Abstract
Gangliogliomas are generally benign tumors, composed of transformed neuronal and glial elements, with rare malignant progression of the glial component. The current study of a rare case of a woman harboring a ganglioglioma with areas of malignant transformation addresses two fundamental questions: (1) Are the ganglioglioma and its malignant component clonal in origin? (2) What are the genetic alterations associated with the initiation and subsequent malignant progression of ganglioglioma? By using the human androgen receptor gene (HUMARA) assay, we found the ganglioglioma and the malignant component to be clonal in origin, suggestive of initial transformation of a single neuroglial precursor cell with subsequent malignant progression. Conventional and array comparative genomic hybridization (approximately 2.5-Mb resolution) analyses found chromosomal losses to be predominant in the benign areas of the ganglioglioma, with gains more prevalent in the malignant component. Regions of chromosomal loss, postulated to harbor genes involved in the initiation of ganglioglioma, included 1p35-36, 2p16-15, 3q13.1-13.3, 3q24-25.3, 6p21.3-21.2, 6q24-25.2, 9p12, Xp11.3-11.22, and Xq22.1-22.3. Direct analysis demonstrated loss of p19 expression and p53 mutation in the malignant areas, highly suggestive of these alterations being involved in the malignant progression of the ganglioglioma. Additional chromosomal alterations specific to the malignancy involved gains on 1p35-34.2, 2q24.1-32.3, 3q13.1-13.3, 6q13-16.2, 7q11.2-31.3, 8q21.1-23, 11q12-31, and 12q13.2-21.3. This molecular-pathological study has provided insight into the pathogenesis of gangliogliomas and associated rare malignant progression. Deciphering the specific genes residing in these chromosomal regions may further our understanding of not only these rare tumors but also the more common gliomas.
Keywords: array CGH (aCGH), comparative genomic hybridization (CGH), ganglioglioma, human androgen receptor gene (HUMARA), malignant ganglioglioma
Gangliogliomas are rare intra-axial tumors, accounting for approximately 1% of all CNS tumors (Johannsson et al., 1981; Kleihues and Cavanee, 2000; Russell and Rubinstein, 1989; Sutton et al., 1983). Although gangliogliomas have been reported in persons of all ages, they are most commonly diagnosed in young adults and children, where they often present as structural epileptogenic lesions of the temporal lobe (Russell and Rubinstein, 1989; Sutton et al., 1983). Histologically, gangliogliomas are mixed neoplasms, in which both neuronal and glial components coexist, with the glial component usually corresponding to a WHO grade I or II glioma (Kleihues and Cavanee, 2000; Russell and Rubinstein, 1989). Rarely will a ganglioglioma undergo malignant transformation in the glial portion of the tumor, with the presence of both a benign and a malignant component being extremely rare. Clonality studies are suggestive of transformation of a common putative neuroglial precursor cell to give rise to a ganglioglioma (Zhu et al., 1997); however, whether there is subsequent clonal progression in those benign tumors that will on rare occasion progress to a more malignant tumor is not known. In addition, the genetic alterations that may underlie this initial transformation and subsequent rare progression are also not known. In this study, we used the occasion of a patient presenting with a rare ganglioglioma with an associated malignant component to undertake molecular-pathological correlative studies to gain initial insights into both the issues of clonality and genetic alterations.
Few molecular and cytogenetic studies have been performed on gangliogliomas to date. The molecular studies that have been undertaken mainly involve specific genetic alterations commonly associated with more prevalent glioma subtypes. These include detection of p53 mutations, without epidermal growth factor receptor (EGFR) amplification in benign gangliogliomas (Fukushima et al., 2005; Hayashi et al., 2001).3 Another study involved analysis of a ganglioglioma with a malignant component (Kim et al., 2003), with p53 mutation and loss of p16 expression secondary to methylation detected in the malignant recurrence.
In the current study, we separated a rare low-grade ganglioglioma from a coexisting malignant component by microdissection to undertake specific analysis of known genetic alterations in gliomas. In addition, we used conventional and high-resolution array comparative genomic hybridization (aCGH) analysis to decipher chromosomal regions of loss and/or gain in the two regions of the tumor. We postulated that these chromosomal regions likely harbor genes involved in the transformation of a neuroglial precursor to a ganglioglioma and subsequent malignant progression.
Materials and Methods
Clinical Summary
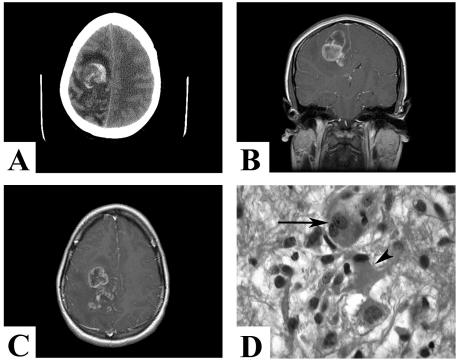
A 20-year-old healthy woman presented with a three-week progressive history of headaches, nausea and vomiting, blurry vision, and left-sided paresthesia. Outpatient examination revealed early papilloedema, without any focal or global neurological deficits. While awaiting her imaging studies, the patient’s condition acutely deteriorated with a decreased level of consciousness and progressive left hemiparesis (grade 3/5), requiring urgent neurosurgical admission. Emergency CT scan without contrast demonstrated a right frontal-parietal lesion with hemorrhage, edema, and shift (Fig. 1A). Emergent MRI study revealed a large, multilobulated, contrast-enhancing right frontal-parietal lesion with perilesional edema and midline shift, suggestive of a glioblastoma (Fig. 1B and C).
Fig. 1.
(A) Presenting CT scan without contrast demonstrating a right frontal-parietal lesion with intralesional hemorrhage, edema, and shift. (B and C) Gadolinium-enhanced coronal (B) and axial (C) MR images demonstrating a large multilobulated, contrast-enhancing right frontal-parietal lesion with perilesional edema and midline shift. (D) The benign component of the tumor, composed of a ganglioglioma containing neurons (arrow), many of them binucleate, as well as large astrocytic cells (arrowhead) arranged in clusters in the neuropil. Hematoxylineosin, 630×.
