Abstract
The disparate lengths of survival among patients with malignant astrocytic gliomas (anaplastic astrocytomas [AAs] and glioblastoma multiforme [GBM]) cannot be adequately accounted for by clinical variables (patient age, histology, and recurrent status). Using real-time quantitative reverse transcription–polymerase chain reaction, we quantified the expression of four genes that were putative prognostic markers (CDK4, IGFBP2, MMP2, and RPS9) in a set of 43 AAs, 41 GBMs, and seven adjacent normal brain tissues. We previously explicated the expression and prognostic value of PAX6, PTEN, VEGF, and EGFR in these glioma tissues and established a comprehensive prognostic model (Zhou et al., 2003). This study attempts to improve that model by including four additional genetic markers, which exhibited a differential expression (P < 0.001) among tumor grades and between tumor and normal tissues. By including eight log-scaled gene expression variables, three clinical variables, and interaction terms among the eight genes, we established a prognostic model that accounted for two thirds of the variation (R2) in survival for this set of patients. To improve the R2 of the model without compromising its clinical utility, our data demonstrated that incorporating genes from different pathways markedly strengthens the model. Spearman rank correlation analysis of gene expression demonstrated a statistically significant positive correlation (P < 0.01) between the expression of IGFBP2–MMP2 and IGFBP2–VEGF in GBMs, but not in AAs. This finding suggests that the expression of IGFBP2 is associated with pathways activated specifically in GBMs that result in enhancing invasiveness and angiogenesis.
Keywords: differential gene expression, astrocytic gliomas, CDK4, IGFBP2, MMP2, RPS9, real-time quantitative RT-PCR, prognostic model
The Central Brain Tumor Registry of the United States reports that 44.4% of primary brain and central nervous system tumors are gliomas, with astrocytic gliomas accounting for 77.5% of all gliomas (CBTRUS, 2002). The WHO classification scheme divides the astrocytic variant of these tumors into four grades (Kleihues, 2000). Grade I is benign pilocytic astrocytoma. Grade II is low-grade malignancy. Grade III is anaplastic astrocytoma (AA),3 and grade IV is glioblastoma multiforme (GBM). Grade IV tumors, whether derived de novo (primary) or from a previously diagnosed lower grade astrocytoma (secondary), are highly malignant and are also the most common. The survival of patients with astrocytic gliomas is highly correlated with their WHO tumor grade. However, even with aggressive treatment using standard multimodality therapies, the overall outlook for patients with WHO grade III and IV tumors remains poor, although the prognosis for individual patients within a given histologic group is variable (Daumas-Duport et al., 1988; Lantos, 1997).
Extensive efforts have been expended to link cytogenetic and genetic dysregulation with the outcomes of patients with glioma, including variable gene expression and gene products involved in multiple pathways. Immunohistochemistry (IHC) is the most commonly used method for such analysis. To date, no comprehensive molecular prognostic model has been developed that is capable of predicting survival or responses to different therapeutic options for individual patients within a histologic subgroup. This deficit is due largely to the heterogeneity of tumors within each diagnostic class and the ineffectiveness and expense of current screening techniques. Genetic screening to identify mutations, amplifications, or deletions of genes and chromosomes can be laborious, costly, and time-consuming when using either genome-wide or locus-by-locus screening techniques. Also, data from IHC staining and gene microarrays are noisy. In addition to difficulties deciphering the comparability of data from microarrays using different analytical platforms (Kuo et al., 2002), statistical complexity is inherent to the use of microarray data for establishing prognostic models with clinical applicability. All the statistical issues faced when looking at a modest list of genetic markers are that much more overwhelming when facing a huge list of unselected markers in an array in that an approximate guideline for developing a prognostic factor model is to have 10 events for each marker (Katz, 1999). An approach that evaluates only a select number of gene expression variables quantified with a reliable technology could provide an efficacious alternative to wide-scale microarray assessment, ensuring the quality of the data generated and satisfying the statistical requirements needed to produce a meaningful analysis.
We exploited a real-time quantitative reverse transcription (QRT)-polymerase chain reaction (PCR) technology to increase the reliability, efficiency, and accuracy of quantifying gene expression in patients with malignant gliomas. With that reliable quantification approach we intend to combine a modest number of biomarkers (n = 8) from a modest size of patients (n = 84) to create an integrated prognostic model. The genes in this prognostic model for patients with WHO grade III or IV gliomas play critical roles in initiating and/or maintaining tumorigenesis. For example, PTEN is a well-studied tumor suppression gene, whereas EGFR, an oncogene, and VEGF, a proangiogenic gene, are instrumental in cancer progression. The CDK4 gene product enhances cell passage through the G1/S checkpoint, leading to cell proliferation. Amplification and expression of CDK4 have been reported in GBMs (Dirks et al., 1997; Lam et al., 2000; Rollbrocker et al., 1996). The MMP2 gene product plays a key role in extracellular matrix degradation, which is associated with invasion and angiogenesis (McCawley and Matrisian, 2001; Noel et al., 1997; Sato et al., 1994). The role of PAX6 in suppressing glioma growth was recently identified (Zhou et al., 2005), which is abrogating cellular invasiveness by blocking the expression of MMP2 (Y.-H. Zhou et al., manuscript in preparation). Although the role of IGFBP2 in cancer is not well known, its expression has been demonstrated in many malignant tumors, including gliomas (Akmal et al., 1995; Fuller et al., 1999; Godard et al., 2003; Richardsen et al., 2003; Sallinen et al., 2000). The only exceptional gene used in the model encodes a ribosomal protein small subunit 9 (RPS9), which was serendipitously discovered because of its inverse expression with glioma grade (Zhou et al., 2003).
Material and Methods
Brain Tumor cDNA Samples
cDNA samples of gliomas as well as adjacent normal brain tissue removed at surgery were created and utilized as previously described by Sano et al. (1999). All specimens were obtained from patients who underwent therapeutic procedures at the University of Texas M.D. Anderson Cancer Center (MDACC). Sections of samples were histologically evaluated by board-certified neuropathologists and classified according to a modified Ringertz grading system. Specimens were snap-frozen in liquid nitrogen and stored at −80°C. The purity of the tumor samples was estimated by using hematoxylin and eosin (H&E) staining to ensure that each sample contained >70% neoplastic cells. mRNAs were isolated from 10 frozen sections of each tumor specimen. We diluted the original cDNA samples with 10 mM of Tris-HCl (pH 7.5), and adjusted all dilutions so that the copy number of β-actin transcripts in each sample remained above 1000 copies per 4 μl per reaction. In our experience, this is the maximal degree of dilution needed to ensure the accurate and reproducible quantification of genes with a low- to mid-level of expression using real-time QRT-PCR.
