 5E-28 |
| 3017BUN48G6 | CV548191 | Type N4 regulatory subunit of protein kinase A [P31319] | A. californica | 1E-50 |
| 3032OTUN57C10 | CV548648 | Testis-enriched protein tyrosine phosphatase [Q9WU22] | M. musculus | 2E-66 |
| 1742HAEMEX7G1 | CK149506 | Integrin alpha 3 [Q86G86] | P. includens | 8E-23 |
| 3032OTUN57G7 | CV548631 | Receptor type protein-tyrosine phosphatase T precursor [O14522] | H. sapiens | 3E-16 |
| 3017HAEMEX19A3 | CK656730 | G protein-coupled receptor kinase type 2 [Q9U756] | H. americanus | 3E-62 |
| 3017HFEX45B5 | CO870422 | Megalin [P98164] | H. sapiens | 6E-20 |
| 1742OTEX5D3 | CK149425 | Activated protein kinase c receptor [Q9W7I1] | X. laevis | 1E-114 |
| 1742OTEX5H8 | CK149451 | Activated protein kinase c receptor [Q9W7I1] | X. laevis | 8E-24 |
| 3032HOUN59A5 | CV548763 | Receptor type guanylyl cyclase [Q9BPR0] | B. mori | 1E-22 |
| 3032BUN58B3 | CV548678 | Soluble guanylyl cyclase alpha [Q7YW37] | L. marginatus | 4E-63 |
| 1742HOEX9C10 | CK149566 | Collagen alpha 1(XIV) chain precusor (Undulin) [P32018] | G. gallus | 7E-15 |
| 1742BEX47A12 | CV548175 | JNK interacting protein 1 [Q9W0K0] | D. melanogaster | 4E-53 |
| 3032BUN58B11 | CV548723 | JNK interacting protein 1 [Q9W0K0] | D. melanogaster | 5E-31 |
| 1742BEX47E10 | CV548166 | JNK interacting protein 1 [Q9W0K0] | D. melanogaster | 2E-27 |
| 3032BUN58E4 | CV548685 | JNK interacting protein 1 [Q9W0K0] | D. melanogaster | 8E-24 |
| 3032BUN58A3 | CV548677 | Serine/threonine kinase receptor type1 [O73801] | T. rubripes | 1E-57 |
| 3032HFEX50H7 | CV548257 | Plectin [Q15149] | H. sapiens | 8E-15 |
| 3032HOEX53F1 | CV548369 | Smad anchor for receptor activation (SARA) [Q9YHB9] | X. laevis | 1E-56 |
| 3017HFEX11A5 | CK149232 | Src-family protein tyrosine kinase [Q8WQM5] | S. purpuratus | 4E-51 |
| 3032HAEMEX55D11 | CV548524 | TNF receptor-associated factor 1 [Q13077] | H. sapiens | 2E-27 |
| 3017HFEX45B8 | CO870431 | Epidermal growth factor precusor [P01133] | H. sapiens | 7E-22 |
| 3017HFEX45A4 | CO870419 | Fibrillin [P35555] | H. sapiens | 8E-26 |
| 3017HOUN32A5 | CO870241 | Macrophage mannose receptor [Q61830] | M. musculus | 2E-14 |
| 3032HOEX53A1 | CV548367 | Macrophage mannose receptor [Q61830] | M. musculus | 9E-23 |
| 1742HOUN8F1 | CK149540 | CYR61 protein precursor [Q9ES72] | R. norvegicus | 4E-29 |
| 3017HAEMUN18E5 | CK656717 | Focal adhesion kinase [Q7Z1D3] | L. variegatus | 1E-45 |
| Cell-cell signalling (GO: 0007267) | ||||
| 3032HAEMUN54B12 | CV548481 | Nicotinic acetylcholine receptor Dalpha6 [Q8T7S2] | D. melanogaster | 1E-49 |
| 1742HOEX9F4 | CK149578 | Afadin (AF-6 protein) [P55196] | H. sapiens | 2E-18 |
| 3017HAEMUN18H12 | CK656729 | Glutamate Receptor 2 [Q10914] | C. elegans | 1E-15 |
| 3017HOUN20E9 | CK656782 | Bone morphogenetic protein 10 preproprotein [Q9R229] | M. musculus | 3E-30 |
| 3017HOEX21A5 | CK656808 | Bone morphogenetic protein 10 preproprotein [Q9R229] | M. musculus | 3E-30 |
| 3032HFUN56E10 | CV548580 | Clathrin heavy chain [P11442] | H. sapiens | 1E-119 |
| 3032BEX52G5 | CV548337 | GABA Transaminase [P50554] | R. norvegicus | 1E-28 |
| 3032BUN58G12 | CV548734 | Cadherin-related tumour suppressor [Q14517] | H. sapiens | 4E-28 |
| 3017HFEX45B11 | CO870445 | Guanylate kinase associated protein [O14490] | H. sapiens | 4E-22 |
| 1742BUN46F12 | CV548100 | Guanylate kinase associated protein [O14490] | H. sapiens | 4E-21 |
| 3032HOUN59G12 | CV548804 | Synaptojanin 2 [O15056] | H. sapiens | 5E-13 |
| 1742BUN46H8 | CV548080 | Kinesin-like protein KIF1A [Q12756] | H. sapiens | 3E-30 |
| 1742BUN46F1 | CV548040 | Kinesin-like protein KIF1B [Q8R524] | R. norvegicus | 1E-108 |
| 1742HFEX1H4 | CK149199 | Lethal giant larvae homolog 1 [O00188] | H. sapiens | 7E-43 |
| 1742BUN46E9 | CV548085 | Prohormone convertase 2 (LPC2) [Q25409] | L. stagnalis | 1E-128 |
| 3032BUN58A9 | CV548712 | Munc13-2 protein [Q62769] | R. norvegicus | 1E-16 |
| 3017HFEX45B3 | CO870416 | Munc13-2 protein [Q62769] | R. norvegicus | 3E-39 |
| 1742HOEX9G9 | CK149583 | Sodium/potassium-transporting ATPase alpha-1 chain [Q9DGL6] | D. rerio | 6E-81 |
| 3017BEX49D12 | CV548229 | Synaptotagmin 11 [O08835] | R. norvegicus | 9E-50 |
| 3017BUN48C2 | CV548182 | Synaptotagmin 11 [O08835] | R. norvegicus | 3E-33 |
| 3032BUN58D7 | CV548701 | Synaptotagmin 11 [O08835] | R. norvegicus | 4E-34 |
| 3032OTUN57B8 | CV548634 | Tyrosine-protein kinase receptor [Q06807] | B. taurus | 6E-61 |
| 3032BUN58C12 | CV548731 | Putative pyrokinin receptor [Q7RTK4] | A. gambiae | 5E-26 |
| 1742BEX47H10 | CV548168 | Tryptophan hydroxylase [Q9NJQ3] | L. stagnalis | 6E-41 |
| 3032BUN58D3 | CV548680 | Tryptophan hydroxylase [Q9NJQ3] | L. stagnalis | 1E-40 |
| Transcription regulator activity (GO: 0030528) | ||||
| 3017BEX49A4 | CV548206 | Eukaryotic translation intiation factor 3 subunit 10 [Q8I5S6] | P. falciparum | 2E-21 |
| 3017OTEX17B6 | CK656654 | Calreticulin [Q26268] | A. californica | 2E-63 |
| 1742HFEX1G4 | CK149194 | Chromodomain helicase DNA binding protein 5 [Q8TDI0] | H. sapiens | 7E-55 |
| 3017OTEX17C2 | CK656662 | C-terminal binding protein [O46036] | D. melanogaster | 1E-79 |
| 1742HOUN8D7 | CK149529 | Cytochrome P450 monooxygenase [O04892] | N. tabacum | 7E-21 |
| 1742HAEMUN10F7 | CK149220 | Cytochrome P450 monooxygenase [O04892] | N. tabacum | 4E-17 |
| 1742HOUN8F12 | CK149546 | Cytochrome P450 monooxygenase [O04892] | N. tabacum | 3E-15 |
| 3017HFEX45H10 | CO870443 | Elongation factor-2 [P13639] | H. sapiens | 3E-49 |
| 3017OTUN16F11 | CK656633 | Embryonic ectoderm development protein [P97462] | M. musculus | 2E-51 |