Emergent right parasagittal awake craniotomy with stereotaxy guidance, cortical stimulation to localize the primary motor and sensory cortex, and entry through an enlarged and electrically silent gyrus were undertaken for the purpose of diagnosis and debulking. A firm, whitish invasive lesion was initially encountered, which on quick section was designated as a glial neoplasm. Subsequent deeper dissection revealed softer and purplish tumor, quick section of which indicated a high-grade glial tumor intermixed with intratumoral hemorrhage. Radical but subtotal resection of the tumor was undertaken, with intraoperative monitoring of left-side motor function. Macroscopic residual tumor was left posteriorly and laterally to minimize the risk of postoperative motor deficit.
The postoperative course in hospital was complicated by the development of left deep venous calf thrombosis, requiring anticoagulation therapy approximately one week after surgery. Over two weeks, the patient’s neurological status improved compared with her preoperative state, with normal cognitive function and almost normal (grade 4+/5) power in her left extremity. External beam radiation (5000 cGy in 25 fractions) was administered, with the tumor controlled for approximately 14 months. Upon documented growth of the posterior-lateral residual tumor accompanied by progressive left-sided hemi-paresis, temozolomide was recommended. However, the patient and family did not wish to pursue additional therapy, and she died of her disease approximately 18 months after surgery.
Neuropathological Analysis
The specimens isolated from the initial whitish and subsequent deeper purplish components of the tumor were fixed in 10% formalin for paraffin embedding and for routine and detailed neuropathological examination.
Extraction of DNA
Regions of benign and malignant components of the tumor were identified in paraffin sections and separated by microdissection. A large volume of tumor was received, and areas of confluent low-grade and high-grade tumor were readily identified. The isolated portions of the tumor were deparaffinized in three washes of xylene, followed by proteinase K digestion at 37°C for three days and then phenol-chloroform extraction of the DNA (Reifenberger et al., 1996; Zielenska et al., 2004).
Analysis of p53 and p16/p14
Extracted genomic DNA was evaluated for the presence of mutations in exons 5–8 of the p53 gene using the p53 Mutation Detection Kit (Panomics, Fremont, Calif.). We used 5'CpG island methylation, using methylation- specific PCR to screen for the methylation status of the p16/p14INK4A promoter region, using standard primers.
HUMARA Assay
As per standard protocols for the human androgen receptor gene (HUMARA) assay (Mutter and Boynton, 1995a, 1995b; Mutter et al., 1995; van Dijk et al., 2002), isolated DNA was digested for at least 16 hours at 37°C in a mixture containing 5 U of DdeI with or without 25 U of concentrated HhaI. After digestion, 5 μ l of the mixture was used for PCR amplification of the HUMARA locus. The PCR mixture contained 300 nmol/liter each of forward and reverse primer, 6% dimethyl sulfoxide, 2.5 mmol/liter dNTP, 1.5 mmol/liter magnesium chloride, and 1.25 U Amplitaq Gold (Applied Biosystems, Foster City, Calif.), in a total reaction mixture of 50 μ l. Primer sequences used were forward 5'-CCCCAGGCACCCAGAGGC-3' and reverse 5'-GAGAACCATCCTCACCCTGCT-3', and the PCR conditions were as follows: 7.5 min at 95°C; followed by three rounds of 2.5 min at 95°C, 30 s at 62°C, and 1 min at 71°C; followed by 32 rounds of 45 s at 95°C, 30 s at 62°C, and 1 min at 72°C; followed by a single step of 10 min at 72°C. Following the PCR, the products were analyzed on a 4% gel.
Comparative Genomic Hybridization
Metaphase spreads from normal human lymphocytes were prepared using standard protocols (Dracopoli, 1999). The slides were aged for 2–3 days before denaturation at 70°C in 70% formamide in 2× saline sodium citrate (SSC), followed by dehydration in a series of ethanol- water mixtures. The slides were treated with 0.1 μ g/ml proteinase K in 20 mM Tris (pH 7.5)–2 mM calcium chloride before hybridization. The comparative genomic hybridization (CGH) procedure was similar to published standard protocols (Kallioniemi et al., 1992; Pandita et al., 1999; Shridhar et al., 2001). Twenty images were captured using a Nikon Labophot-2 microscope equipped with an automatic filter wheel and a 83,000 filter set (Chroma, Brattleboro, Vt.) with single bandpass exciter filters for UV/fluorescein isothiocyanate (490 nm), DAPI (360 nm), and rhodamine (570 nm), and the images were analyzed using the QUIPS CGH software, version 3.12 (Vysis, Des Plaines, Ill.). Using this software, the rhodamine–fluorescein isothiocyanate signal ratio is expressed as a red:green ratio, with deviations from a 1:1 ratio indicative of gain or loss of chromosomal material. The lower and upper limits for gain and loss were established by performing control CGH experiments with a well-characterized tumor cell line (IMR32) and DNA derived from male or female normal tissue. On the basis of these findings, 95% confidence intervals for gain and loss were set at 1.20 and 0.80, respectively, with gene amplification defined as gain > 1.5 (Pandita et al., 1999; Shridhar et al., 2001).
Array Comparative Genomic Hybridization
cDNA Microarrays
The 19k6 microarray chip (Micro-array Centre, Toronto, Canada) used for CGH contains a total of 19,200 features or arrayed spots (Research Genetics, Invitrogen, Huntsville, Ala.), including 18,980 human cDNAs and 220 positive and negative control spots. The spots of the 19k6 microarray are arranged on one glass slide, with 32 subgrids spotted over an area of 18 × 36 mm2. A complete list of the cDNA collection and protocols used for microarray construction can be found at the University Health Network Clinical Genomics Center Web site (www.microarrays.ca).