Real-Time QRT-PCR
Standard
We used our previously reported approach for preparing real-time QRT-PCR standards for all eight genes (Zhou et al., 2003). Briefly, the cDNA fragments (200–500 base pairs) used as standards for each gene were amplified from the first-strand cDNA of glioma cell lines and then purified by using a QIAquick Gel Extraction kit (Qiagen, Valencia, Calif.) after running them through a 1.6% agarose gel. Each amplified DNA fragment was verified by sequencing, and the molecular weight was calculated based on the molecular weight of the four nucleotide bases and the composition of each base in the double-strand sequence. The DNA concentration was measured by reading optical density at 260 nm by using a spectrophotometer, and the molar concentration of the DNA was calculated. Each gene-specific standard DNA fragment was diluted to 4.15 amol/μl (107 molecules per 4 μl), from which six 10-fold serial dilutions were made. Four (102–105 molecules per 4 μl) dilutions were used in each run to calculate the copy number of the transcripts in the sample, which was based on the cycle number when PCR entered the stage of log-phase amplification. We normally obtained 1010 to 1011 copies of standard DNA amplified in four 50-μl PCR reactions for 33 cycles and stored them in aliquots of serial dilutions in 10 mM of Tris-HCl (pH 7.5) at −20°C. Thus, the standard for each gene that we developed for real-time QRT-PCR can be used for millions of reactions or to screen millions of samples.
Primers
When using SYBR Green I to quantify the cDNA copy number of a given gene in real-time PCR, it is critical for the primer set (forward and reverse primers) to specifically amplify the transcripts of candidate marker genes. By searching and comparing the primer sequences against the human genome database, we ensured that the primer set was designed to amplify only the cDNA of the selected gene, not the corresponding genomic fragment of that gene and its pseudogene(s), which commonly exist in the human genome. This technique eliminates deriving false positive results from real-time QRT-PCR of genes with a low level of expression. Although RNA samples can be treated by RNase-free DNase before reverse transcription to cDNA to avoid DNA contamination, working on large numbers of samples can be highly laborious. Additionally, cDNA samples from different laboratories could have been made without DNase treatment to the RNA samples, such as the glioma cDNA we used in this study. To achieve this goal, the primers were designed to anneal to different exons with intervening intron(s) that were greater than approximately 1 kb. If this criterion was not met, at least one primer was designed to anneal to the exon/intron boundary region. To avoid amplifying pseudogene(s), we ensured that the regions used for annealing primers had at least 30% mismatches over the entire primer sequences or 20% mismatches in the 3′ end of the primer sequence as compared with the homologous regions in the pseudogene(s). Table 1 lists the sequence and PCR condition for QRT-PCR for each gene described here. For each gene, we prepared 1 ml of 50× primer stock containing the forward and reverse primers and MgCl2, which can be used for 50,000 reactions.
Table 1.
Primer sequences and parameters for quantifying the expression of human genes using real-time QRT-PCR
| Gene | Primer set (5′-3′) | Amplicon | MgCl2(mM) | Parameter for amplification cycle1 |
|---|---|---|---|---|
| β-actin | TCCTTCCTGGGCATGGAGT
AAAGCCATGCCAATCTCATC |
419 bp | 3.5 | 95°C, 0″; 62°C, 5″; 72°C, 15″; 83°C, 2″ |
| CDK4 | TGGTGACAAGTGGTGGAAC
CAGAAGAGAGGCTTTCGAC |
205 bp | 3.5 | 95°C, 0″; 60°C, 5″; 72°C, 10″ |
| IGFBP2 | AGCAGGTTGCAGACAATGG
TCATCCGGAAGGCGCAT |
307 bp | 1.5 | 95°C, 0″; 63°C, 5″; 72°C, 12″; 84°C, 2″ |
| MMP2 | CAAAAACAAGAAGACATACATCTT
GCTTCCAAACTTCACGCTC |
232 bp | 3 | 95°C, 0″; 60°C, 5″; 72°C, 10″; 80°C, 2″ |
| RPS9 | GAAGCTGATCGGCGAGTATG
AGAGAAGTCAAGTGCTTCTG |
393 bp | 3.5 | 95°C, 0″; 62°C, 5″; 72°C, 15″; 83°C, 2″ |
Fluorescent signal was collected at the end of each amplification cycle. Parameters are expressed as degrees Celsius (°C) and seconds (″).
Real-Time QRT-PCR Procedure
Real-time QRT-PCR was conducted by using Roche’s LightCycler Instrument in 10 μl volume in glass capillaries (Roche, Indianapolis, Ind.), which contained 4 μl of standard/cDNA template, 1 μl of 10× primer-MgCl2 mix (final 0.5 μM primers, 1.5–3.5 mM of MgCl2), 4 μl of water, and 1 μl of Fast-Start DNA Master SYBR Green I mix (Roche). Fluorescence was measured at the end of each cycle during amplification. A melting-curve cycle was used to check the quality of the amplification to ensure that no primer dimer was formed. When there was a primer dimer in a reaction with a low copy number of the template (e.g., 10 copies of the molecule), we avoided picking up a fluorescent signal from the primer dimer DNA by collecting data at a higher temperature than 78°C, the temperature at which primer dimer DNAs will melt. The correlation coefficients, R = 1.00, and the standard error of the regression, <0.1, were typically achieved for crossing points and log concentrations of five 10-fold dilutions (101–105 copy number) of the standard DNA. The number of target copies in each diluted cDNA sample was normally within the range of 101 to 104, ensuring accurate determination. To further ensure accuracy, each reaction was repeated at least two times. When a variation between the two repeats was >2-fold, four to five repeats were performed for that sample. The mean value of the repeated copy numbers of each transcript was used to calculate the ratio between the marker gene and the internal control gene.