| 3032OTUN57E7 | CV548630 | HemK methyltransferase family member [Q9Y5R4] | H. sapiens | 3E-30 |
| 3017OTUN34G10 | CO870315 | High mobility group protein [P40618] | G. gallus | 2E-11 |
| 1742OTEX5D12 | CK149429 | Homeodomain interacting protein kinase 2 [Q9H2×6] | H. sapiens | 1E-57 |
| 3017OTEX17A8 | CK656647 | Fragile-chorion membrane protein [P13709] | D. melanogaster | 6E-27 |
| 1742BUN46A9 | CV548081 | LIM domain protein BX (BEADEX protein). [P91608] | Drosophila sp. | 2E-60 |
| 1742BEX47F11 | CV548173 | LIM domain protein BX (BEADEX protein). [P91608] | Drosophila sp. | 1E-60 |
| 3017HFEX45E11 | CO870447 | Max dimerization protein 1; mad [Q05195] | H. sapiens | 7E-24 |
| 3032OTUN57C3 | CV548602 | Myeloid/lymphoid or mixed-lineage leukemia protein 2 [O14686] | H. sapiens | 4E-59 |
| 3017HAEMUN28B11 | CO870190 | Bifunctional aminoacyl-tRNA synthetase [P28668] | D. melanogaster | 2E-56 |
| 3017OTUN16C3 | EG030744 | Nuclear factor NF-kB1 [P25799] | M. musculus | 1E-17 |
| 3017HFUN44C3 | CO870383 | LReO_3 protein [Q8UUM8] | O. latipes | 4E-16 |
| 3032HOEX53E9 | CV548405 | LReO_3 protein [Q8UUM8] | O. latipes | 1E-18 |
| 3032HFEX50B11 | CV548268 | LReO_3 protein [Q8UUM8] | O. latipes | 2E-18 |
| 1742HOUN8F2 | CK149541 | LReO_3 protein [Q8UUM8] | O. latipes | 2E-31 |
| 3017OTEX35D8 | CO870357 | Transcriptional activator protein Pur-alpha [P42669] | M. musculus | 7E-31 |
| 1742HAEMUN10D9 | CK149214 | Retinoblastoma binding protein 5 [Q15291] | H. sapiens | 1E-61 |
| 3032HFEX50F6 | CV548253 | Smad4 type2 [Q9W639] | X. laevis | 7E-76 |
| 1742HAEMEX7F10 | CK149504 | Transcription elongation factor DSIF [O00267] | H. sapiens | 1E-94 |
| 3017HOUN32D1 | CO870235 | Transcription elongation factor DSIF [O00267] | H. sapiens | 8E-32 |
| 3032HOEX53F10 | CV548409 | RUSH-1 [Q95216] | O. cuniculus | 9E-68 |
| 1742HFEX1H6 | CK149201 | Tis11 family protein [P47974] | H. sapiens | 3E-36 |
| 3017HOEX33B8 | CO870263 | Transcription factor IID p80 chain homolog [Q91857] | X. laevis | 2E-55 |
| 1742HFEX1C3 | CK149163 | Tropomyosin [O97192] | H. aspersa | 6E-39 |
| 3017HOEX21A2 | CK656807 | Tropomyosin [O97192] | H. aspersa | 1E-40 |
| 3017HFUN44E7 | CO870396 | Tropomyosin [O97192] | H. aspersa | 7E-77 |
| 3032HAEMUN54H4 | CV548439 | Staphylococcal nuclease domain-containing protein 1 [Q7K2F4] | H. sapiens | 3E-26 |
| 3032HAEMUN54E4 | CV548436 | Winged-helix repressor FOXP4 [Q8CIS1] | M. musculus | 3E-12 |
| 3032HAEMUN54C3 | CV548430 | Jumonji domain containing protein 2C [Q9H3R0] | H. sapiens | 5E-73 |
| Signal transducers and transcription factor regulators (both GO: 0007165 and GO: 0030528) | ||||
| 1742OTEX5C1 | CK149418 | WDR9protein [Q9NSI6] | H. sapiens | 1E-80 |
| 3017HFEX11G11 | CK149281 | Pliotropic regulator 1 [Q9WUC8] | R. norvegicus | 2E-29 |
| 1742BUN46G3 | CV548048 | Transcriptional regulator ATRX protein [P46100] | H. sapiens | 7E-90 |
| 1742BEX47D9 | CV548159 | Beta-catenin [P35224] | U. caupo | 2E-34 |
| 3032HOEX53H9 | CV548408 | Beta-catenin [P35224] | U. caupo | 2E-83 |
| 1742BUN46H7 | CV548074 | Fibropellin-1 [P10079] | S. purpuratus | 1E-17 |
| 3032OTUN57B7 | CV548627 | HIRA protein [P54198] | H. sapiens | 3E-68 |
| 1742OTEX5A12 | EG030742 | IKAP [O95163] | H. sapiens | 4E-41 |
| 1742HFUN2A2 | CK149289 | Nuclear hormone receptor FTZ-F1 beta [Q05192] | D. melanogaster | 2E-17 |
| 3032HOUN59B7 | CV548771 | Orphan nuclear receptor NR1D2 [Q14995] | H. sapiens | 4E-25 |
| 1742OTEX5C2 | CK149419 | Histone-binding protein RBBP4 [Q09028] | H. sapiens | 1E-131 |
| Immune response (GO: 0006955) | ||||
| 3032BUN58C5 | CV548690 | Alpha 2-macroglobulin [O01717] | Limulus sp. | 5E-20 |
| 1742OTEX6D7 | CK149467 | Alpha-1 inhibitor III [Q62591] | R. sordidus | 3E-16 |
| 1742BEX47H4 | CV548126 | ATP-binding cassette sub-family F member 1 [Q8NE71] | H. sapiens | 5E-56 |
| 1742HOUN8H9 | CK149553 | Paramyosin [O96064] | M. galloprovincialis | 7E-61 |
| 3017OTUN16E12 | CK656627 | Exosome complex exonuclease RRP45 [Q9JHI7] | M. musculus | 5E-73 |
| 3032BEX52B6 | CV548339 | Exosome complex exonuclease RRP45 [Q9JHI7] | M. musculus | 2E-77 |
| 1742OTUN3F6 | CK149346 | J kappa-recombination signal binding protein [P31266] | M. musculus | 2E-24 |
| 3017OTUN34C5 | CO870295 | Rho guanine nucleotide exchange factor 4 [Q9NR80] | H. sapiens | 4E-36 |
| 1742BEX47G7 | CV548147 | Soma ferritin [P42577] | L. stagnalis | 7E-60 |
| 3032HFUN56G3 | CV548545 | Soma ferritin [P42577] | L. stagnalis | 3E-59 |
| Response to stress (GO: 0006950) | ||||
| 3017HOUN20F4 | CK656786 | 60S acidic ribosomal protein P0 [Q9NHP0] | S. crassipalpis | 9E-73 |
| 1742HOEX9H9 | CK149589 | Cdc7-related kinase [Q9Z2Y5] | M. musculus | 4E-45 |
| 3032HAEMUN54A6 | CV548444 | ATP-dependent RNA helicase Ddx1 [Q9VNV3] | D. melanogaster | 1E-60 |
| 3017OTUN34C6 | CO870298 | 78 kDa glucose-regulated protein precursor [Q16956] | A. californica | 2E-86 |
| 3017BUN48H5 | CV548188 | Helicase-like protein NHL [Q9NZ71] | H. sapiens | 1E-25 |
| 3032OTUN57E6 | CV548623 | Hypoxia up-regulated 1 [Q9Y4L1] | H. sapiens | 1E-38 |
| 3032HOEX53A9 | CV548403 | Mismatch repair protein pms1 homologue [Q8IBJ3] | P. falciparum | 1E-14 |
| 3017HOEX33D3 | CO870252 | DNA mismatch repair protein Mlh1 [P40692] | H. sapiens | 2E-25 |
| 1742OTEX6D1 | CK149464 | Polyubiquitin [P62988] | H. sapiens | 2E-55 |
| 3017OTUN34G6 | CO870301 | Polyubiquitin [P62988] | H. sapiens | 6E-85 |
| 1742HOUN8C5 | CK149524 | Polyubiquitin [P62988] | H. sapiens | 8E-44 |
GenBank accession number, organism and E value given for the top match.