DNA Labeling and CGH Hybridization
The protocol used was based on published methods for CGH analysis with cDNA glass microarray targets (Pollack et al., 1999). Briefly, 2 μ g each of genomic tumor and normal male reference DNA were labeled by random priming with Cy3-dCTP or Cy5-dCTP (Amersham, Baie D’Urfe, Canada) by use of the Bioprime labeling kit (Invitrogen, Burlington, Canada) in a 50-μ l reaction volume containing dATP, dGTP, and dTTP (120 μ M each), dCTP (60 μ M), and Cy5-dCTP or Cy3-dCTP (60 μ M). The Cy3-labeled and Cy5-labeled probes were purified with a Microcon 30 filter (Millipore, Bedford, Mass.) and combined with human Cot-1 DNA (40 μ g, Invitrogen), yeast tRNA (100 μ g, Invitrogen), and poly(dA-dT) (20 μ g; Sigma, Oakville, Canada). The hybridization mixture was concentrated by use of a Microcon 30 filter and reconstituted in 80 μ l of 3.4× SSC–0.3% sodium dodecyl sulfate. After denaturation at 100°C for 90 s and a 30-min Cot-1 preannealing step at 37°C, the probes were allowed to hybridize to the microarray surface at 65°C overnight in a humidified chamber. After hybridization, the arrays were washed in 2× SSC–0.03% sodium dodecyl sulfate at 65°C for 5 min, followed by a 5-min wash each with 1× and 0.2× SSC.
Microarray CGH Imaging and Analytical Software
The microarrays were scanned with the Axon GenePix 4000A confocal scanner, and the intensity of each fluorescence signal was collected separately and quantified with GenePix Pro 3.0 software (Axon Instruments, Union City, Calif.). Experimental variables of individual spot ratios were addressed by use of a sliding-window median normalization method (Beheshti et al., 2003). This algorithm utilizes a user-determined window of median calculation that is defined as the number of contiguous linear array spots along each chromosome. Starting at the pter of a chromosome, the median value of the intensity ratios of the window of spots normalized is determined. The window subsequently slides a single spot position toward the chromosome qter and repeats the median value determination for the next window. This process is reiterated until the end of the chromosome is reached, and all median values for each window along the chromosome are plotted. For each aCGH profile in this study, a sliding-window size of 15 spots was found to provide the optimal minimization of signal- to-noise ratio normalization for determining regions of imbalance. Copy-number gain was assigned when the mean normalization ratio for 10 features mapping to a contiguous genomic region was 1.5. Similarly, genomic loss was assigned when a contiguous gain had a ratio of 0.7.
The software suite used for the analysis included three PC-DOS–based components: normalize, project, and profiler (Beheshti et al., 2003). The normalize software normalizes quantified Cy5 and Cy3 fluorescence intensities, with or without background subtraction, across the entire array, as well as across individual sub-grids. Additionally, preceding normalization, filters are applied to remove spots below user-specified thresholds for fluorescence intensity, foreground-to-background ratio, and/or spot diameter size. Using project, normalized replicates of an experiment, including reverse labeling, can be grouped together as a project. Finally, profiler uses the updated chromosome localization information to generate composite chromosome plots of normalized fluorescence intensity ratios from selected projects, generating images analogous to karyotype profiles from chromosome CGH. Only results for cDNA sequences with known cytoband and megabase position were plotted.
Results
Neuropathological Examination
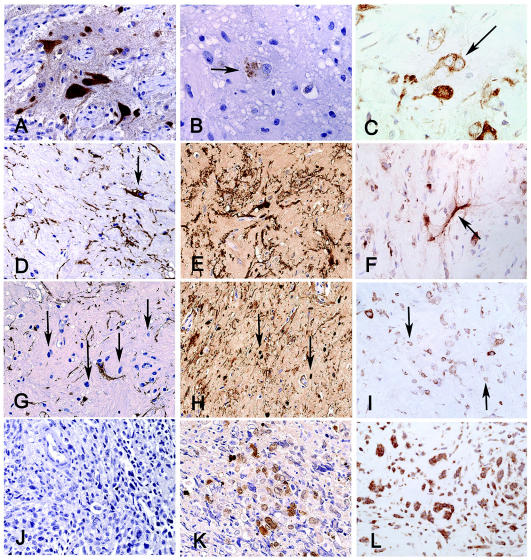
The superficial whitish, firm tumor corresponded to a WHO grade I ganglioglioma with abundant stromal collagen deposition. This part of the tumor was composed of irregular groups of large multipolar neurons (Fig. 1D, arrow), intermixed with large (Fig. 1D, arrowhead) and small glial cells. Immunohistochemical characterization demonstrated that both the neuronal and glial cellular elements were present in the benign portion of the ganglioglioma. The large multipolar neurons were neuronal nuclei (NeuN) positive (Fig. 2A), contained cytoplasmic chromogranin (Fig. 2B), and were often binucleate and stained for nestin (Fig. 2C). The large pleomorphic glial cells were positive for glial fibrillary acid protein (GFAP), S-100 protein, and nestin (Fig. 2D–F), whereas smaller stromal cells were negative for GFAP but positive for S-100 and nestin (Fig. 2G–I). The malignant component of the tumor was composed of confluent areas of small, cytologically malignant cells arranged in a loose myxoid stroma. The malignant cells were negative for GFAP but were variably and strongly positive for S-100 and nestin, respectively (Fig. 2J–L). MIB1 staining ranged from 9% to 20% in this component of the tumor. We found no morphological evidence of a neuronal population in the malignant component, nor did we detect immunoreactivity for neuron-specific epitopes (including NeuN, synaptophysin, chromogranin, and neurofilament).
Fig. 2.
Benign component of the lesion, containing (A) large NeuN-positive neurons (400×), which occasionally stained for (B) chromogranin (arrow) within the neuronal soma (630×), with some (C) being binucleate and nestin positive (arrow, 400×). (D) The large GFAP-positive astrocytes (400×) were also (E) S-100 positive (400×) and (F) nestin positive (400×). (G) Between the large glial and neuronal cells were additional stromal cells with small rounded nuclei (arrows) that did not stain for GFAP (400×) but did stain positive for both (H) S-100 and (I) nestin. The malignant component of the tumor was composed of small cells in a loose myxoid stroma and did not stain for (J) GFAP (400×) but demonstrated (K) variable nuclear and cytoplasmic reactivity for S-100 (400×) and (L) strong reactivity for nestin (400×).