Statistical Methods for Prognostic Modeling
Prognostic data for each patient were collected from the Brain Tumor Center database at MDACC. Tumor recurrence data were obtained from the Department of Neurosurgery at MDACC. Twelve of 43 AAs and 10 out of 41 GBMs were recurrent tumors. Three of 10 recurrent GBMs were from lower grade gliomas, whereas the rest were recurrences of GBMs. The recurrent tumor histology was diagnosed at the time of recurrence. Survival was calculated from the date of surgery to the last contact date and includes patients with recurrent tumors. The last contact date occurred no later than February 25, 2003, for all patients in the study. All patients with GBMs were deceased, including two patients with whom we lost contact after 1995. Thirty-five of 43 patients with AAs were deceased, including six patients with whom we lost contact after 2001. Thus, our survival data are sufficiently mature for drawing meaningful predictions of long-term survival with our model. Among the 84 tumors considered in this study, 92% were collected before 1995, with only seven tumor samples collected between 1996 and 1998. All patients underwent similar treatment regimens, which included surgery followed by radiation therapy, with or without chemotherapy. Because of the lack of demonstrated efficacy for chemotherapy in glioma treatment, we did not subdivide our patients by differing responses to chemotherapy.
We compared the relative expression of genes among different WHO grades using the Wilcoxon rank sum test and correlated the results with survival in a Cox proportional hazards (Cox PH) regression analysis. In our earlier prognostic model, gene expression variables were dichotomized by optimal cutpoints derived from recursive partitioning analysis (RPA). Driven by concerns about the reproducibility of the RPA-derived model, we changed our strategy in this study. The new model uses log-scaled gene expression data but no optimal cutpoints for the four previously studied genes (PAX6, PTEN, VEGF, and EGFR). It also incorporates four new genes (CDK4, IGFBP2, MMP2, and RPS9) that have potential prognostic significance. Compared with a log-scaled continuous expression variable model, dichotomizing expression variables produced a model with a higher and biased R2 value. Thus, we elected to use log-scaled gene expression data in our prognostic model.
We used a backward variable elimination by applying the Akaike information criterion (AIC) (Akaike, 1973; Harvey, 1981). This method allowed us to minimize the number of variables in our survival model through the use of a likelihood-based measurement of R2 as a gauge of predictive accuracy to quantify the approximate proportion of variation in survival explained by a particular statistical model. While this R2 statistic is not truly equal to the proportion of explained variation for the Cox PH model, it is nonetheless a useful measure and has the same general interpretation. The Spearman rank correlation test assessed the possibility of a paired correlation existing among gene expression data. All statistical analyses were performed with S-Plus 2000 computer software (MathSoft Inc., Seattle, Wash.).
Results
Differential Expression of CDK4, MMP2, IGFBP2, and RPS9 in Malignant Astrocytic Gliomas
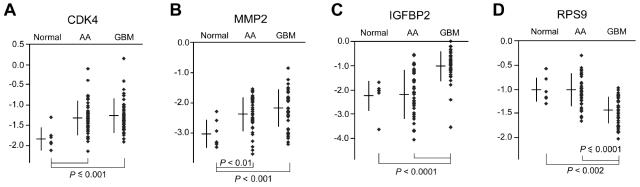
We used a set of gliomas (45 AAs and 42 GBMs) to establish beforehand a statistical prognostic model based on seven variables (4 gene expression variables [PAX6, PTEN, EGFR, and VEGF] and three clinical variables [patient age, tumor histology, and tumor recurrence status]). From these, we used 43 AA and 41 GBM cDNA samples to evaluate an additional four genes, CDK4, MMP2, IGFBP2, and RPS9. Except for RPS9, these additional genes are potential prognostic markers for malignant astrocytic gliomas identified through DNA analysis, microarray, and IHC approaches. Using real-time QRT-PCR, we quantified the expression of these new genetic markers. β-actin expression was characterized in our previous study in this set of samples, in which we quantified the degree of transcription from the same aliquot of cDNA samples for three housekeeping genes (β-actin, GAPDH, and enolase-α). We determined that the differences in their expression in AA versus GBM were negligible and arbitrarily selected β-actin for normalizing the expression of genetic markers and comparing their relative levels of expression. Figures 1A and B and Table 2 show that the expression of CDK4 and MMP2 was markedly upregulated in both AAs and GBMs compared with adjacent normal tissues (P < 0.001). The expression of IGFBP2 was strikingly elevated in GBMs (Fig. 1C and Table 2) as compared with its expression in normal tissues and in AAs (P < 0.0001). We previously evaluated RPS9 as an internal control and unexpectedly found that its expression was reduced significantly in GBMs as compared to normal tissues and AAs (P < 0.001) (Fig. 1D).
Fig. 1.
Analysis of the relative levels of expression of CDK4, MMP2, IGFBP2, and RPS9 by real-time QRT-PCR in 43 AAs, 41 GBMs, and seven samples of adjacent normal tissue, normalized to β-actin. The y-axis is a log 10–transformed ratio of target gene versus β-actin. The bar and vertical line for each class indicate the mean and standard deviation, respectively. Statistical significance is shown by P values from a Wilcoxon rank sum test based on gene expression ratio.
Table 2.
Median levels of relative gene expression in gliomas and normal tissues detected by real-time QRT-PCR after normalizing data to β-actin
| Marker | Normal n = 7 | AA n = 43 | GBM n = 41 |
|---|---|---|---|
| RPS9 | 0.0681 | 0.0898 | 0.0418 |
| CDK4 | 0.0120 | 0.0455 | 0.0462 |
| IGFBP2 | 0.0088 | 0.0078 | 0.1111 |
| MMP2 | 0.0005 | 0.0042 | 0.0067 |
Abbreviations: AA, anaplastic astrocytoma; GBM, glioblastoma multiforme
We compared the differences in expression of the four genes in the old model and the four new genes in the present model by quantifying expression levels in tumors and adjacent normal tissues from the same patients (4 with GBMs and 2 with AAs). We have shown that PAX6 expression was low in all four GBMs and in one AA (Zhou et al., 2003) and have included it in Table 3 for comparison with the other seven gene expressions. In comparison to the adjacent normal tissues, RPS9 and PTEN expression was low in three out of four GBMs and in the 2 AAs (the patient number is the same as earlier reported, but we did not have enough cDNA from the GBM of patient #5 to include in this study). The expression levels of MMP2, IGFBP2, VEGF, and CDK4 were high in the four GBMs. Elevated expressions for these four genes were not observed in the two AAs. In fact, VEGF expressions in the two AAs were decreased.
Table 3.
