Abstract
Intracellular expression of gene products that inhibit viral replication have the potential to complement current antiviral approaches to the treatment of AIDS. We previously have shown that a mutant inhibitory form of an essential viral protein, Rev M10, prolongs the survival of T cells transduced with a nonviral vector in HIV-infected individuals. Because these gene-modified cells were not observed in patients beyond 8 weeks, efforts were made to improve the duration of engraftment. In this study, we used retroviral vector delivery of Rev M10 to CD4+ cells and analyzed relevant immune responses in a pilot study of three HIV-seropositive patients. DNA and RNA PCR analyses revealed that cells transduced with Rev M10 retroviral vectors survived and expressed the recombinant gene for significantly longer time periods than those transduced with a negative control vector in all three patients. Immune responses were not detected either to Rev M10 or to Moloney murine leukemia virus gp70 envelope protein. Rev M10-transduced cells were detected for an average of 6 months after retroviral gene transfer compared with ≈3 weeks for the previously reported nonviral vector delivery. These findings suggest that retroviral delivery of an antiviral gene may potentially contribute to immune reconstitution in AIDS and could provide a more effective vector to prolong survival of CD4+ cells in HIV infection.
The inhibition of HIV replication by interference with essential structural or regulatory viral genes has been investigated to understand the molecular pathogenesis of HIV infection and to explore its potential clinical applications (1–15). Among viral products that may be targeted genetically, Rev is a regulatory protein required for productive viral replication. Rev represents a 118-aa protein encoded by a fully spliced mRNA synthesized early after viral infection of host cells, which facilitates the accumulation of unspliced and partially spliced viral mRNAs in the cytoplasm (1, 16–18) through its interaction with host cell factors (19–21) and plays an important role in the activation of virus in infected cells (22). Introduction of two point mutations in a highly conserved region of Rev gives rise to a defective protein, Rev M10, which acts as a potent inhibitor of virus replication that does not adversely affect a variety of normal T cell functions (1–3, 23–25). We previously have evaluated the potential of Rev M10 gene delivery to inhibit the replication of both cloned and clinical isolates of HIV in primary T cells transduced with plasmid or retroviral vectors expressing Rev M10 protein (4). A human clinical study also has examined the potential of Rev M10 to prolong the survival of transduced CD4+ T cells in vivo after ex vivo transduction, expansion, and reinfusion into HIV-seropositive patients. By using plasmid vectors with gold particle-mediated gene delivery, a 4- to 5-fold survival advantage was found in cells that expressed Rev M10 compared with negative control transduced cells (26); however, the duration of engraftment was limited. Although genetically modified cells were detected in one patient up to 2 months after gene transfer, the recombinant gene was not detectable in two patients after 2 weeks. To determine whether more durable engraftment could be achieved, alternative gene transfer vectors and protocols for T cell stimulation thus have been developed (27). We now have analyzed lymphocyte survival in HIV-seropositive individuals whose peripheral blood CD4+ lymphocytes were stimulated with anti-CD3 and interleukin 2 and transduced with retroviral vectors that express Rev M10 or a negative control frameshifted vector, which transcribes a similar mRNA but encodes no functional Rev M10 protein. We have found that genetically modified cells are detected for longer time periods with these vectors.
MATERIALS AND METHODS
Isolation and Culture of Human Peripheral Blood Lymphocytes (PBLs).
Blood for these studies was obtained from normal or HIV-seropositive donors. PBLs were isolated by using Ficoll-Hypaque separation (28). The cells were stimulated in flasks coated with immobilized OKT3 mAb and soluble interleukin 2 (50 units/ml) for 48–72 hr. Cells were recovered and resuspended at 5 × 105/ml in X-Vivo-15 medium (BioWhittaker) containing 50 units/ml of interleukin 2 and 5 μM delavirdine. Cells were maintained at 2 × 105 to 1.5 × 106/ml throughout the experiments.
Retroviral Transduction.
Freshly isolated human PBLs from donors were purified by centrifugation on Ficoll gradients. Retroviral vectors were derived from the pLJ plasmid (29). After stimulation, cells were infected for 6–12 hr with ψ-Crip supernatants containing pLJ-Rev M10 or frameshift pLJ-ΔRev M10 retroviruses (1). Cells (1 × 106/ml) were inoculated in medium consisting of equal volumes of ψ-Crip supernatant and X-Vivo medium in the presence of 5 μg/ml protamine sulfate (28). After infection, cells were washed once by centrifugation and resuspended at 5 × 105 cells/ml in conditioned X-Vivo-15 medium + 50 units/ml of interleukin 2.
HIV Challenge and Reverse Transcriptase (RT) Assays.