3.5. Functional classification based on KEGG pathway analysis
As an alternative method of categorising ESTs by biochemical function, clones were assigned to biochemical pathways using the KEGG website. Four hundred and thirteen ESTs were assigned to metabolic, genetic information processing, environmental information processing and cellular pathways (Tables 4 and 5). Thirty-one enzymes (38 clones) from 7 out of a possible 11 signal transduction pathways were identified as well as 25 enzymes (31 clones) from 7 out of a possible 9 immune-related pathways.
Table 4.
KEGG pathways identified by B. glabrata ESTs
| KEGG categories represented | Enzymesa | Clonesb | |
|---|---|---|---|
| 1 | Metabolism | ||
| 1.1 | Carbohydrate metabolism | ||
| 1.1.1 | Glycolysis/gluconeogenesis | 7 | 12 |
| 1.1.3 | Pentose phosphate pathway | 4 | 5 |
| 1.1.5 | Fructose and mannose metabolism | 1 | 2 |
| 1.1.6 | Galactose metabolism | 2 | 2 |
| 1.1.7 | Ascorbate and aldarate metabolism | 3 | 4 |
| 1.1.8 | Starch and sucrose metabolism | 6 | 12 |
| 1.1.9 | Aminosugars metabolism | 1 | 1 |
| 1.1.11 | Pyruvate metabolism | 4 | 5 |
| 1.1.12 | Glyoxylate and dicarboxylate metabolism | 1 | 1 |
| 1.1.13 | Propanoate metabolism | 2 | 2 |
| 1.1.14 | Butanoate metabolism | 5 | 5 |
| 1.1.17 | Inositol phosphate metabolism | 1 | 1 |
| 1.2 | Energy metabolism | ||
| 1.2.1 | Oxidative phosphorylation | 7 | 7 |
| 1.2.2 | ATP synthesis | 1 | 1 |
| 1.2.4 | Carbon fixation | 3 | 7 |
| 1.2.6 | Methane metabolism | 2 | 2 |
| 1.2.7 | Nitrogen metabolism | 1 | 1 |
| 1.3 | Lipid metabolism | ||
| 1.3.1 | Fatty acid biosynthesis | 1 | 1 |
| 1.3.2 | Fatty acid elongation in mitochondria | 1 | 2 |
| 1.3.3 | Fatty acid metabolism | 2 | 2 |
| 1.3.4 | Synthesis and degradation of ketone bodies | 1 | 1 |
| 1.3.6 | Bile acid biosynthesis | 1 | 1 |
| 1.3.9 | Glycerolipid metabolism | 1 | 1 |
| 1.3.10 | Glycerophospholipid metabolism | 2 | 2 |
| 1.4 | Nucleotide metabolism | ||
| 1.4.1 | Purine metabolism | 6 | 8 |
| 1.4.2 | Pyrimidine metabolism | 3 | 4 |
| 1.5 | Amino acid metabolism | ||
| 1.5.1 | Glutamate metabolism | 2 | 2 |
| 1.5.2 | Alanine and aspartate metabolism | 1 | 1 |
| 1.5.3 | Glycine, serine and threonine metabolism | 4 | 6 |
| 1.5.4 | Methionine metabolism | 1 | 2 |
| 1.5.6 | Valine, leucine and isoleucine degradation | 2 | 2 |
| 1.5.7 | Valine, leucine and isoleucine biosynthesis | 2 | 3 |
| 1.5.8 | Lysine biosynthesis | 1 | 1 |
| 1.5.9 | Lysine degradation | 2 | 2 |
| 1.5.10 | Arginine and proline metabolism | 5 | 7 |
| 1.5.11 | Histidine metabolism | 2 | 3 |
| 1.5.12 | Tyrosine metabolism | 2 | 3 |
| 1.5.13 | Phenylalanine metabolism | 2 | 2 |
| 1.5.14 | Tryptophan metabolism | 5 | 6 |
| 1.5.15 | Phenylalanine, tyrosine and tryptophan biosynthesis | 2 | 3 |
| 1.5.16 | Urea cycle and metabolism of amino groups | 3 | 4 |
| 1.6 | Metabolism of other amino acids | ||
| 1.6.1 | β-alanine metabolism | 3 | 4 |
| 1.6.3 | Aminophosphonate metabolism | 1 | 2 |
| 1.6.4 | Selenoamino acid metabolism | 1 | 2 |
| 1.7 | Glycan biosynthesis and metabolism | ||
| 1.7.1 | N-Glycan biosynthesis | 1 | 1 |
| 1.7.3 | N-Glycan degradation | 1 | 1 |
| 1.7.4 | O-Glycan biosynthesis | 1 | 1 |
| 1.7.12 | Glycosphingolipid metabolism | 1 | 1 |
| 1.8 | Biosynthesis of polyketides and nonribosomal peptides | ||
| 1.8.3 | Biosynthesis of ansamycins | 1 | 1 |
| 1.9 | Metabolism of cofactors and vitamins | ||
| 1.9.3 | Vitamin B6 metabolism | 2 | 2 |
| 1.9.6 | Biotin metabolism | 1 | 1 |
| 1.9.7 | Folate biosynthesis | 3 | 8 |
| 1.9.10 | Porphyrin and chlorophyll metabolism | 3 | 4 |
| 1.9.11 | Ubiquinone biosynthesis | 4 | 4 |
| 1.10 | Biosynthesis of secondary metabolites | ||
| 1.10.4 | Limonene and pinene degradation | 2 | 2 |
| 1.10.6 | Stilbene, coumarine and lignin biosynthesis | 3 | 4 |
| 1.10.8 | Alkaloid biosynthesis I | 1 | 2 |
| 1.10.12 | Streptomycin biosynthesis | 1 | 1 |
| 1.11 | Biodegradation of xenobiotics | ||
| 1.11.4 | γ-hexachlorocyclohexane degradation | 2 | 2 |
| 1.11.5 | 3-chloroacrylic acid degradation | 1 | 2 |
| 1.11.6 | 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) degradation | 1 | 2 |
| 1.11.8 | 1,2-dichloroethane degradation | 1 | 1 |
| 1.11.14 | Fluorene degradation | 2 | 3 |
| 1.11.17 | Benzoate degradation via hydroxylation | 1 | 2 |
| 1.11.18 | Atrazine degradation | 1 | 1 |
| 1.11.20 | 1- and 2-methylnaphthalene degradation | 1 | 2 |
| 2 | Genetic information processing | ||
| 2.1 | Transcription | ||
| 2.1.2 | RNA polymerase | 1 | 1 |
| 2.1.3 | Basal transcription factors | 2 | 2 |
| 2.2 | Translation | ||
| 2.2.2 | Ribosome | 20 | 27 |
| 2.2.3 | Aminoacyl-tRNA biosynthesis | 16 | 35 |
| 2.3 | Folding, sorting and degradation | ||
| 2.3.1 | Protein export | 2 | 3 |
| 2.3.2 | Type II secretion system | 1 | 1 |
| 2.3.7 | Ubiquitin mediated proteolysis | 3 | 4 |
| 2.3.8 | Proteasome | 7 | 11 |
| 3 | Environmental information processing | ||
| 3.1 | Membrane transport | ||
| 3.1.1 | ABC transporters | 15 | 23 |
| 3.2 | Signal transduction | ||
| 3.2.1 | Two-component system | 3 | 3 |
| 3.2.2 | MAPK signalling pathway | 4 | 7 |
| 3.2.3 | Wnt signalling pathway | 6 | 6 |
| 3.2.4 | Notch signalling pathway | 1 | 1 |
| 3.2.6 | TGF-β signalling pathway | 5 | 5 |
| 3.2.9 | Calcium signalling pathway | 10 | 14 |
| 3.2.10 | Phosphatidylinositol signalling system | 2 | 2 |
| 3.3 | Signalling molecules and interaction | ||
| 3.3.1 | Neuroactive ligand–receptor interaction | 17 | 20 |
| 3.3.2 | Cytokine–cytokine receptor interaction | 3 | 3 |
| 3.3.3 | ECM–receptor interaction | 5 | 7 |
| 3.3.4 | Cell adhesion molecules (CAMs) | 25 | 36 |
| 4 | Cellular processes | ||
| 4.1 | Cell motility | ||
| 4.1.3 | Regulation of actin cytoskeleton | 15 | 37 |
| 4.2 | Cell growth and death | ||
| 4.2.1 | Cell cycle | 5 | 5 |
| 4.2.2 | Apoptosis | 6 | 11 |
| 4.3 | Cell communication | ||
| 4.3.1 | Focal adhesion | 13 | 26 |
| 4.3.2 | Adherens junction | 8 | 16 |
| 4.3.3 | Tight junction | 9 | 21 |
| 4.3.4 | Gap junction | 6 | 25 |
| 4.4 | Endocrine system | ||
| 4.4.1 | Insulin signalling pathway | 6 | 8 |
| 4.4.2 | Adipocytokine signalling pathway | 3 | 3 |
| 4.5 | Immune system | ||
| 4.5.1 | Hematopoietic cell lineage | 2 | 2 |
| 4.5.2 | Complement and coagulation cascades | 5 | 7 |
| 4.5.3 | Toll-like receptor signalling pathway | 1 | 1 |
| 4.5.4 | Natural killer cell mediated cytotoxicity | 2 | 2 |
| 4.5.6 | T cell receptor signalling pathway | 2 | 2 |
| 4.5.7 | B cell receptor signalling pathway | 6 | 7 |
| 4.5.9 | Leukocyte transendothelial migration | 7 | 10 |
| 4.8 | Development | ||
| 4.8.2 | Axon guidance | 7 | 10 |
| 4.9 | Behaviour | ||
| 4.9.1 | Circadian rhythm | 3 | 7 |
Enzymes—the number of enzymes from each pathway that were identified.