p53 Mutation, p16/p14 Methylation, and Clonality Analysis
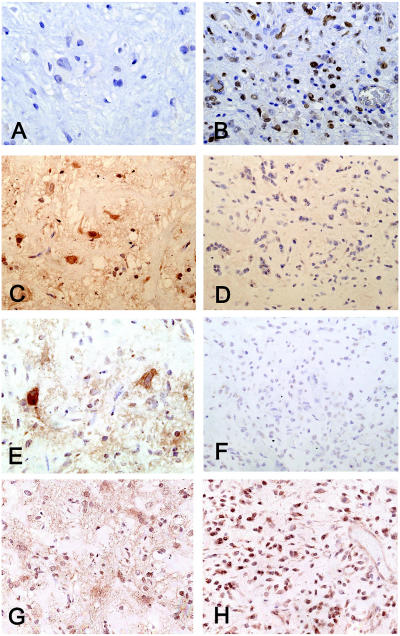
Analysis for alterations in p53, p16, p14, and EGFR status was undertaken in the benign and malignant areas of the tumor. p53 immunoreactivity was negative in both the neuronal and glial elements of the ganglioglioma (Fig. 3A), but the majority of the nuclei in the malignant component were labeled (Fig. 3B). p14 and EGFR immunoreactivity was strong in both cellular elements of the ganglioglioma (Fig. 3C and E) but negative in the malignant component (Fig. 3D and F). In contrast, p16 expression was readily detected in the benign and malignant areas of the tumor portions of the tumor (Fig. 3G and H).
Fig. 3.
(A) p53 protein was not detectable in the nuclei of the benign component of the ganglioglioma but was detected (B) in more than 60% of the nuclei in the malignant component. (C) p14 immunoreactivity was present in all cell types of the benign areas, but (D) immunoreactivity was essentially absent in the malignant component. (E) EGFR reactivity was positive in the benign areas but (F) negative in the malignant component. p16 immunoreactivity was present in both the benign (G) and malignant (H) components.
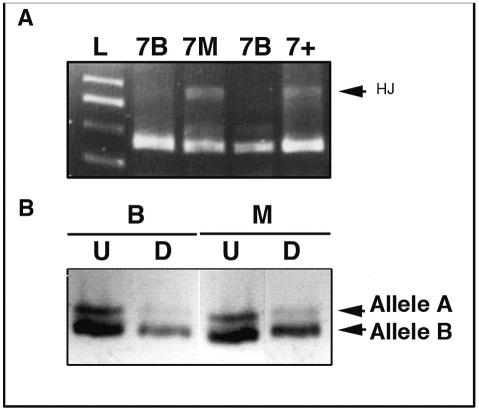
p53 mutational analysis of exons 5–8 revealed a band at approximately 800 bp, indicative of Holliday junction formation in exon 7 of the DNA isolated from the malignant component of the tumor (Fig. 4A, lane 7M). DNA from the benign ganglioglioma did not reveal any mutations in any of the exons analyzed (lanes 7B). The mutational result was consistent with the noted increased p53 immunoreactivity in the malignant portion of the tumor (Fig. 3B). Methylation-specific PCR analysis did not reveal any deletion or methylation of the p16/p14INK4A promoter region in either the benign or malignant component of the tumor, consistent with the positive p16 staining (Fig. 3G and H). To test for clonal evolution of the benign and malignant areas of the tumor, the HUMARA assay was used to detect the pattern of X chromosome inactivation using the human androgen receptor gene. The near absence of the higher-molecular-weight band (allele A) in both the HhaI-digested benign (lanes marked B) and malignant (lanes marked M) components was indicative of a clonal origin (Fig. 4B).
Fig. 4.
(A) Screening for the p53 mutation, based on the formation of Holliday junctions (HJs) when a nucleotide mismatch is present, was undertaken for exons 5–8. p53 mutations were not detected in any of the exons in the low-grade areas of tumor (7B); however, p53 mutation in exon 7 was observed in the malignant component (7M), denoted by the presence of HJs (arrow). The p53 mutation was consistent with the detection of p53 protein in the malignant component (Fig. 3B) (L, ladder; 7+, positive control for exon 7). (B) HUMARA analysis to determine clonality between the benign (B) and malignant (M) components. The near loss of the band for allele A in the digested (D) versus undigested (U) lanes for both the benign (B) and the malignant (M) components indicates they are likely clonal. The faint band in the digested lanes likely represents contamination of bystander nonclonal stromal cells.
Comparative Genomic Hybridization
To screen for additional genetic alterations in the benign and malignant areas of the tumor, we analyzed the microisolated genomic DNA by using CGH. The ganglioglioma harbored losses on chromosomal regions 1p35-pter and 4p16, whereas a gain was observed on 4q13-28 (Table 1A and Fig. 5A, bars). The +X and − Y were expected internal controls, because the analyzed tumor was from a female and the normal control DNA was male in origin. In comparison, the malignant component did harbor the chromosomal alterations noted in the aforementioned benign areas, which we postulate to contain genes involved in initiation or formation of the benign tumor. In addition to the common alterations, the malignancy-specific chromosomal alterations included loss of 12p13 and all of chromosome 13, while gains were observed on 2q22-31, 6q12-16, 12q13.3-21, and the whole of chromosome 7 (Table 1A and Fig. 5B, bars). We postulate these chromosomal regions harbor genes involved in clonal malignant progression.
Table 1A.
Genetic alterations detected by conventional comparative genomic hybridization of the benign and malignant components of the ganglioglioma
| Chromosome No. | Benign Component of the Tumor | Malignant Component of the Tumor |
|---|---|---|
| 1 | –1p35-pter | –1p35-pter |
| 2 | NAD | +2q22-31 |
| 4 | –4p16, +4q13-28 | –4p16, +4q13-28 |
| 6 | NAD | +6q12-16 |
| 7 | NAD | +7 |
| 12 | NAD | –12p13, +12q13.3-21 |
| 13 | NAD | –13 |
| X | +X | +X |
| Y | –Y | –Y |
Abbreviation: N AD, no abnormality detected.
Fig. 5.