Comparison of gene expression in tumors and that in the surrounding normal tissues
| Patient | Grade | PAX6 | PTEN | RPS9 | EGFR | MMP2 | IGFBP2 | VEGF | CDK4 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | GBM | 0.09 | 0.30 | 0.24 | 0.14 | 10.67 | 22.44 | 2.69 | 2.96 |
| 2 | GBM | 0.12 | 5.97 | 0.27 | 1.47 | 46.29 | N/N | 8.54 | 2.23 |
| 3 | GBM | 0.52 | 0.25 | 0.83 | 18.15 | 67.95 | 21.23 | 2.54 | 3.29 |
| 4 | GBM | 0.25 | 0.03 | 0.99 | 0.95 | 54.56 | 4.83 | 5.25 | 2.71 |
| 6 | AA | 0.52 | 0.26 | 0.56 | 0.45 | 0.54 | 2.38 | 0.41 | 0.71 |
| 7 | AA | 1.04 | 0.46 | 0.70 | 3.87 | N/B | 0.39 | 0.25 | 1.89 |
Abbreviations: N/B, not expressed in both tumor and the normal; N/N, not expressed in the normal tissue.
The alteration on the expression of EGFR, however, differed in the six tumors compared to adjacent normal tissue. The primer set used to amplify and quantify EGFR expression in our real-time QRT-PCR assay anneals to exon 8 sequences and the boundaries of exons 9 and 10 and is thus able to identify most EGFR transcripts. Assay results in our previous work revealed only a slight difference in the upregulation of EGFR in GBM as compared to that in AA (P = 0.046). These gene expression data in combination with new gene expression data from six paired tumor/normal tissue samples and 84 tumors revealed that EGFR dysregulation is an uncommon event that occurs in only a subset of gliomas. In contrast, the expression of PAX6, PTEN, and RPS9 is typically downregulated, whereas the opposite is true for MMP2, IGFBP2, VEGF, and CDK4 in GBMs. Only two AA tumors were paired with normal tissues, which confounded definitive comparisons of gene expression in AA versus normal tissue.
Establishing a Prognostic Model Based on Continuous Log-Scaled Gene Expression Variables
We previously used dichotomized four-gene expression variables (PAX6, PTEN, VEGF and EGFR) to establish a prognosis model. Although a widely used approach, cutpoint selection creates models that may not generalize well and could produce P values that are problematic because they do not reflect the increased uncertainty due to the selection process (i.e., using the same data to test the model that was used to select the cutpoint and variables). To assess this possibility, we tested the model in our 84 glioma patients using log-scaled data without dichotomization for the four previously reported genes. The model that evaluated the expression of PAX6, PTEN, EGFR, and VEGF in the 84 patients using log-scaled data without dichotomization yielded a lower R2 value (39%, model 2 in Table 4) than that from the seven-variable model that used RPA cutpoints (55%) (Zhou et al., 2003), which demonstrates that RPA data were biased in favor of a high R2 value.
Table 4.
Models based on clinical and gene expression variables on log scales
Three clinical variables (patient age, histology, and recurrence status).
Three clinical variables plus four genes (PAX6, PTEN, EGFR, and VEGF)
Three clinical variables plus four AIC-selected genes (PTEN, EGFR, CDK4, and RPS9).
Eleven variables plus eight gene-gene interactions (PTEN-EGFR, P = 0.0024; PTEN-MMP2, P = 0.00002; PTEN-RPS9, P = 0.00063; EGFR-VEGF, P = 0.13; VEGF-MMP2, P = 0.007; VEGF-RPS9, P = 0.019; VEGF-CDK4, P = 0.047; MMP2-CDK4, P = 0.00013).
Using a cutpoint implies that the marker has a step function or threshold relationship with the outcome in question (in this case, survival) (Altman et al., 1994). We found that none of the eight gene expressions have such an ideal relationship, although several confirmed a strong, nonlinear, sigmoidal relationship that approximates a step function. The distributions of the marker values present another potential problem in that are all unimodal and right-skewed with a long right tail. Outliers are on the high side, and the mode is near zero. Thus, we believe that the lower value based on data without dichotomization may be more realistic because it is not severely influenced by overfitting as compared to the R2 value from a method that uses selected cut-points. We focused on prognostic modeling using log-scaled continuous variables composed of data from the expression of eight genes. As shown in Table 4, by including all eight genes and three clinical variables and AIC backward variable elimination on this 11-variable model, forcing the three clinical variables to remain in the model, we obtained a model that contained four genes PTEN (P = 0.023), EGFR (P = 0.094), RPS9 (P = 0.0056), and CDK4 (P = 0.13) with an R2 = 41% (model 3), a significantly improved result over the model with only the three clinical variables (model 1) (P = 0.013). In comparison to model 2, with the same number of variables, model 3 was improved by including the four AIC-selected genes.
Improving the R2 of the Model by Including Interaction Terms Between Gene-Gene and Gene-Clinical Variables
We selected the genes to be used as candidate markers in our prognostic model on the basis of their differential expression in malignant glioma subtypes and for their potential involvement in key suppression/proliferative pathways. Because interactions between genes as well as between genes and clinical variables could contribute to the validity of our prognostic model, we included all two-way interactions in the model and analyzed their prognostic significance by AIC backward elimination. As shown in Table 4, by utilizing this approach, eight out of 28 gene-gene interactions for the eight genes remained in the model (model 4). The R2 of this model, with 19 variables, was 66%, significantly improved (P < 0.0001) over the four-gene model after AIC elimination (model 3). Of the eight genes, only PAX6 (P = 0.066) and IGFBP2 (P = 0.84) did not contribute significantly to improving the model after including gene-gene interactions terms (P > 0.05).
We performed bootstrap cross-validation on this set of patients to correct overfitting, and the corrected R2 of each model is shown in Table 4. In Table 5, we included the estimated parameter values, their estimated standard errors, and the P values. For a Cox model, the parameter values are adjusted log hazard ratios. Negative (positive) values indicate that increasing values of a covariate are associated with lower (higher) rates of death (i.e., better survival). The last eight parameters are products representing the interactions. Negative (positive) values indicate that the joint effect (i.e., as measured by the hazard ratio) of the two variables is less (more) than the product of the hazard ratios for the separate effects.
Table 5.