Clones—the number of ORESTES clones that identified enzymes.
Table 5.
Summary of KEGG pathways identified by B. glabrata ESTs
| Pathways represented | Total possible | Enzymes identified | No. EST clones | ||
|---|---|---|---|---|---|
| Metabolic pathway | |||||
| 1.1 | Carbohydrate metabolism | 13 | 17 | 37 | 52 |
| 1.2 | Energy metabolism | 5 | 8 | 14 | 18 |
| 1.3 | Lipid metabolism | 7 | 12 | 9 | 10 |
| 1.4 | Nucleotide metabolism | 2 | 2 | 9 | 12 |
| 1.5 | Amino acid metabolism | 15 | 16 | 36 | 47 |
| 1.6 | Metabolism of other amino acids | 3 | 9 | 5 | 8 |
| 1.7 | Glycan biosynthesis and metabolism | 4 | 19 | 4 | 4 |
| 1.8 | Biosynthesis of polyketides and nonribosomal peptides | 1 | 9 | 1 | 1 |
| 1.9 | Metabolism of cofactors and vitamins | 5 | 11 | 13 | 19 |
| 1.10 | Biosynthesis of secondary metabolites | 4 | 16 | 7 | 9 |
| 1.11 | Biodegradation of xenobiotics | 8 | 21 | 10 | 15 |
| 67 | 140 | 145 | 195 | ||
| Genetic information processing | |||||
| 2.1 | Transcription | 2 | 3 | 3 | 3 |
| 2.2 | Translation | 2 | 3 | 36 | 62 |
| 2.3 | Folding, sorting and degradation | 4 | 8 | 13 | 19 |
| 2.4 | Replication and repair | 0 | 1 | 0 | 0 |
| 8 | 15 | 52 | 84 | ||
| Environmental information processing | |||||
| 3.1 | Membrane transport | 1 | 2 | 15 | 23 |
| 3.2 | Signal transduction | 7 | 11 | 31 | 38 |
| 3.3 | Signalling molecules and interaction | 4 | 4 | 50 | 66 |
| 12 | 17 | 96 | 127 | ||
| Cellular processes | |||||
| 4.1 | Cell motility | 1 | 3 | 15 | 37 |
| 4.2 | Cell growth and death | 2 | 2 | 11 | 16 |
| 4.3 | Cell communication | 4 | 4 | 36 | 88 |
| 4.4 | Endocrine system | 2 | 3 | 9 | 11 |
| 4.5 | Immune system | 7 | 9 | 25 | 31 |
| 4.6 | Nervous system | 0 | 2 | 0 | 0 |
| 4.7 | Sensory system | 0 | 2 | 0 | 0 |
| 4.8 | Development | 1 | 2 | 7 | 10 |
| 4.9 | Behaviour | 1 | 1 | 3 | 7 |
| 18 | 28 | 106 | 200 | ||
4. Discussion
Use of the ORESTES approach generated 1843 ESTs from different tissues and strains of B. glabrata. Only 3.4% of these had been previously characterized in B. glabrata and cluster analysis with other B. glabrata ESTs identified less than 300 clusters of overlapping sequences. Over half of the sequences showed no matches to previously sequenced genes in the non-redundant section of GenBank. Functional analysis of those with sequence similarity to previously characterized genes, using gene ontologies and KEGG assignments identified a number of antioxidant, signalling and transcriptional regulatory genes, molecules that may potentially be involved in snail/parasite interactions, as well as several immune and stress response proteins.
4.1. Antioxidant proteins
Four different genes were identified that were similar to molecules that demonstrate antioxidant functions in other organisms. Peroxinectin (CV548486) is a cell adhesion protein with peroxidase activity, which has been identified in other invertebrates including the crayfish Pacifastacus leniusculus [25], the black tiger shrimp Penaeus monodon [26], Drosophila melanogaster [27] and the white shrimp Litopenaeus vannamei [28], and is a functional equivalent of the vertebrate myeloperoxidase [25,29]. Peroxidasin (CV548777) is a similar protein with peroxidase activity associated with developmental processes in both Drosophila [30] and Xenopus tropicalis [31]. Dual oxidase 1 (Duox1) (CK149203), possesses a peroxidase domain and is thus categorized with antioxidant function; interestingly though, these transmembrane proteins also have a superoxide-generating subunit homologous to glycoprotein p91phox [32], a host defence molecule that generates reactive oxygen species (ROS) [33,34]. Another Duox1 sequence has been identified in B. glabrata [17] but our sequence (CK149203) seems to be a paralog of this gene as it shows no similarity at the nucleotide level with the other sequences (CK989379, CK990069) and did not identify these sequences in BlastN searches, despite all matching the same section of translated protein in tBlastX searches.
4.2. Signalling molecules and transcriptional regulators
Based on our knowledge of other, well-characterised, biological systems, some of the identified signalling molecules and transcriptional regulators play a part in pathways that are likely to be involved in the innate immune response of snails. In a few cases (as described below), functional studies have shown that such signalling pathways contribute to the regulation of molluscan defence reactions.
One clone (EG030744) matched the transcription factor nuclear factor-κB1 (NF-κB1), a p105 NF-κB subunit that is proteolytically processed to yield NF-κB p50 [35]. The NF-κB/Rel transcription factors comprise a family of evolutionarily conserved and structurally related proteins identified in a variety of vertebrates and invertebrates including the beetle Allomyrina dichotoma [36], the sea squirt Ciona intestinalis [37] and the bivalve mollusc Crassostrea gigas [38]. Such transcription factors are central to the NF-κB pathway, a key intracellular pathway that co-ordinates the induction of defence genes in both mammals and Drosophila, and plays a pivotal role in vertebrate and invertebrate innate immunity [39,40]. IκB kinase (IKK) complex associated protein (IKAP) [41] was also identified (EG030742). This protein contains potential IKK association sites and was thus originally thought to play a role in NF-κB signalling by scaffolding the IKK signalsome [41]. Although this now seems unlikely (as discussed in [42]) IKAP seems to associate with stress-activated protein kinase/c-jun NH2-terminal kinase (SAPK/JNK) and regulate its activity in mammals [42]. Activation of SAPK/JNK occurs via the transmission of extracellular stress signals, and aside from the role that this protein plays in processes such as development, apoptosis, and proliferation, it can regulate immune responses in Drosophila [43,44]. Interestingly, SAPK/JNK is activated by recombinant human TNF-α in defence cells (haemocytes) of the bivalve mollusc Mytilus galloprovincialis [45] and, in the present study, we identified a homologue (clones CV548175, CV548723, CV548166, CV548685) of Drosophila JNK interacting protein 1 [46], a scaffold protein that aggregates specific components to form a functional SAPK/JNK signalling module in mammals [47].