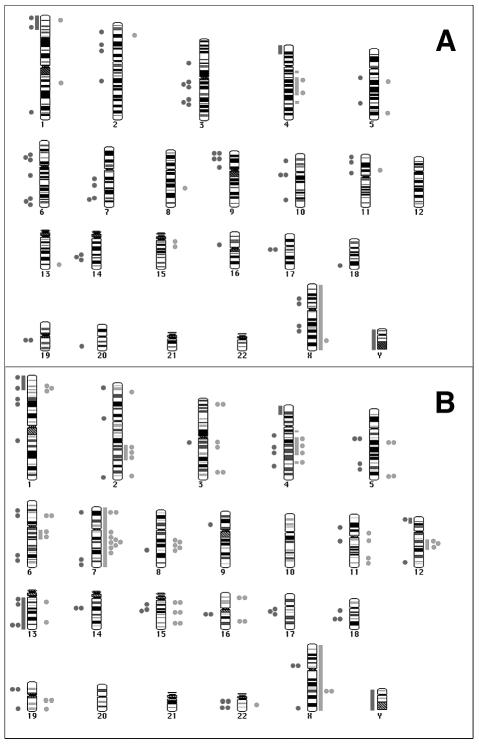
CGH and aCGH karyograms representing the gains and losses observed in the benign (A) and malignant (B) components. The bars represent the alterations observed by conventional CGH, whereas the dots (1 dot < 4 clones and 2 dots > 4 clones) represent the changes observed by high-resolution aCGH. Red indicates losses, whereas green indicates overall gains. The specific chromosomal regions and candidate genes within these regions are categorized in detail in the tables.
Array Comparative Genomic Hybridization
Higher-resolution screening was possible with aCGH (approximately 2–5 Mb) compared with the aforementioned CGH results (approximately 10–20 Mb), using the in-house cDNA 19k6 array. Of the 19,008 spots on the array, data from 18,300 (96%) were used for the analysis, with the rest being discarded because of insufficient quality. Of these 18,300 spots, 7000 were of unknown location on the genome, allowing identification of additional chromosomal alterations not detected by conventional CGH. The majority of the 7000 unknown spots had information regarding the chromosomal origin; however, specific band locations were not available. Similarly, some expressed sequence tags present on the array did not have any chromosomal band specific location. Only the data points that had complete chromosomal information were used for defining genetic alterations.
Alterations in the ganglioglioma as per aCGH were present throughout the genome, with more prevalent losses on chromosomes 1p35-36, 2p16-15, 3q13.1-13.3, 3q24-25.3, 6p21.3-21.2, 6q24-25.2, 9p12, Xp11.3-11.22, and Xq22.1-22.3 (Table 1B and Fig. 5A, dots). The malignant component revealed more gains, clustered on chromosomes 1p35-34.2, 2q24.1-32.3, 3q13.1-13.3, 6q13-16.2, 7q11.2-31.3, 8q21.1-23, 11q12-31, and 12q13.2-21.3, with some losses on chromosomes 1p, 4q, 6, 13, 15, 17q, and 19q (Table 1B and Fig. 5B, dots). Chromosomal alterations common to both the benign and malignant components of the tumor, with potential candidate linked genes involved in initiation and formation of the tumor, are listed in Table 2. Chromosomal alterations unique to only the malignant component, with potential candidate genes involved in the progression of the tumor, are listed in Table 3.
Table 1B.
Genetic alterations detected by array comparative genomic hybridization of the benign and malignant components of the ganglioglioma
| Chromosome No. | Benign Component of the Tumor | Malignant Component of the Tumor |
|---|---|---|
| 1 | –1p36.3, +1p36.2, –1p35, +1q23, –1q44.2 | –1p36.3, –1p35, +1p35, +1p34.2, –1p32.3, –1p31.1, –1q22 |
| 2 | –2p23, +2p22, –2p16, –2p15, –2q22 | –2p24, +2p23, –2p11.2, +2q24.1-24.3, +2q32.1-32.3, +2q37.1, |
| –2q37.3 | ||
| 3 | –3p14.3-14.1, –3q13.1-13.3, –3q24-25.3 | +3p25.1, –3q13.1, +3q13.1-13.3, +3q26.3-27 |
| 4 | +4q21 | –4q21.2, +4q21.3-24, –4q22.23, –4q31.2 |
| 5 | –5q13.2, +5q14, –5q31.2, +5q34 | –5q13.2, +5q14, –5q31.1-33.2, +5q34 |
| 6 | –6p21.3-21.2, –6q14, –6q24-25.2 | –6p21.3-21.2, +6p21.2-21.1, +6q13-16.2, –6q24-25.2 |
| 7 | –7q21.1, –7q22, –7q33 | –7p21, +7q11.22-31.3, –7q33.34 |
| 8 | +8q22.2 | +8q21.1-23, –8q22.1 |
| 9 | –9p24, –9p21, –9p12 | –9p12 |
| 10 | –10p13, –10q11.2, –10q25.1 | NAD |
| 11 | –11p15.4, +11p13-12, –11p11.2 | –11p13, +11p11.2, +11q12, –11q12-13.1 |
| 12 | NAD | –12p13.2, +12q13.2-21.3, –12q24.3 |
| 13 | +13q33 | –13q13, +13q13-14.1, –13q32-33, +13q22 |
| 14 | –14q21 | –14q21 |
| 15 | +15q12 | +15q12, –15q13, –15q21.1-21.3, +15q22.2, +15q25 |
| 16 | –16p11.2 | +16p13.2, –16q12.2-13, +16q22 |
| 17 | –17q21.2 | –17q21.2-21.3 |
| 18 | –18q23 | –18q11.2, –18q21.1-21.2 |
| 19 | –19q13.1 | –19p13.2-q13.1, +19q13.1-13.2, +19q13.2, –19q13.3 |
| 20 | –20q13.2 | NAD |
| 21 | NAD | NAD |
| 22 | NAD | –22q11.2, +22q12.2-12.3, 22q13.2 |
| X | –Xp11.3-11.22, –Xq22.1-22.3, +Xq26-27 | –Xp11.3-11.22, +Xq24, –Xq28 |
| Y | NAD | NAD |
Abbreviation: NAD, no abnormality detected.
Table 2.