After retroviral transduction, cells were selected in 300 μg/ml of G418 (active) for 7 days before HIV challenge. Cells (1 × 106 cells/ml) were incubated with HIV-1 at a multiplicity of infection of 0.05 for 2–4 hr at 37°C. After incubation, cells were centrifuged with a 20× volume of fresh medium to remove the virus and resuspended at 5 × 105 cells/ml in medium without G-418. Cells were maintained at a density of 0.2 to 1.5 × 106 cells/ml throughout the infection. Culture supernatants were assayed for RT activity as described previously (30).
Cell Proliferation Assays.
PBL cells were resuspended in X-Vivo-15 medium (BioWhittaker, 106 cells/ml) in 96-well flat bottom cell culture plates (200 μl/well). Cells were cultured for 6 days at 37°C, 5% CO2 either in the presence (2 μg/ml) or absence of purified HIV-1 Rev protein. In parallel, irradiated (2,000 rads) PBLs from normal volunteers were added at a 1:1 ratio as a positive control. After 6 days of incubation, 0.5 μCi of 3H-thymidine was added to each well and incubation continued for an additional day. Cells were harvested and washed twice with PBS, and the radioactive counts were determined by using a scintillation counter.
Quantitative DNA PCR analysis.
To analyze gene transfer frequencies, limiting dilution PCR was performed. Briefly, cells carrying Rev M10 or ΔRev M10 were diluted into CEM cells at progressively lower cell numbers. Genomic DNA was prepared from a total of 105 or 106 cells by using a quick lysis method described previously (28). The cell pellet was suspended in 10 mM KCl/1 mM Tris⋅HCl, pH 8.3/0.25 mM MgCl2 (50 μl) and an equal volume of solution B (1 mM Tris⋅HCl, pH 8.3/0.25 mM MgCl2/0.1% Tween-20/0.1% Nonidet P-40/50 mg/ml proteinase-K). The reaction mixture was incubated at 56°C for 1 hr and at 95°C for 20 min. Cell lysate (5 μl) was used in the PCR without additional purification of the DNA, to minimize the risk of false positive results. Cell lysate (5 μl) equivalent to 1,000, 100, 10, 5, 2, or 0.1 cells was analyzed with the target DNA in a background of DNA derived from 25,000 CEM cells. The PCR mixture was comprised of 50 mM KCl, 10 mM Tris⋅HCl (pH 8.8), 0.01% gelatin, 250 nM primers, 200 μM dNTPs, 1.5 mM MgCl2, and 1.25 units of Taq polymerase (Promega) in a final volume of 50 μl. The amplification was performed in a thermal cycler (Perkin–Elmer Cetus) for a total of 36 cycles, each cycle consisting of melting at 91°C for 1 min, annealing at 68°C for 10 sec, and extension at 72°C for 20 sec. The nucleotide sequences of the primers and the probe that are common for both Rev M10 and ΔRev M10 vectors are as follows: the forward primer 5′-GGAGGGATATGTGGTTCTGGTAGGAGACGAG-3′, the reverse primer 5′-CGGGATTGGGAGGTGGGTTGCTTTGATAG-3′, and the probe 5′-ACCTAAAACAGTTCCCGCCTCCGTCTGAATT-3′. The primers, which are common for both vectors, amplify fragments that differ in size. The expected fragment sizes were 218 and 331 bp for pLJ-Rev M10 and pLJ-ΔRev M10, respectively. The PCR products were resolved on an 8% polyacrylamide gel supplemented with 8 M urea and 30% formamide and probed as previously described (28). The intensity of the PCR bands was quantitated with the help of the Image Quantitation software (Molecular Dynamics).
RNA PCR Analysis.
The RT-based PCR analysis was performed as described previously (26) with modifications. Briefly, previously frozen patient samples of PBLs were activated by incubation of cells with Dynal beads coated with antibodies against CD28 and CD3, which selectively promotes the growth of naive and memory CD4+ cells (31). Although gene-modified cells can be selected in the presence of G418, the nonspecific toxic effects of this drug make the process less efficient, and it was more difficult to propagate these cells. Therefore, the rescue procedure was routinely performed in the absence of G418. After activation for 4 days, RNA was extracted, and the RNA was reverse-transcribed with a specially designed primer (RTP, Fig. 1) and the cDNA was amplified with a combination of forward primer and RTP. After RT, the conditions used for amplification were denaturation at 91°C for 1 min, annealing at 68°C for 10 sec, and extension at 72°C for 50 sec for a total of 40 cycles. The amplified products, 384 and 497 bp for pLJ Rev M10 and pLJ ΔRev M10, respectively, were visualized as described above.