Estimated parameter values, their estimated standard errors, and the P values in a Cox model
| Variables | Adjusted log HR | SE | Pvalue |
|---|---|---|---|
| Age at diagnosis | −0.0038 | 0.0100 | 0.700000 |
| Histology | 0.0053 | 0.2067 | 0.980000 |
| Recurrent status | 1.2131 | 0.3718 | 0.001100 |
| log (PAX6) | −0.2555 | 0.1389 | 0.066000 |
| log (PTEN) | −2.4570 | 0.6003 | 0.000043 |
| log (EGFR) | 0.8277 | 0.4662 | 0.076000 |
| log (VEGF) | −0.0884 | 0.5488 | 0.870000 |
| log (MMP2) | −0.3643 | 0.5308 | 0.490000 |
| log (RPS9) | −2.4381 | 0.7619 | 0.001400 |
| log (CDK4) | 4.3633 | 1.0069 | 0.000015 |
| log (IGFBP2) | 0.0191 | 0.0959 | 0.840000 |
| log (PTEN) : log (EGFR) | 0.2280 | 0.0752 | 0.002400 |
| log (PTEN) : log (MMP2) | −0.3012 | 0.0706 | 0.000020 |
| log (PTEN) : log (RPS9) | −0.4504 | 0.1318 | 0.000630 |
| log (EGFR) : log (VEGF) | −0.1084 | 0.0713 | 0.130000 |
| log (VEGF) : log (MMP2) | −0.2559 | 0.0950 | 0.007000 |
| log (VEGF) : log (RPS9) | 0.2801 | 0.1194 | 0.019000 |
| log (VEGF) : log (CDK4) | 0.2214 | 0.1114 | 0.047000 |
| log (MMP2) : log (CDK4) | 0.6121 | 0.1602 | 0.000130 |
Abbreviations: HR, hazard ratio; SE, standard error.
The corrected R2 for models 3 and 4 do not include the AIC selection in the bootstrapping. Bootstrapping could not be done with the model containing all the pairwise interactions. (The model just had too many parameters.) Thus model 4 is almost certainly overfit to the data. Although AIC variable selection is better than P-value-driven selection, it still leads to P values biased low and coefficients biased away from zero (i.e., too big in absolute value).
The Strength of a Model Is Augmented by Incorporating Genes from Different Pathways
Excluding IGFBP2 from the model without losing statistical predictive power suggests that the genes remaining in the model may be upstream or downstream of IGFBP2 in the functional pathway(s), which correlates with the pattern of IGFBP2 expression. Our results from eight gene expression values in 84 glioma specimens including or excluding seven normal tissues, as assessed by using a Spearman rank correlation test, support this hypothesis. As shown in Table 6, the expression of IGFBP2 in the entire set of gliomas is significantly and positively correlated with the expression of MMP2 (R = 0.45) and VEGF (R = 0.43), with P < 0.0001, and it is negatively correlated with the expression of RPS9 (R = −0.33, P = 0.0022) and PAX6 (R = −0.26, P = 0.0186).
Table 6.
Significant pairwise correlation coefficients derived from a Spearman rank correlation test of the expressions of CDK4, IGFBP2, MMP2, PAX6, PTEN, RPS9, and VEGF in gliomas and normal tissuesa
| R (P) | IGFBP2-MMP2 | IGFBP2-VEGF | IGFBP2-RPS9 | IGFPB2-PAX6 | PAX6-PTEN | PTEN-MMP2 | MMP2-CDK4 |
|---|---|---|---|---|---|---|---|
| n = 91b | 0.48 (<0.0001) | 0.42 (<0.0001) | −0.36 (0.0004) | −0.29 (0.0047) | 0.31 (0.0026) | 0.23 (0.0289) | 0.32 (0.0021) |
| n = 84c | 0.45 (<0.0001) | 0.43 (<0.0001) | −0.33 (0.0022) | −0.26 (0.0186) | 0.31 (0.0035) | 0.26 (0.0169) | 0.21 (0.0519) |
| n = 43d | 0.36 (0.0178) | 0.21 (0.1747) | −0.18 (0.2380) | −0.10 (0.5147) | 0.13 (0.4170) | 0.52 (0.0004) | 0.31 (0.0397) |
| n = 41e | 0.41 (0.0078) | 0.32 (0.0387) | 0.29 (0.0646) | 0.09 (0.5636) | 0.032 (0.8431) | 0.21 (0.1686) | 0.10 (0.5253) |
Correlations, with P ⩽ 0.005, were shown in at least one group and are presented in bold.
43 AAs, 41 GBMs, and 7 normal tissues.
43 AAs and 41 GBMs.
43 AAs.
41 GBMs.
Table 6 also demonstrates a statistically significant positive correlation between the expressions of tumor suppressor genes PTEN and PAX6 (R = 0.31, P < 0.005) and between those of MMP2 and CDK4 (R = 0.32, P = 0.0021) in the entire set of gliomas, including normal tissues (n = 91). However, the MMP2-CDK4 positive correlation exists in AA and not in GBM. Interestingly, of the eight genes, the expression of only the oncogenic gene EGFR was not significantly correlated with that of any of the other genes in the entire glioma set, with only moderate correlation with the expression of MMP2 in GBMs (R = 0.34, P = 0.0311). This may explain why EGFR remained in the four-gene model (EGFR, CDK4, PTEN, and RPS9) after AIC backward selection of the eight genes, although its prognostic value was not shown to be significant in a univariate analysis (Zhou et al., 2003).
As demonstrated in Table 6, when looking at tumor grade, the significance and R value for positive correlation for IGFBP2-MMP2 and IGFBP2-VEGF in AA and GBM decreased because of decrease of sample size. However, even with smaller sample size, positive correlations for IGFBP2-MMP2 and IGFBP2-VEGF occur more in GBMs than in AAs. When data for seven normal tissues adjacent to five GBMs and two AAs in this study was included in the correlation analysis with each grade, the positive correlation IGFBP2-MMP2 became more striking for GBM (R = 0.56, P < 0.0001) than for AA (R = 0.35, P = 0.0108). This indicates that over-expression of IGFBP2 occurred specifically in GBMs, and it correlates positively with MMP2 overexpression.
In contrast to the expression of IGFBP2, MMP2 expression was associated with genes expressed in different WHO III or IV glioma grades. For example, MMP2 associates specifically with IGFBP2 in GBM, and specifically with PTEN and CDK4 in AA. This finding also suggests that the expression of MMP2 in AA and GBM is regulated by different molecular mechanisms, although it is overexpressed in both. Taken together, our data suggest that the strength of a prognostic model is augmented by incorporating genes that have functions in diverse suppressive or tumor progression pathways.