Homologues of invertebrate integrin α3 [48] (CK149506), focal adhesion kinase (FAK) [49] (CK656717) and mammalian protein tyrosine kinase Src [50] (CK149232) were also identified. Integrins are a family of heterodimeric, transmembrane adhesion receptors whose ligand specificities are determined by the specific α and β subunits; integrins are crucial to cell adhesion and organisation of the actin cytoskeleton and they serve as important receptors in immune cell responses, cell migration and tissue integrity. Expressed in all metazoans, integrins have been characterized in several invertebrates [51], with β1 integrin subunits reported from haemocytes of the molluscs C. gigas [52] and B. glabrata [53]. Integrins nucleate the formation of focal adhesions and focal complexes and these events rely on the co-ordinated actions of signalling proteins that include FAK and Src. In mammals, integrin clustering is known to lead to autophosphorylation of FAK at Tyr397, FAK then associates with the SH2 domain of Src, which in turn phosphorylates FAK at Tyr925 [54]. In some cell types this can result in downstream signalling to the extracellular signal-regulated kinase (ERK) pathway [55], a signalling module that has been shown to regulate phagocytosis and nitric oxide production in haemocytes from L. stagnalis [56,57]. A recent study has demonstrated that integrin engagement results in increased phosphorylation of a FAK-like protein in L. stagnalis haemocytes and that integrin blockade inhibits phagocytosis and spreading by these cells [58]. Since integrins are also known to regulate cell spreading by haemocytes of B. glabrata [59] it appears that integrin binding is crucial to the defence responses of snails, particularly those involving actin remodelling. Thus, signalling through the identified FAK/Src proteins is likely to regulate such defence reactions, as has been shown in insects [60].
Protein kinase C (PKC) is known to play a role in regulating innate defences in mammals; this has also been documented for snails in which PKC seems to regulate nitric oxide (NO) and hydrogen peroxide (H2O2) production, phagocytosis and cell spreading by haemocytes [56,57,61,62]. In this context, it is interesting that we have now identified homologues of two proteins, activated protein kinase C receptor (RACK) (CK149425, CK149451) from Xenopus [63] and 14-3-3 γ (CO870195) from humans [64], which are known to interact with PKC. RACK has previously been characterized in B. glabrata by other workers [65], however, the nucleotide sequence fragment we identified differs significantly from that previously published, with only three short stretches being identified in a BlastN search with an E value of 3e-5. BlastX searches did identify (amongst other RACK sequences) the amino acid sequence of the previously sequenced B. glabrata RACK, with 89% similarity. Our nucleotide sequence also identified 12 other B. glabrata ESTs with close homology, so it seems likely that we have identified a second gene for RACK. 14-3-3 γ appears to be phosphorylated by PKC and might facilitate signalling to the ERK pathway via Raf [64]. Biomphalaria glabrata RACK might serve to direct the translocation of PKC isoforms to specific cellular compartments as it does in higher organisms [66].
We also identified a homologue of the B (regulatory) subunit of serine threonine protein phosphatase 2A (PP2A) [67] (CV548346), a heterotrimeric holoenzyme that either positively or negatively regulates the activities of wide variety of cellular signal transduction pathways including those involving IKK and ERK discussed above (for reviews see [68,69]). Also of interest are genes that were found to be homologous to those coding for proteins involved in protein kinase A and cAMP signalling, namely: adenylyl cyclase [70] (CV548064), the enzyme that generates the second messenger cAMP; cAMP-specific 3′,5′-cyclic phosphodiesterase (CV548523), an enzyme involved in cyclic nucleotide metabolism [71]; and the type N4 regulatory subunit of PKA [72] (CV548191). These proteins likely play a role in mollusc defence since the catecholamine noradrenaline modulates the phagocytic activity of C. gigas haemocytes via a β-adrenergic receptor-cAMP signalling pathway [73]. Finally, genes were found which matched to those of the transmembrane glycoprotein macrophage mannose receptor [74] (CO870241, CV548367), the Ras-related GTPase protein Rab 21 [75] (CK149518, CK149287) and hemolectin [76] (CV548237, CV548539, CV548566). The macrophage mannose receptor is a phagocytic receptor that targets pathogens such as bacteria and yeast which express mannose-rich glycoproteins [74] and Rab 21 has been recently found to interact with two LIM domain proteins in the slime mould Dictyostelium to collectively regulate phagocytosis [77]. The identified hemolectin showed homology to Drosophila hemolectin which is a major clot constituent in these flies [76].
4.3. Immune and stress response genes
Examination of the gene ontologies revealed 8 immune response genes and 10 response-to-stress genes. Of particular interest are α 2-macroglobulin (α2 M) and Rho-guanine nucleotide-exchange factor 4 (Rho-GEF 4). The identified α2 M (CV548690) is similar to that from the horseshoe crab Limulus polyphemus [78], a proteinase inhibitor similar to mammalian α2 M with broad reactivity towards proteinases; a similar inhibitor with activity towards serine, cysteine and metalloproteinase has been purified from B. glabrata plasma [79]. Such inhibitors could be important to defence, since they may be expressed during the humoral immune response in order to inactivate proteinases produced by invading pathogens [80]. The Rho-GEF 4 homologue (CO870295) was similar to that sequenced in humans [81]. This GEF is operational towards the small GTPases Rho A and Rac 1 and is thought to play a role in cell migration and cell–cell adhesion [82]. Given the universal nature of these cellular processes it is likely that Rho GEF 4 has a similar role in snails and thus may be important in the snail defence response towards pathogens.
5. Conclusions
The genes described above represent a set of those identified that might play important roles in molluscan defence. To eliminate pathogens such as parasites, the molluscan immune system must be able to mount a co-ordinated response to the invading organism, with processes such as cell adhesion and the production of reactive oxygen and nitrogen intermediates being crucial to the outcome of infection. Despite a parasite-mediated interference theory being proposed 25 years ago [83], the precise mechanism(s) by which schistosomes evade the defence response of their snail intermediate hosts remain largely unknown. A recent hypothesis paper has explored the idea that parasites might blunt the defence response of susceptible snails by interfering with key signal transduction pathways in their defence cells [84]; such a strategy could serve to alter gene expression and functional defence responses.
This EST gene discovery project has provided a significant number of genes for the first version of a custom B. glabrata cDNA microarray. A detailed investigation of the transcriptome in response to trematode infection in this snail intermediate host, in order to identify and understand the role of specific genes involved in the snail internal defence system can therefore now be carried out. Thus, we anticipate that through application of the microarray, we will move closer to gaining a comprehensive understanding of snail–schistosome interactions and the complex nature of the biological interplay that exists between snail and schistosome parasite.
Acknowledgements
This work was carried out with funding from the Wellcome Trust (068589/Z/02/Z). We would like to thank Jayne King and Mike Anderson, NHM, for snail and parasite culture and Julia Llewellyn-Hughes and Claire Griffin, NHM, for sequencing assistance. Emmanuel Dias-Neto thanks Associacao Beneficente Alzira Denise Hertzog Silva (ABADHS) for their support to the Laboratory of Neurosciences (LIM27). Cathy Jones was in receipt of a Royal Society of Edinburgh Sabbatical Fellowship whilst co-authoring this article.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dci.2006.11.004.