Genetic alterations identified in both the benign and the malignant components of the ganglioglioma and candidate genes involved in the initiation or formation of the tumor in these chromosomal regions
| Chromosome No. | Conventional CGH | Array CGH | Candidate Genes |
|---|---|---|---|
| 1 | –1p35-pter, –1p35 | –1p36.3, –1p35 | TP73, DFFB, MYCL1 |
| 3 | No common alterations | –3q13.1 | |
| 4 | –4p16, +4q13–28 | +4q21.3-24 | SEPT11 , CCN1 |
| 5 | No common alterations | –5q13.2, +5q14, –5q31.2, +5q34 | FGF1, RHOBTB3 , GLRX |
| 6 | No common alterations | –6p21.3, –6q24-25.2 | PPP1R2P1 , TAP1 , SOD2 , WTAP |
| 7 | No common alterations | –7q36 | TRIM24 |
| 8 | No common alterations | +8q22.2 | OAZIN |
| 9 | No common alterations | –9p12 | ADFP |
| 14 | No common alterations | –14q21 | RBM25 |
| 15 | No common alterations | +15q12 | RASGRP1 |
| 17 | No common alterations | –17q21.2 | UBTF |
| X | No common alterations | –Xp11.3-11.22 |
Abbreviation: CGH, comparative genomic hybridization.
Table 3.
Genetic alterations identified as unique to the malignant component of the ganglioglioma and putative candidate genes involved in malignant progression within these chromosomal regions
| Chromosome No. | Conventional CGH | Array CGH | Candidate Genes |
|---|---|---|---|
| 1 | — | +1p35, +1p34.2, –1p32.3, –1p31.1, –1q22 | FGR, MYCL , SYDN3, CDKN2C, FAF1 |
| 2 | +2q22-31 | +2p23, –2p11.2, +2q24.1-24.3, | ASXL2 , FAP , COL3A |
| +2q32.1-32.3, +2q37.1, –2q37.3 | |||
| 3 | — | +3q25.1, +3q13.1-13.3, +3q26.3-27 | RAF1 , FSTL1 , PIK3CA |
| 4 | — | –4q22.23, –4q31.2 | |
| 5 | — | –5q33.2 | GEMIN5 |
| 6 | — | +6p21.2-21.1, +6q13-16.2 | PTK7, KNSL8 |
| 7 | +7 | +7q11.2-31.3 | FZD1, VIK , NRF1, CMET |
| 8 | — | +8q21.1-23, –8q22.1 | |
| 11 | — | +11q11.2-12 | API5 |
| 12 | –12p13, +12q13-21 | –12p13.2, +12q13.2-21.3, –12q24.3 | CCND2, GLI, MDM2 , CDK4 |
| 13 | –13 | –13q13, +13q13-14.1, –13q32-33 | WB4, GTF2F2 , LCP1 |
| 15 | — | –15q13, –15q21.1-21.3, +15q22.2, +15q25 | BMF, BUB1B , ITGA11 , AKAP13 |
| 18 | — | –18q11.2, –18q21.1-21.2 | |
| 19 | — | –19q13.1-13.2, +19q13.3 | INK4D |
| 22 | — | –22q11.2, +22q12.2-12.3, –22q13.2 | RBX1, BCR, PDGFB |
Abbreviation: CGH, comparative genomic hybridization.
Discussion
Gangliogliomas are rare benign mixed neuronal and glial tumors of the brain, likely arising from transformation of a neuroglial precursor cell, which on rare occasion progress to a higher-grade glial malignancy (Kim et al., 2003; Kleihues and Cavanee, 2000; Sutton et al., 1983; van Dijk et al., 2002). A rare presentation of a ganglioglioma with well-defined benign and malignant areas in the same patient provided an excellent opportunity to query whether there is a clonal progression of the tumor to a more malignant form and to further identify the potential genes involved in this progression. A previous report had shown the neuronal and glial components of the tumor to be of clonal origin (Zhu et al., 1997). Our HUMARA analysis (Fig. 4B) indicated a clonal progression of the benign component to a more malignant form of the tumor. We do, however, appreciate that the cells used for this analysis included not only transformed cells but also a small amount of contaminating nontransformed stromal and bystander cells (approximately 5%–7%), hence the near absence, rather than complete absence, of allele A in the HUMARA assay for clonality (Fig. 4B). Isolating different cell types by using laser capture microdissection within the tumor would theoretically be helpful in avoiding this contamination, but this was not technologically possible in these samples. However, in further support of the HUMARA results, the common chromosomal alterations between the benign and malignant areas of the tumor illustrated by CGH or aCGH were also indicative of a clonal origin of both components of the tumor (Table 2).
The genetic events involved in the initiation and formation of gangliogliomas and the rare progression to malignancy are not well known (Fukushima et al., 2005; Kim et al., 2003; Kleihues and Cavanee, 2000). Genetic alterations prevalent in both the benign and malignant components of the tumor can be postulated to be of relevance toward initiation and formation of the ganglioglioma. Using conventional and high-resolution aCGH, several regions of chromosomal alterations that are common were noted (Table 2 and Fig. 5). Some of the candidate genes, such as TP73 and MYCL within these regions, have already been linked to tumor formation, and their role in glial transformation merits additional study. In contrast to these common genetic alterations, those that were unique to the malignant component would likely be involved in progression. Aberrations in cell-cycle regulatory genes are one such group of progression factors, including loss of p14 and p53 mutation, with associated increased p53 immunoreactivity found specifically in the malignant component (Fig. 3). Loss of p16 expression by direct mutation or promoter methylation has been detected in high-grade gangliogliomas and sporadic glioblastomas (Hulleman and Helin, 2005; Kleihues and Cavanee, 2000; Moulton et al., 1995; Ueki et al., 1996). In the current study, we did not detect any chromosomal deletion of the 9p21 loci, methylation of the p16/p14INK4A promoter, or loss of p16 expression in the malignant component (Fig. 3). However, there was loss of chromosomal region 19p13.2, which harbors INK4D or CDKN2D, another cyclin-dependent kinase inhibitor family member. Of interest, although we identified loss of chromosomal regions on 13q by aCGH (Table 3), this was proximal to the Rb1 locus. These cumulative data do suggest that alterations in a variety of cell-cycle regulatory proteins are involved in progression of benign ganglioglioma to increased malignancy.