Discussion
Our study was designed to improve an earlier prognostic model for patients with WHO grade III or IV gliomas by including a greater number of gene expression variables than had been previously used (Zhou et al., 2003). The four new gene variables in this study, selected because of their prognostic value for patients with malignant astrocytic gliomas, each have prognostic value as univariates. However, we found that the prognostic model can be strengthened by including genes with roles in different pathways associated with the intrinsic aggressiveness of the tumor(s) of interest.
Functional Genomic Information from Gene Expression Derived Through Correlation Analysis
Our study shows that using a pairwise correlation analysis on real-time-PCR-quantified gene expressions from a large set of tumor specimens provides new information regarding their functionality in tumor formation and progression. Our finding that the expression of IGFBP2 had a statistically significant positive correlation with the expression of MMP2 and VEGF in GBM suggests that IGFBP2 is involved in enhancing invasiveness and angiogenesis, processes that are specific to formation and progression in GBM. Concurring with this finding, IGFBP2’s ability to enhance glioma cell invasion by activating the expression of MMP2 was reported in an in vitro study (Wang et al., 2003), and the coexpression of IGFBP2 and VEGF in pseudopalisading cells surrounding necrotic areas in GBM was also demonstrated (Godard et al., 2003).
However, our finding of a statistically significant positive correlation between PTEN and MMP2 in AA (R = 0.51, P = 0.0002) is in contradistinction to the identified suppressive effect of PTEN on the expression of MMP2 in glioma cells shown in an earlier in vitro study (Koul et al., 2001). This disparity could be explained by the diverse genetic alterations found in AA versus GBM. The expression of PTEN in GBM might be downregulated because the gene is frequently lost, together with a loss of heterozygosity of chromosome 10, a common event in GBM (Kon et al., 1998; Liu et al., 1997; Ransom et al., 1992; Rasheed et al., 1992; von Deimling et al., 1993; Wiltshire et al., 2000). If mutated PTEN, a frequent occurrence in both AA and GBM, enhances the overall expression of a mutant gene, that event could account for the positive correlation between the expression of PTEN (mutant) and MMP2. Although not included in this model, the prognostic significance of PTEN mutation, as reported earlier (Smith et al., 2001), will be assessed as an improvement of our current model in a future study.
Controversial results have been reported regarding the prognostic value of EGFR amplification/overexpression in GBM, paradoxically demonstrating both prognostic significance (Shinojima et al., 2003) and no significance (Kordek et al., 1995; Newcomb et al., 1998; Rainov et al., 1997). Our finding in a previous univariate Cox analysis showed that EGFR by itself has no outstanding prognostic value, but contributes significantly to the prognostic model with the other expression variables. We also found that it strongly trended toward a positive correlation with the expression of MMP2 in GBM. It thus appears that EGFR exerts a synergistic effect in different underlying pathways in GBMs that involve other genes in our model that are associated with a poor prognosis. Although gross rearrangements of the EGFR gene do occur in the genome of glioma cells and these should be included for consideration in a prognostic model, amplification of the EGFR gene and amplification of chromosome 7 are the most frequent genetic alterations in de novo GBMs and are related to an unsatisfactory patient survival.
Our gene expression correlation analysis revealed a negative correlation for RPS9-IGFBP2 (R = −0.36, P = 0.0004) and RPS9-VEGF (R = −0.28, P = 0.0068) in the entire set of gliomas examined in this study, including normal tissues. However, these two relationships did not hold up when we looked at tumor grade, which suggests that they were confounded by diverse tumor histologies. Inclusion of RPS9 in the four-gene model after AIC elimination (model 3 in Table 5) indicated that RPS9 is a significant favorable prognostic marker in gliomas, independent of other genes in this study. Recent reports showed that RPS9 expression increased in both a PC12 pheochromocytoma cell line and Neuro-A2 neuroblastoma cell line in response to hydrogen peroxide treatment in vitro, and these reports showed that RPS9 plays an antiapoptotic role that protects neuronal cells from oxidative injury (Kim et al., 2003). The expression of RPS9 was also downregulated in pancreatic cancer tissue as compared to the normal pancreas (Crnogorac-Jurcevic et al., 2001). One hypothesis is that RPS9 has an antiapoptosis function in glioma cells in response to reactive oxygen species, products of a hypoxic and inflammatory microenvironment. This is also a potential explanation for the upregulation of RPS9 expression in AA, a histology devoid of necrosis (Table 2). The precise consequences of increasing of RPS9 expression in GBM cells in response to reactive oxygen species is not yet known, nor is it known whether its expression is correlated with a pseudopalisading structure, a hallmark pathologic feature of GBM, which is composed of a high percentage of apoptotic cells (Brat et al., 2004).
Statistical Considerations on Improving Prognostic Models
In this study, we paid considerable attention to how best to model each variable to improve prognostication as well as how best to combine data from the variables to improve the model’s predictive power. The standard prognostic model is a so-called main-effects model, which implies that each variable has the same predictive capacity regardless of the values of the other variables. However, we can augment this approach by including variable-to-variable interaction terms that permit drawing parameter estimates, thereby intensifying the predictive capacity of a variable to vary with values of the other variables. Data from our study showed that the predictive accuracy of the model based on log-scaled gene expression data and clinical variables may be significantly increased by considering interactions between genes.
Excessive numbers of variables and complexity in combining them may compromise the model’s clinical utility. However, statistically, the problem comes down to how complex a model can be and still be supported by the data without overfitting such that a fitted model will perform well on training data but not on independent test data. There is no general answer to this question, but we can attempt to address it by performing cross-validation. The key is to include all the model selection processes as part of the cross-validation analysis. In our case, the model with all the gene-gene interactions is too big for performing bootstrap cross-validation. For the model generated with AIC backward elimination, the corrected R2 for model 4 after bootstrap cross-validation is 43%, down from the naive estimate of 66% (and this does not account for overfitting due to selection). This shift in value indicates that we may experience the problem associated with limited sample size. Thus, it is important to increase sample size and retest the model. The new set would ideally be combined with the current training set to form a larger set of samples for bootstrap cross-validation. It is also widely held that whatever internal validation is performed, final validation must be performed in a large independent data set, preferably at a different institution than the one that generated the original data set.