Appendix A. Supplementary Materials
Online Supplementary Materials
Online Supplementary Materials
References
- 1.LoVerde P.T., Hirai H., Merrick J.M., Lee N.H., El-Sayed N. Schistosoma mansoni genome project: an update. Parasitol Int. 2004;53(2):183–192. doi: 10.1016/j.parint.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Rollinson D., Johnston D.A. Genome analysis: helping to solve the molecular mysteries of schistosomes. Helminthologia. 1999;36(3):147–158. [Google Scholar]
- 3.Tanaka M., Tanaka T., Inazawa J., Nagafuchi S., Mutsui Y., Kaukas A. Proceedings of the schistosome genome project. Mem Inst Oswaldo Cruz. 1997;92(6):829–834. doi: 10.1590/s0074-02761997000600019. [DOI] [PubMed] [Google Scholar]
- 4.Dias-Neto E., Harrop R., Correa-Oliveira R., Pena S.D.J., Wilson R.A., Simpson A.J.G. The schistosome genome project: RNA arbitrarily primed PCR allows the accelerated generation of expressed sequence tags. Mem Inst Oswaldo Cruz. 1996;91(5):655–657. doi: 10.1590/s0074-02761996000500020. [DOI] [PubMed] [Google Scholar]
- 5.Verjovski-Almeida S., DeMarco R., Martins E.A.L., Guimaraes P.E.M., Ojopi E.P.B., Paquola A.C.M. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nat Genet. 2003;35(2):148–157. doi: 10.1038/ng1237. [DOI] [PubMed] [Google Scholar]
- 6.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Boguski M.S., Lowe T.M.J., Tolstoshev C.M. dbEST—database for expressed sequence tags. Nat Genet. 1993;4(4):332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 8.Jenny M.J., Ringwood A.H., Lacy E.R., Lewitus A.J., Kempton J.W., Gross P.S. Potential indicators of stress response identified by expressed sequence tag analysis of hemocytes and embryos from the American oyster, Crassostrea virginica. Mar Biotechnol. 2002;4(1):81–93. doi: 10.1007/s10126-001-0072-8. [DOI] [PubMed] [Google Scholar]
- 9.Gueguen Y., Cadoret J.P., Flament D., Barreau-Roumiguiere C., Girardot A.L., Garnier J. Immune gene discovery by expressed sequence tags generated from hemocytes of the bacteria-challenged oyster, Crassostrea gigas. Gene. 2003;303:139–145. doi: 10.1016/s0378-1119(02)01149-6. [DOI] [PubMed] [Google Scholar]
- 10.Davison A., Blaxter M.L. An expressed sequence tag survey of gene expression in the pond snail Lymnaea stagnalis, an intermediate vector of Fasciola hepatica. Parasitology. 2005;130:539–552. doi: 10.1017/s0031182004006791. [DOI] [PubMed] [Google Scholar]
- 11.Knight M., Miller A.N., Geoghagen N.S.M., Lewis F.A., Kerlavage A.R. Expressed sequence tags (ESTs) of Biomphalaria glabrata, an intermediate snail host of Schistosoma mansoni: use in the identification of RFLP markers. Malacologia. 1998;39(1-2):175–182. [Google Scholar]
- 12.Lockyer A.E., Jones C.S., Noble L.R., Rollinson D. Use of differential display to detect changes in gene expression in the intermediate snail host Biomphalaria glabrata upon infection with Schistosoma mansoni. Parasitology. 2000;120:399–407. doi: 10.1017/s0031182099005624. [DOI] [PubMed] [Google Scholar]
- 13.Schneider O., Zelck U.E. Differential display analysis of hemocytes from schistosome-resistant and schistosome-susceptible intermediate hosts. Parasitol Res. 2001;87(6):489–491. doi: 10.1007/s004360100394. [DOI] [PubMed] [Google Scholar]
- 14.Miller A.N., Raghavan N., FitzGerald P.C., Lewis F.A., Knight M. Differential gene expression in haemocytes of the snail Biomphalaria glabrata: effects of Schistosoma mansoni infection. Int J Parasitol. 2001;31(7):687–696. doi: 10.1016/s0020-7519(01)00133-3. [DOI] [PubMed] [Google Scholar]
- 15.Raghavan N., Miller A.N., Gardner M., FitzGerald P.C., Kerlavage A.R., Johnston D.A. Comparative gene analysis of Biomphalaria glabrata hemocytes pre- and post-exposure to miracidia of Schistosoma mansoni. Mol Biochem Parasitol. 2003;126(2):181–191. doi: 10.1016/s0166-6851(02)00272-4. [DOI] [PubMed] [Google Scholar]
- 16.Nowak T.S., Woodards A.C., Jung Y.H., Adema C.M., Loker E.S. Identification of transcripts generated during the response of resistant Biomphalaria glabrata to Schistosoma mansoni infection using suppression subtractive hybridization. J Parasitol. 2004;90(5):1034–1040. doi: 10.1645/GE-193R1. [DOI] [PubMed] [Google Scholar]
- 17.Mitta G., Galinier R., Tisseyre P., Allienne J.F., Girerd-Chambaz Y., Guillou F. Gene discovery and expression analysis of immune-relevant genes from Biomphalaria glabrata hemocytes. Dev Comp Immunol. 2005;29(5):393–407. doi: 10.1016/j.dci.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Dias-Neto E., Correa R.G., Verjovski-Almeida S., Briones M.R.S., Nagai M.A., da Silva W. Shotgun sequencing of the human transcriptome with ORF expressed sequence tags. Proc Natl Acad Sci USA. 2000;97(7):3491–3496. doi: 10.1073/pnas.97.7.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camargo A.A., Samaia H.P.B., Dias-Neto E., Simao D.F., Migotto I.A., Briones M.R.S. The contribution of 700,000 ORF sequence tags to the definition of the human transcriptome. Proc Natl Acad Sci USA. 2001;98(21):12103–12108. doi: 10.1073/pnas.201182798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Souza S.J., Camargo A.A., Briones M.R.S., Costa F.F., Nagai M.A., Verjovski-Almeida S. Identification of human chromosome 22 transcribed sequences with ORF expressed sequence tags. Proc Natl Acad Sci USA. 2000;97(23):12690–12693. doi: 10.1073/pnas.97.23.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dias-Neto E., Harrop R., Correa-Oliveira R., Wilson R.A., Pena S.D.J., Simpson A.J.G. Minilibraries constructed from cDNA generated by arbitrarily primed RT-PCR: an alternative to normalized libraries for the generation of ESTs from nanogram quantities of mRNA. Gene. 1997;186(1):135–142. doi: 10.1016/s0378-1119(96)00699-3. [DOI] [PubMed] [Google Scholar]
- 22.Paraense W.L., Correa L.R. Variation in susceptibility of populations of Austrolorbis glabratus to a strain of Schistosoma mansoni. Rev Inst Med Trop Sao Paulo. 1963;5:15–22. [PubMed] [Google Scholar]
- 23.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Weston D.S., Kemp W.M. Schistosoma mansoni—comparison of cloned tropomyosin antigens shared between adult parasites and Biomphalaria glabrata. Exp Parasitol. 1993;76(4):358–370. doi: 10.1006/expr.1993.1044. [DOI] [PubMed] [Google Scholar]
- 25.Johansson M.W., Lind M.I., Holmblad T., Thornqvist P.O., Soderhall K. Peroxinectin, a novel cell-adhesion protein from crayfish blood. Biochem Biophys Res Commun. 1995;216(3):1079–1087. doi: 10.1006/bbrc.1995.2731. [DOI] [PubMed] [Google Scholar]
- 26.Sritunyalucksana K., Wongsuebsantati K., Johansson M.W., Soderhall K. Peroxinectin, a cell adhesive protein associated with the proPO system from the black tiger shrimp, Penaeus monodon. Dev Comp Immunol. 2001;25(5-6):353–363. doi: 10.1016/s0145-305x(01)00009-x. [DOI] [PubMed] [Google Scholar]
- 27.Vazquez M., Rodriguez R., Zurita M. A new peroxinectin-like gene preferentially expressed during oogenesis and early embryogenesis in Drosophila melanogaster. Dev Genes Evol. 2002;212(11):526–529. doi: 10.1007/s00427-002-0265-9. [DOI] [PubMed] [Google Scholar]