Conventional and high-resolution aCGH (approximately 2–5 Mb) revealed additional alterations of chromosomal regions, specifically in the malignant component putatively containing candidate genes involved in progression (Table 3 and Fig. 5). We were interested in determining the similarities and differences between alterations found in the malignant component and those described for sporadic glioblastoma multiforme (GBM) (Hui et al., 2001; Kleihues and Cavanee, 2000; Nishizaki et al., 1998; Roversi et al., 2006), with both being pathologically classified by WHO as grade IV gliomas. The chromosomal regions observed to be common in the malignant component of this ganglioglioma and sporadic GBM included +1p35-34, +3q26-28, +7, +12q13-21, − 13q32-34, − 15q31, − 18q21, +19q, and − 22q13. Some common putative candidate genes within these regions are FGR, MYCL, CMET, PIK3CA, MDM2, GLI, and CDK4, with some of these genes already implicated in the GBM biology. However, there were several alterations on chromosomal regions 6q13-16.2, 2q22-31, 17q21, and 11q that were unique to the malignant component of the ganglioglioma and that have not yet been associated with sporadic GBM, although they have been linked to other human cancers (Eclache et al., 2005; Roy et al., 2006; Sakakura et al., 1999; Shadeo and Lam, 2006; Shibata et al., 2005; Uribe et al., 2005). Specific differences include lack of EGFR amplification or overexpression in the malignant component, which was present in the benign ganglioglioma. Of note, the gain of chromosome 7 by CGH in the malignant component was localized by aCGH to a region harboring genes such as CMET and not the region containing EGFR (Table 3). This is in contrast to sporadic gliomas, in which aberrations of EGFR expression and activation are the most common gain-of-function alterations in GBM but are not associated with low-grade gliomas (Kleihues and Cavanee, 2000). A second specific difference is the mutation and/or loss of expression of PTEN seen in approximately 90% of sporadic GBM, leading to activation of the PI3-kinase signaling pathway (Kleihues and Cavanee, 2000). Alterations of PTEN or other direct members of the PI3-kinase pathway were not observed in either the low- or high-grade elements of this tumor and have not been previously reported in the literature with these gliomas.
In summary, this molecular-pathological study of a rare presenting ganglioglioma with distinct benign and malignant components led to several molecular insights that require further detailed verification studies. Our results are in support of a clonal transformation of a neuroglial precursor cell leading to the formation of the benign ganglioglioma and its subsequent malignant progression, resulting from additional genetic alterations. These genetic alterations include cell-cycle regulatory genes, including p53. Many of these progression alterations are similar to those of sporadic GBM; however, significant differences, such as in EGFR amplification and PTEN loss, exist. Admittedly, these postulates are based on the analysis of a single patient tumor; however, such a detailed study can be highly informative in our overall understanding of the molecular pathogenesis of the diverse tumor types that compose the pathological entity known as glioma.
Acknowledgment
The authors thank Dr. Jeremy Squire and Jane Bayani for the help provided in the CGH analysis.
Footnotes
This work was supported by grants from National Cancer Institute of Canada (NCIC) to A.G. A.P. is the recipient of a postdoctoral fellowship from the American Brain Tumor Association.
Abbreviations used are as follows: aCGH, array CGH; CGH, comparative genomic hybridization; EGFR, epidermal growth factor receptor; GBM, glioblastoma multiforme; GFAP, glial fibrillary acid protein; HUMARA, human androgen receptor gene; NeuN, neuronal nuclei; SSC, saline sodium citrate.
References
- Beheshti B, Braude I, Marrano P, Thorner P, Zielenska M, Squire JA. Chromosomal localization of DNA amplifications in neuroblastoma tumors using cDNA microarray comparative genomic hybridization. Neoplasia. 2003;5:53–62. doi: 10.1016/s1476-5586(03)80017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracopoli, N. (1999) Current Protocols in Human Genetics New York: John Wiley and Sons.
- Eclache V, Viguie F, Frocrain C, Cassinat B, Chomienne C, Cymbalista F, Fenaux P. A new variant t(6;15;17)(q25;q22;q21) in acute promyelocytic leukemia: Fluorescence in situ hybridization confirmation. Cancer Genet Cytogenet. 2005;159:69–73. doi: 10.1016/j.cancergencyto.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Fukushima T, Katayama Y, Watanabe T, Yoshino A, Komine C, Yokoyama T. Aberrant TP53 protein accumulation in the neuronal component of ganglioglioma. J Neurooncol. 2005;72:103–106. doi: 10.1007/s11060-004-3549-2. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Iwato M, Hasegawa M, Tachibana O, von Deimling A, Yamashita J. Malignant transformation of a gangliocytoma/ganglioglioma into a glioblastoma multiforme: A molecular genetic analysis. Case report J Neurosurg. 2001;95:138–142. doi: 10.3171/jns.2001.95.1.0138. [DOI] [PubMed] [Google Scholar]
- Hui AB, Lo KW, Yin XL, Poon WS, Ng HK. Detection of multiple gene amplifications in glioblastoma multiforme using array-based comparative genomic hybridization. Lab Invest. 2001;81:717–723. doi: 10.1038/labinvest.3780280. [DOI] [PubMed] [Google Scholar]
- Hulleman E, Helin K. Molecular mechanisms in gliomagenesis. Adv Cancer Res. 2005;94:1–27. doi: 10.1016/S0065-230X(05)94001-3. [DOI] [PubMed] [Google Scholar]
- Johannsson JH, Rekate HL, Roessmann U. Gangliogliomas: Pathological and clinical correlation. J Neurosurg. 1981;54:58–63. doi: 10.3171/jns.1981.54.1.0058. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Kim NR, Wang KC, Bang JS, Choe G, Park Y, Kim SK, Cho BK, Chi JG. Glioblastomatous transformation of ganglioglioma: Case report with reference to molecular genetic and flow cytometric analysis. Pathol Int. 2003;53:874–882. doi: 10.1046/j.1440-1827.2003.01574.x. [DOI] [PubMed] [Google Scholar]
- Kleihues, P., and Cavenee, W. (2000) Pathology and Genetics of the Tumours of the Nervous System Lyon: IARC.