Consideration for Future Clinical Application
To ensure reproducibility of a prognostic model for clinical application, it is important to use biomarkers that can be standardized. For gene expression markers, the reproducible data can be achieved by use of the absolute expression ratio between a marker gene and an internal control gene. This can be accomplished by using a multigene DNA standard for real-time PCR, which can be made by ligating the marker gene standard DNA and the standards for internal control genes together in one piece and in a one-to-one ratio.
Another issue is the stability of the internal control gene’s expression. An example would be a patient with GBM who has a differential expression of the internal control gene β-actin because of chromosome 7 trisomy. It is important for that reason to establish multiple prognostic models with marker genes normalized to different internal control genes spanning different biologic pathways and different chromosome locations. The predicted outcome for an individual patient should be analyzed with such diverse models to avoid drawing misleading conclusions from the variable expression of an internal control gene.
Acknowledgment
We thank Joann Aaron in the Department of Neuro-Oncology at M.D. Anderson for editing this manuscript.
Footnotes
This work was supported by the National Brain Tumor Foundation Pediatric Brain Tumor Grant, the Gilliland Foundation for Brain Tumor Research, the Arkansas Cancer Research Center Tobacco Settlement Fund, and the Fraternal Order of Eagles.
Abbreviations used are as follows: AA, anaplastic astrocytoma; AIC, Akaike information criterion; GBM, glioblastoma multiforme; IHC, immunohistochemistry; PH, proportional hazards; MDACC, M.D. Anderson Cancer Center; PCR, polymerase chain reaction; QRT, quantitative reverse transcription; RPA, recursive partitioning analysis.
References
- Akaike, H. (1973) Information theory and an extension of the maximum likelihood principle. In Petrov, B.N., and Csáki, F. (Eds.), 2nd International Symposium on Information Theory. Budapest: Akadémiai Kiadó, 267–281.
- Akmal SN, Yun K, MacLay J, Higami Y, Ikeda T. Insulin-like growth factor 2 and insulin-like growth factor binding protein 2 expression in hepatoblastoma. Hum Pathol. 1995;26:846–851. doi: 10.1016/0046-8177(95)90005-5. [DOI] [PubMed] [Google Scholar]
- Altman DG, Lausen B, Sauerbrei W, Schumacher M. Dangers of using “optimal” cutpoints in the evaluation of prognostic factors. J Natl Cancer Inst. 1994;86:829–835. doi: 10.1093/jnci/86.11.829. [DOI] [PubMed] [Google Scholar]
- Brat DJ, Castellano-Sanchez AA, Hunter SB, Pecot M, Cohen C, Hammond EH, Devi SN, Kaur B, Van Meir EG. Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer Res. 2004;64:920–927. doi: 10.1158/0008-5472.can-03-2073. [DOI] [PubMed] [Google Scholar]
- CBTRUS (2002) Primary Brain Tumors in the United States: Statistical Report, 1995–1999. Central Brain Tumor Registry of the United States. Available at http://www.cbtrus.org/reports/reports.html
- Crnogorac-Jurcevic T, Efthimiou E, Capelli P, Blaveri E, Baron A, Terris B, Jones M, Tyson K, Bassi C, Scarpa A, Lemoine NR. Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene. 2001;20:7437–7346. doi: 10.1038/sj.onc.1204935. [DOI] [PubMed] [Google Scholar]
- Daumas-Duport C, Scheithauer B, O’Fallon J, Kelly P. Grading of astrocytomas. A simple and reproducible method. Cancer. 1988;62:2152–2165. doi: 10.1002/1097-0142(19881115)62:10<2152::aid-cncr2820621015>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Dirks PB, Hubbard SL, Murakami M, Rutka JT. Cyclin and cyclin-dependent kinase expression in human astrocytoma cell lines. J Neuropathol Exp Neurol. 1997;56:291–300. doi: 10.1097/00005072-199703000-00008. [DOI] [PubMed] [Google Scholar]
- Fuller GN, Rhee CH, Hess KR, Caskey LS, Wang R, Bruner JM, Yung WK, Zhang W. Reactivation of insulin-like growth factor binding protein 2 expression in glioblastoma multiforme: A revelation by parallel gene expression profiling. Cancer Res. 1999;59:4228–4232. [PubMed] [Google Scholar]
- Harvey, A.C. (1981) Time Series Models New York: John Wiley & Sons, Inc.
- Godard S, Getz G, Delorenzi M, Farmer P, Kobayashi H, Desbaillets I, Nozaki M, Diserens AC, Hamou MF, Dietrich PY, Regli L, Janzer RC, Bucher P, Stupp R, de Tribolet N, Domany E, Hegi ME. Classification of human astrocytic gliomas on the basis of gene expression: A correlated group of genes with angiogenic activity emerges as a strong predictor of subtypes. Cancer Res. 2003;63:6613–6625. [PubMed] [Google Scholar]
- Katz, M.H. (1999) Multivariable Analysis Cambridge: Cambridge University Press.
- Kim SY, Lee MY, Cho KC, Choi YS, Choi JS, Sung KW, Kwon OJ, Kim HS, Kim IK, Jeong SW. Alterations in mRNA expression of ribosomal protein S9 in hydrogen peroxide–treated neurotumor cells and in rat hippocampus after transient ischemia. Neurochem Res. 2003;28:925–931. doi: 10.1023/a:1023283628454. [DOI] [PubMed] [Google Scholar]
- Kleihues, P., and Cavenee, W.K. (2000) Pathology and Genetics of Tumours of the Nervous System 2nd ed. Lyon: IARC Press.