- 28.Gross P.S., Bartlett T.C., Browdy C.L., Chapman R.W., Warr G.W. Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific white shrimp, Litopenaeus vannamei, and the Atlantic white shrimp, L. setiferus. Dev Comp Immunol. 2001;25(7):565–577. doi: 10.1016/s0145-305x(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 29.Johansson M.W., Patarroyo M., Oberg F., Siegbahn A., Nilsson K. Myeloperoxidase mediates cell adhesion via the alpha(M)beta(2) integrin (Mac-1, CD11b/CD18) J Cell Sci. 1997;110:1133–1139. doi: 10.1242/jcs.110.9.1133. [DOI] [PubMed] [Google Scholar]
- 30.Nelson R.E., Fessler L.I., Takagi Y., Blumberg B., Keene D.R., Olson P.F., Parker C.G., Fessler J.H. Peroxidasin—a novel enzyme-matrix protein of Drosophila development. EMBO J. 1994;13(15):3438–3447. doi: 10.1002/j.1460-2075.1994.tb06649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tindall A.J., Pownall M.E., Morris I.D., Isaacs H.V. Xenopus tropicalis peroxidasin gene is expressed within the developing neural tube and pronephric kidney. Dev Dyn. 2005;232(2):377–384. doi: 10.1002/dvdy.20226. [DOI] [PubMed] [Google Scholar]
- 32.Edens W.A., Sharling L., Cheng G.J., Shapira R., Kinkade J.M., Lee T. Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91 phox. J Cell Biol. 2001;154(4):879–891. doi: 10.1083/jcb.200103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forteza R., Salathe M., Miot F., Conner G.E. Regulated hydrogen peroxide production by Duox in human airway epithelial cells. Am J Respir Cell Mol Biol. 2005;32(5):462–469. doi: 10.1165/rcmb.2004-0302OC. [DOI] [PubMed] [Google Scholar]
- 34.Geiszt M., Witta J., Baffi J., Lekstrom K., Leto T.L. Dual oxidases represent novel hydrogen peroxide sources supporting mucosal surface host defense. FASEB J. 2003;17(9) doi: 10.1096/fj.02-1104fje. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh S., Gifford A.M., Riviere L.R., Tempst P., Nolan G.P., Baltimore D. Cloning of the p50 DNA binding subunit of NFκB: Homology to rel and dorsal. Cell. 1990;62(5):1019–1029. doi: 10.1016/0092-8674(90)90276-k. [DOI] [PubMed] [Google Scholar]
- 36.Sagisaka A., Tanaka H., Furukawa S., Yamakawa M. Characterization of a homologue of the Rel/NF-kappa B transcription factor from a beetle, Allomyrina dichotoma. Biochim Biophys Acta-Gene Struct Express. 2004;1678(2-3):85–93. doi: 10.1016/j.bbaexp.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Yagi K., Satou Y., Mazet F., Shimeld S.M., Degnan B., Rokhsar D. A genomewide survey of developmentally relevant genes in Ciona intestinalis—III. Genes for Fox, ETS, nuclear receptors and NF kappa B. Dev Genes Evol. 2003;213(5-6):235–244. doi: 10.1007/s00427-003-0322-z. [DOI] [PubMed] [Google Scholar]
- 38.Montagnani C., Kappler C., Reichhart J.M., Escoubas J.M. Cg-Rel, the first Rel/NF-kappa B homolog characterized in a mollusk, the Pacific oyster Crassostrea gigas. FEBS Lett. 2004;561(1-3):75–82. doi: 10.1016/S0014-5793(04)00124-3. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann J.A., Kafatos F.C., Janeway C.A., Ezekowitz R.A.B. Phylogenetic perspectives in innate immunity. Science. 1999;284(5418):1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 40.Khush R.S., Leulier F., Lemaitre B. Drosophila immunity: two paths to NF-kappa B. Trends Immunol. 2001;22(5):260–264. doi: 10.1016/s1471-4906(01)01887-7. [DOI] [PubMed] [Google Scholar]
- 41.Cohen L., Henzel W.J., Baeuerle P.A. IKAP is a scaffold protein of the I kappa B kinase complex. Nature. 1998;395(6699):292–296. doi: 10.1038/26254. [DOI] [PubMed] [Google Scholar]
- 42.Holmberg C., Katz S., Lerdrup M., Herdegen T., Jaattela M., Aronheim A. A novel specific role for I kappa B kinase complex-associated protein in cytosolic stress signalling. J Biol Chem. 2002;277(35):31918–31928. doi: 10.1074/jbc.M200719200. [DOI] [PubMed] [Google Scholar]
- 43.Kallio J., Leinonen A., Ulvila J., Valanne S., Ezekowitz R.A., Ramet M. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes Infect. 2005;7(5–6):811–819. doi: 10.1016/j.micinf.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 44.Zhuang Z.H., Sun L., Kong L., Hu J.H., Yu M.C., Reinach P. Drosophila TAB2 is required for the immune activation of JNK and NF-kappaB. Cell Signal. 2006;18(7):964–970. doi: 10.1016/j.cellsig.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Betti M., Ciacci C., Lorusso L.C., Canonico B., Falcioni T., Gallo G. Effects of tumour necrosis factor alpha (TNF-α) on Mytilus haemocytes: role of stress-activated mitogen-activated protein kinases (MAPKs) Biol Cell. 2006;98(4):233–244. doi: 10.1042/BC20050049. [DOI] [PubMed] [Google Scholar]
- 46.Taru H., Iijima K., Hase M., Kirino Y., Yagi Y., Suzuki T. Interaction of Alzheimer's beta-amyloid precursor family proteins with scaffold proteins of the JNK signaling cascade. J Biol Chem. 2002;277(22):20070–20078. doi: 10.1074/jbc.M108372200. [DOI] [PubMed] [Google Scholar]
- 47.Yasuda J., Whitmarsh A.J., Cavanagh J., Sharma M., Davis R.J. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol Cell Biol. 1999;19(10):7245–7254. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lavine M.D., Strand M.R. Haemocytes from Pseudoplusia includens express multiple alpha and beta integrin subunits. Insect Mol Biol. 2003;12(5):441–452. doi: 10.1046/j.1365-2583.2003.00428.x. [DOI] [PubMed] [Google Scholar]
- 49.Garcia M.G., Toney S.J., Hille M.B. Focal adhesion kinase (FAK) expression and phosphorylation in sea urchin embryos. Gene Expression Patterns. 2004;4(2):223–234. doi: 10.1016/j.modgep.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Giusti A.F., O’Neill F.J., Yamasu K., Foltz K.R., Jaffe L.A. Function of a sea urchin egg Src family kinase in initiating Ca2+ release at fertilization. Dev Biol. 2003;256(2):367–378. doi: 10.1016/s0012-1606(03)00043-5. [DOI] [PubMed] [Google Scholar]
- 51.Huhtala M., Heino J., Casciari D., de Luise A., Johnson M.S. Integrin evolution: Insights from ascidian and teleost fish genomes. Matrix Biol. 2005;24(2):83–95. doi: 10.1016/j.matbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Terahara K., Takahashi K.G., Nakamura A., Osada M., Yoda M., Hiroi T. Differences in integrin-dependent phagocytosis among three hemocyte subpopulations of the Pacific oyster Crassostrea gigas. Dev Comp Immunol. 2006;30(8):667–683. doi: 10.1016/j.dci.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Davids B.J., Wu X.J., Yoshino T.P. Cloning of a beta integrin subunit cDNA from an embryonic cell line derived from the freshwater mollusc, Biomphalaria glabrata. Gene. 1999;228(1-2):213–223. doi: 10.1016/s0378-1119(99)00008-6. [DOI] [PubMed] [Google Scholar]
- 54.Calalb M.B., Zhang X.E., Polte T.R., Hanks S.K. Focal adhesion kinase tyrosine-861 is a major site of phosphorylation by Src. Biochem Biophys Res Commun. 1996;228(3):662–668. doi: 10.1006/bbrc.1996.1714. [DOI] [PubMed] [Google Scholar]
- 55.Schlaepfer D.D., Hunter T. Focal adhesion kinase overexpression enhances Ras-dependent integrin signaling to ERK2/mitogen-activated protein kinase through interactions with and activation of c-Src. J Biol Chem. 1997;272(20):13189–13195. doi: 10.1074/jbc.272.20.13189. [DOI] [PubMed] [Google Scholar]