- Moulton T, Samara G, Chung WY, Yuan L, Desai R, Sisti M, Bruce J, Tycko B. MTS1/p16/CDKN2 lesions in primary glioblastoma multiforme. Am J Pathol. 1995;146:613–619. [PMC free article] [PubMed] [Google Scholar]
- Mutter GL, Boynton KA. PCR bias in amplification of androgen receptor alleles, a trinucleotide repeat marker used in clonality studies. Nucleic Acids Res. 1995a;23:1411–1418. doi: 10.1093/nar/23.8.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutter GL, Boynton KA. X chromosome inactivation in the normal female genital tract: Implications for identification of neoplasia. Cancer Res. 1995b;55:5080–5084. [PubMed] [Google Scholar]
- Mutter GL, Chaponot ML, Fletcher JA. A polymerase chain reaction assay for non-random X chromosome inactivation identifies monoclonal endometrial cancers and precancers. Am J Pathol. 1995;146:501–508. [PMC free article] [PubMed] [Google Scholar]
- Nishizaki T, Ozaki S, Harada K, Ito H, Arai H, Beppu T, Sasaki K. Investigation of genetic alterations associated with the grade of astrocytic tumor by comparative genomic hybridization. Genes Chromosomes Cancer. 1998;21:340–346. doi: 10.1002/(sici)1098-2264(199804)21:4<340::aid-gcc8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Pandita A, Zielenska M, Thorner P, Bayani J, Godbout R, Greenberg M, Squire JA. A pplication of comparative genomic hybridization, spectral karyotyping, and microarray analysis in the identification of subtype-specific patterns of genomic changes in rhabdomyosarcoma. Neoplasia. 1999;1:262–275. doi: 10.1038/sj.neo.7900036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- Reifenberger J, Ring GU, Gies U, Cobbers L, Oberstrass J, An HX, Niederacher D, Wechsler W, Reifenberger G. Analysis of p53 mutation and epidermal growth factor receptor amplification in recurrent gliomas with malignant progression. J Neuropathol Exp Neurol. 1996;55:822–831. doi: 10.1097/00005072-199607000-00007. [DOI] [PubMed] [Google Scholar]
- Roversi G, Pfundt R, Moroni RF, Magnani I, van Reijmersdal S, Pollo B, Straatman H, Larizza L, Schoenmakers EF. Identification of novel genomic markers related to progression to glioblastoma through genomic profiling of 25 primary glioma cell lines. Oncogene. 2006;25 :1571–1583. doi: 10.1038/sj.onc.1209177. [DOI] [PubMed] [Google Scholar]
- Roy D, Calaf GM, Hande MP, Hei TK. Allelic imbalance at 11q23–q24 chromosome associated with estrogen and radiation-induced breast cancer progression. Int J Oncol. 2006;28:667–674. doi: 10.3892/ijo.28.3.667. [DOI] [PubMed] [Google Scholar]
- Russell, D., and Rubinstein, L. (1989) Tumours of Central Neuroepithelial Origin Baltimore: Williams and Wilkins.
- Sakakura C, Hagiwara A, Taniguchi H, Yamaguchi T, Yamagishi H, Takahashi T, Koyama K, Nakamura Y, Abe T, Inazawa J. Chromosomal aberrations in human hepatocellular carcinomas associated with hepatitis C virus infection detected by comparative genomic hybridization. Br J Cancer. 1999;80:2034–2039. doi: 10.1038/sj.bjc.6690638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadeo A, Lam WL. Comprehensive copy number profiles of breast cancer cell model genomes. Breast Cancer Res. 2006;8:R9. doi: 10.1186/bcr1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Uryu S, Kokubu A, Hosoda F, Ohki M, Sakiyama T, Matsuno Y, Tsuchiya R, Kanai Y, Kondo T, Imoto I, Inazawa J, Hirohashi S. Genetic classification of lung adenocarcinoma based on array-based comparative genomic hybridization analysis: Its association with clinicopathologic features. Clin Cancer Res. 2005;11:6177–6185. doi: 10.1158/1078-0432.CCR-05-0293. [DOI] [PubMed] [Google Scholar]
- Shridhar V, Lee J, Pandita A, Iturria S, Avula R, Staub J, Morrissey M, Calhoun E, Sen A, Kalli K, Keeney G, Roche P, Cliby W, Lu K, Schmandt R, Mills GB, Bast RC, Jr, James CD, Couch FJ, Hartmann LC, Lillie J, Smith DI. Genetic analysis of early- versus late-stage ovarian tumors. Cancer Res. 2001;61:5895–5904. [PubMed] [Google Scholar]
- Sutton LN, Packer RJ, Rorke LB, Bruce DA, Schut L. Cerebral gangliogliomas during childhood. Neurosurgery. 1983;13:124–128. doi: 10.1227/00006123-198308000-00003. [DOI] [PubMed] [Google Scholar]
- Ueki K, Ono Y, Henson JW, Efird JT, von Deimling A, Louis DN. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res. 1996;56:150–153. [PubMed] [Google Scholar]
- Uribe P, Wistuba II, Solar A, Balestrini C, Perez-Cotapos ML, Gonzalez S. Comparative analysis of loss of heterozygosity and microsatellite instability in adult and pediatric melanoma. Am J Dermatopathol. 2005;27:279–285. doi: 10.1097/01.dad.0000171599.40562.7c. [DOI] [PubMed] [Google Scholar]
- van Dijk JP, Heuver LH, van der Reijden BA, Raymakers RA, de Witte T, Jansen JH. A novel, essential control for clonality analysis with human androgen receptor gene polymerase chain reaction. Am J Pathol. 2002;161:807–812. doi: 10.1016/S0002-9440(10)64240-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JJ, Leon SP, Folkerth RD, Guo SZ, Wu JK, Black PM. Evidence for clonal origin of neoplastic neuronal and glial cells in gangliogliomas. Am J Pathol. 1997;151:565–571. [PMC free article] [PubMed] [Google Scholar]
- Zielenska M, Marrano P, Thorner P, Pei J, Beheshti B, Ho M, Bayani J, Liu Y, Sun BC, Squire JA, Hao XS. High-resolution cDNA microarray CGH mapping of genomic imbalances in osteosarcoma using formalin-fixed paraffin-embedded tissue. Cytogenet Genome Res. 2004;107:77–82. doi: 10.1159/000079574. [DOI] [PubMed] [Google Scholar]