- Kon H, Sonoda Y, Kumabe T, Yoshimoto T, Sekiya T, Murakami Y. Structural and functional evidence for the presence of tumor suppressor genes on the short arm of chromosome 10 in human gliomas. Oncogene. 1998;16:257–263. doi: 10.1038/sj.onc.1201488. [DOI] [PubMed] [Google Scholar]
- Kordek R, Biernat W, Alwasiak J, Maculewicz R, Yanagihara R, Liberski PP. p53 protein and epidermal growth factor receptor expression in human astrocytomas. J Neurooncol. 1995;26:11–16. doi: 10.1007/BF01054764. [DOI] [PubMed] [Google Scholar]
- Koul D, Parthasarathy R, Shen R, Davies MA, Jasser SA, Chintala SK, Rao JS, Sun Y, Benvenisite EN, Liu TJ, Yung WK. Suppression of matrix metalloproteinase-2 gene expression and invasion in human glioma cells by MMAC/PTEN. Oncogene. 2001;20:6669–6678. doi: 10.1038/sj.onc.1204799. [DOI] [PubMed] [Google Scholar]
- Kuo WP, Jenssen TK, Butte AJ, Ohno-Machado L, Kohane IS. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- Lam PY, Di Tomaso E, Ng HK, Pang JC, Roussel MF, Hjelm NM. Expression of p19INK4d, CDK4, CDK6 in glioblastoma multiforme. Br J Neurosurg. 2000;14:28–32. doi: 10.1080/02688690042870. [DOI] [PubMed] [Google Scholar]
- Lantos, P.L., VandenBerg, S.R., and Kleihues, P (1997) Tumours of the nervous system. In Graham, D.I., and Lantos, P.L., Eds., Greenfield’s Neuropathology, Sixth Edition London: Arnold.
- Liu W, James CD, Frederick L, Alderete BE, Jenkins RB. PTEN/MMAC1 mutations and EGFR amplification in glioblastomas. Cancer Res. 1997;57:5254–5257. [PubMed] [Google Scholar]
- McCawley LJ, Matrisian LM. Matrix metalloproteinases: They’re not just for matrix anymore! Curr Opin Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- Newcomb EW, Cohen H, Lee SR, Bhalla SK, Bloom J, Hayes RL, Miller DC. Survival of patients with glioblastoma multiforme is not influenced by altered expression of p16, p53, EGFR, MDM2 or Bcl-2 genes. Brain Pathol. 1998;8:655–667. doi: 10.1111/j.1750-3639.1998.tb00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel A, Gilles C, Bajou K, Devy L, Kebers F, Lewalle JM, Maquoi E, Munaut C, Remacle A, Foidart JM. Emerging roles for proteinases in cancer. Invasion Metastasis. 1997;17:221–239. [PubMed] [Google Scholar]
- Rainov NG, Dobberstein KU, Bahn H, Holzhausen HJ, Lautenschlager C, Heidecke V, Burkert W. Prognostic factors in malignant glioma: Influence of the overexpression of oncogene and tumor-suppressor gene products on survival. J Neurooncol. 1997;35:13–28. doi: 10.1023/a:1005841520514. [DOI] [PubMed] [Google Scholar]
- Ransom DT, Ritland SR, Moertel CA, Dahl RJ, O‘Fallon JR, Scheithauer BW, Kimmel DW, Kelly PJ, Olopade OI, Diaz MO, Jenkins RB. Correlation of cytogenetic analysis and loss of heterozygosity studies in human diffuse astrocytomas and mixed oligo-astrocytomas. Genes Chromosomes Cancer. 1992;5:357–374. doi: 10.1002/gcc.2870050412. [DOI] [PubMed] [Google Scholar]
- Rasheed BK, Fuller GN, Friedman AH, Bigner DD, Bigner SH. Loss of heterozygosity for 10q loci in human gliomas. Genes Chromosomes Cancer. 1992;5:75–82. doi: 10.1002/gcc.2870050111. [DOI] [PubMed] [Google Scholar]
- Richardsen E, Ukkonen T, Bjornsen T, Mortensen E, Egevad L, Busch C. Overexpression of IGBFB2 is a marker for malignant transformation in prostate epithelium. Virchows Arch. 2003;442:329–335. doi: 10.1007/s00428-003-0786-2. [DOI] [PubMed] [Google Scholar]
- Rollbrocker B, Waha A, Louis DN, Wiestler OD, von Deimling A. Amplification of the cyclin-dependent kinase 4 (CDK4) gene is associated with high cdk4 protein levels in glioblastoma multiforme. Acta Neuropathol. 1996;92:70–74. doi: 10.1007/s004010050491. [DOI] [PubMed] [Google Scholar]
- Sallinen SL, Sallinen PK, Haapasalo HK, Helin HJ, Helen PT, Schraml P, Kallioniemi OP, Kononen J. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000;60:6617–6622. [PubMed] [Google Scholar]
- Sano T, Lin H, Chen X, Langford LA, Koul D, Bondy ML, Hess KR, Myers JN, Hong YK, Yung WK, Steck PA. Differential expression of MMAC/PTEN in glioblastoma multiforme: Relationship to localization and prognosis. Cancer Res. 1999;59:1820–1824. [PubMed] [Google Scholar]
- Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, Seiki M. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, Makino K, Saya H, Hirano H, Kuratsu J, Oka K, Ishimaru Y, Ushio Y. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, O’Fallon JR, Schaefer PL, Scheithauer BW, James CD, Buckner JC, Jenkins RB. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–1256. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- von Deimling A, von Ammon K, Schoenfeld D, Wiestler OD, Seizinger BR, Louis DN. Subsets of glioblastoma multiforme defined by molecular genetic analysis. Brain Pathol. 1993;3:19–26. doi: 10.1111/j.1750-3639.1993.tb00721.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang H, Shen W, Huang H, Hu L, Ramdas L, Zhou Y-H, Liao WS-L, Fuller GN, Zhang W. Insulin-like growth factor binding protein 2 enhances glioblastoma invasion by activating invasion-enhancing genes. Cancer Res. 2003;63:4315–4321. [PubMed] [Google Scholar]
- Wiltshire RN, Rasheed BK, Friedman HS, Friedman AH, Bigner SH. Comparative genetic patterns of glioblastoma multiforme: Potential diagnostic tool for tumor classification. Neuro-Oncology. 2000;2:164–173. doi: 10.1093/neuonc/2.3.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YH, Tan F, Hess KR, Yung WK. The expression of PAX6, PTEN, vascular endothelial growth factor, and epidermal growth factor receptor in gliomas: Relationship to tumor grade and survival. Clin Cancer Res. 2003;9:3369–3375. [PubMed] [Google Scholar]
- Zhou Y-H, Wu X, Tan F, Shi Y-X, Glass T, Liu TJ, Wathen K, Hess KR, Gumin J, Lang F, Yung WKA. PAX6 suppresses growth of human glioblastoma cells. J Neurooncol. 2005;71:223–229. doi: 10.1007/s11060-004-1720-4. [DOI] [PubMed] [Google Scholar]