- 56.Plows L.D., Cook R.T., Davies A.J., Walker A.J. Activation of extracellular-signal regulated kinase is required for phagocytosis by Lymnaea stagnalis haemocytes. Biochim Biophys Acta-Mol Cell Res. 2004;1692(1):25–33. doi: 10.1016/j.bbamcr.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Wright B., Lacchini A.H., Davies A.J., Walker A.J. Regulation of nitric oxide production in snail (Lymnaea stagnalis) defence cells: a role for PKC and ERK signalling pathways. Biol Cell. 2006;98(5):265–278. doi: 10.1042/BC20050066. [DOI] [PubMed] [Google Scholar]
- 58.Plows L.D., Cook R.T., Davies A.J., Walker A.J. Integrin engagement modulates the phosphorylation of focal adhesion kinase, cell spreading, and phagocytosis in molluscan defence cells. Biochim Biophys Acta—Mol Cell Res. 2006;1763:779–786. doi: 10.1016/j.bbamcr.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 59.Davids B.J., Yoshino T.P. Integrin-like RGD-dependent binding mechanism involved in the spreading response of circulating molluscan phagocytes. Dev Comp Immunol. 1998;22(1):39–53. doi: 10.1016/s0145-305x(97)00039-6. [DOI] [PubMed] [Google Scholar]
- 60.Metheniti A., Paraskevopoulou N., Lambropoulou M., Marmaras V.J. Involvement of FAK/Src complex in the processes of Escherichia coli phagocytosis by insect hemocytes. FEBS Lett. 2001;496(1):55–59. doi: 10.1016/s0014-5793(01)02405-x. [DOI] [PubMed] [Google Scholar]
- 61.Humphries J.E., Elizondo L., Yoshino T.P. Protein kinase C regulation of cell spreading in the molluscan Biomphalaria glabrata embryonic (Bge) cell line. Biochim Biophys Acta—Mol Cell Res. 2001;1540(3):243–252. doi: 10.1016/s0167-4889(01)00136-7. [DOI] [PubMed] [Google Scholar]
- 62.Lacchini A.H., Davies A.J., Mackintosh D., Walker A.J. β-1,3-glucan modulates PKC signalling in Lymnaea stagnalis defence cells: a role for PKC in H2O2 production and downstream ERK activation. J Exp Biol. 2006;209(24):4829–4840. doi: 10.1242/jeb.02561. [DOI] [PubMed] [Google Scholar]
- 63.Kwon H.J., Bae S., Son Y.H., Chung H.M. Expression of the Xenopus homologue of the receptor for activated C-kinase 1 (RACK1) in the Xenopus embryo. Dev Genes Evol. 2001;211(4):195–197. doi: 10.1007/s004270100144. [DOI] [PubMed] [Google Scholar]
- 64.Autieri M.V., Carbone C.J. 14-3-3gamma interacts with and is phosphorylated by multiple protein kinase C isoforms in PDGF-stimulated human vascular smooth muscle cells. DNA Cell Biol. 1999;18(7):555–564. doi: 10.1089/104454999315105. [DOI] [PubMed] [Google Scholar]
- 65.Lardans V., Serra E., Capron A., Dissous C. Characterization of an intracellular receptor for activated protein kinase C (RACK) from the mollusc Biomphalaria glabrata, the intermediate host for Schistosoma mansoni. Exp Parasitol. 1998;88(3):194–199. doi: 10.1006/expr.1998.4236. [DOI] [PubMed] [Google Scholar]
- 66.Mochly-Rosen D., Gordon A.S. Anchoring proteins for protein kinase C: a means for isozyme selectivity. FASEB J. 1998;12(1):35–42. [PubMed] [Google Scholar]
- 67.Stone S.R., Hofsteenge J., Hemmings B.A. Molecular cloning of cDNAs encoding 2 isoforms of the catalytic subunit of protein phosphatase 2A. Biochemistry. 1987;26(23):7215–7220. doi: 10.1021/bi00397a003. [DOI] [PubMed] [Google Scholar]
- 68.Millward T.A., Zolnierowicz S., Hemmings B.A. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem Sci. 1999;24(5):186–191. doi: 10.1016/s0968-0004(99)01375-4. [DOI] [PubMed] [Google Scholar]
- 69.Adams D.G., Coffee R.L., Zhang H., Pelech S., Strack S., Wadzinski B.E. Positive regulation of Raf1-MEK1/2-ERK1/2 signalling by protein serine/threonine phosphatase 2A holoenzymes. J Biol Chem. 2005;280(52):42644–42654. doi: 10.1074/jbc.M502464200. [DOI] [PubMed] [Google Scholar]
- 70.Glatt C.E., Snyder S.H. Cloning and expression of an adenylyl cyclase localized to the corpus striatum. Nature. 1993;361(6412):536–538. doi: 10.1038/361536a0. [DOI] [PubMed] [Google Scholar]
- 71.Qiu Y.H., Chen C.N., Malone T., Richter L., Beckendorf S.K., Davis R.L. Characterization of the memory gene dunce of Drosophila melanogaster. J Mol Biol. 1991;222(3):553–565. doi: 10.1016/0022-2836(91)90496-s. [DOI] [PubMed] [Google Scholar]
- 72.Bergold P.J., Beushausen S.A., Sacktor T.C., Cheley S., Bayley H., Schwartz J.H. A regulatory subunit of the cAMP-dependent protein kinase down-regulated in Aplysia sensory neurons during long-term sensitization. Neuron. 1992;8(2):387–397. doi: 10.1016/0896-6273(92)90304-v. [DOI] [PubMed] [Google Scholar]
- 73.Lacoste A., Malham S.K., Cueff A., Poulet S.A. Noradrenaline modulates oyster hemocyte phagocytosis via a beta-adrenergic receptor-cAMP signaling pathway. Gen Comp Endocrinol. 2001;122(3):252–259. doi: 10.1006/gcen.2001.7643. [DOI] [PubMed] [Google Scholar]
- 74.Harris N., Super M., Rits M., Chang G., Ezekowitz R. Characterization of the murine macrophage mannose receptor: demonstration that the downregulation of receptor expression mediated by interferon-gamma occurs at the level of transcription. Blood. 1992;80(9):2363–2373. [PubMed] [Google Scholar]
- 75.Opdam F.J.M., Kamps G., Croes H., van Bokhoven H., Ginsel L.A., Fransen J.A.M. Expression of Rab small GTPases in epithelial Caco-2 cells: Rab21 is an apically located GTP-binding protein in polarised intestinal epithelial cells. Eur J Cell Biol. 2000;79(5):308–316. doi: 10.1078/S0171-9335(04)70034-5. [DOI] [PubMed] [Google Scholar]
- 76.Scherfer C., Karlsson C., Loseva O., Bidla G., Goto A., Havemann J. Isolation and characterization of hemolymph clotting factors in Drosophila melanogaster by a pullout method. Curr Biol. 2004;14(7):625–629. doi: 10.1016/j.cub.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 77.Khurana T., Brzostowski J.A., Kimmel A.R. A Rab21/LIM-only/CH-LIM complex regulates phagocytosis via both activating and inhibitory mechanisms. EMBO J. 2005;24(13):2254–2264. doi: 10.1038/sj.emboj.7600716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iwaki D., Kawabata S., Miura Y., Kato A., Armstrong P.B., Quigley J.P. Molecular cloning of Limulus alpha(2)-macroglobulin. Eur J Biochem. 1996;242(3):822–831. doi: 10.1111/j.1432-1033.1996.0822r.x. [DOI] [PubMed] [Google Scholar]
- 79.Bender R.C., Bayne C.J. Purification and characterization of a tetrameric alpha- macroglobulin proteinase inhibitor from the gastropod mollusc Biomphalaria glabrata. Biochem J. 1996;316:893–900. doi: 10.1042/bj3160893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanost M.R. Serine proteinase inhibitors in arthropod immunity. Dev Comp Immunol. 1999;23(4-5):291–301. doi: 10.1016/s0145-305x(99)00012-9. [DOI] [PubMed] [Google Scholar]
- 81.Thiesen S., Kubart S., Ropers H.H., Nothwang H.G. Isolation of two novel human RhoGEFs, ARHGEF3 and ARHGEF4, in 3p13-21 and 2q22. Biochem Biophys Res Commun. 2000;273(1):364–369. doi: 10.1006/bbrc.2000.2925. [DOI] [PubMed] [Google Scholar]
- 82.Kawasaki Y., Sato R., Akiyama T. Mutated APC and Asef are involved in the migration of colorectal tumour cells. Nature Cell Biol. 2003;5(3):211–215. doi: 10.1038/ncb937. [DOI] [PubMed] [Google Scholar]
- 83.Lie K.J. Survival of Schistosoma mansoni and other trematode larvae in the snail Biomphalaria glabrata—a discussion of the interference theory. Trop Geogr Med. 1982;34(2):111–122. [PubMed] [Google Scholar]
- 84.Walker A.J. Do trematode parasites disrupt defence-cell signalling in their snail hosts? Trends Parasitol. 2006;22(4):154–159. doi: 10.1016/j.pt.2006.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Supplementary Materials
Online Supplementary Materials