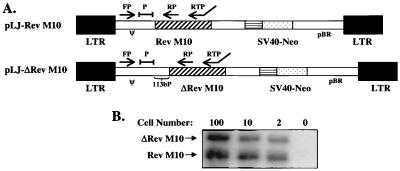
Figure 1.
Schematic diagrams of the retroviral vectors and PCR analysis of DNA in genetically modified cells. (A) Structures of the pLJ-Rev M10 expression vector and a nearly identical negative control vector pLJ-ΔRev M10, which contained a 2-bp deletion in the translation initiation codon and an additional 113 bp at the 3′ end of the packaging/encapsidation region. The maps are not drawn to scale. Arrows denote the sites of primers and their orientation. A combination of the forward primer (FP) and the reverse primer (RP) was used to amplify 218-bp (pLJ-Rev M10) and 331-bp (pLJ-ΔRev M10) fragments in DNA PCR. The primer used to reverse-transcribe RNA has been described previously (26). This primer in combination with the FP was used in RT-PCR, to amplify 384- and 497-bp fragments with pLJ-Rev M10 and pLJ-ΔRev M10, respectively. P = probe. (B) DNA PCR standards. CEM cells at the indicated numbers (0–1000), stably transduced with pLJ-Rev M10 or pLJ-ΔRev M10, were mixed with 105 untransduced CEM cells. DNA-PCR was performed, and the products were visualized as described in Materials and Methods.
Western Blot for the Detection of Antibodies to Murine Amphotropic Retroviruses.
Western blot analysis was performed by Microbiological Associates (Gaithersburg, MD). Briefly, serum from patients or retroviral particles were mixed with SDS loading buffer, resolved on a 8–16% acrylamide gel (Novex), and transferred to a nitrocellulose for probing with relevant anti-gp70 control antibodies, followed by secondary goat anti-human H + L antibodies diluted from 10−3 to 10−4 (Kirkegaard & Perry Laboratories) and appropriate developing reagents.
RESULTS
Three patients, with CD4 counts between 400 and 500 cells/mm3 on enrollment, satisfied the entry criteria of the study (27) and were treated in the General Clinical Research Center at the University of Michigan Medical Center. These patients were designated 5, 6, and 7 as four patients had been studied in a previous nonviral gene transfer study of Rev M10 (26). They were asymptomatic, HIV-seropositive, and maintained on azidothymidine (AZT), but with low viral burdens. Each patient tolerated the treatments well, with no complications or adverse events.
CD4+ cells were enriched by depletion of CD8+ cells and infected separately with either a Rev M10 or a control murine amphotropic retroviral vector. The viral vector pLJ-Rev M10 expressed Rev M10, the inhibitory mutant form of Rev, whose expression was regulated by the enhancer of the long terminal repeat. The negative control vector, pLJ-ΔRev M10, was nearly identical to pLJ-Rev M10 but differed in two respects (Fig. 1A). First, the control vector contained a 2-bp deletion in the translation start codon of the Rev M10 and hence produces no protein. Second, there was additional sequence of 113 bp at the 3′ end of the packaging/encapsidation region. These differences in sequence were used to distinguish between these vectors by using a PCR detection assay, which used limited cell dilution and quantitation of different-sized fragments as described previously (26), to confirm differences between samples (Fig. 1B).
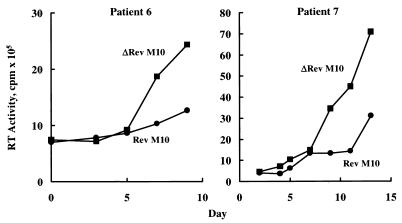
To determine whether the pLJ-Rev M10 retroviral vector conferred protection to CD4+ cells from HIV-infected individuals, we first performed viral challenge studies in vitro. CD4+ enriched peripheral blood mononuclear cells (PBMCs) from two of the patients were stimulated in vitro, infected with either of the vectors, maintained under selection separately, and challenged with HIV-1LAI. Cell supernatants were collected for up to 10–15 days, and RT levels were determined. The time course of infection in two patients revealed a significant reduction in RT levels in cells transduced with the Rev M10 retroviral vector compared with the control (Fig. 2). Although not all cells were gene-modified at the time of exposure to virus (≈30–50%), a 2- to 3-fold reduction in RT levels indicated that expression of Rev M10 in the PBLs of these patients could inhibit HIV replication in vitro, as demonstrated previously in T cells from normal, uninfected individuals (4). These cells had been maintained in the presence of delavirdine before infection, and no endogenous was detected before HIV challenge, indicating that Rev M10 protected against infection by exogenous HIV.
Figure 2.
Antiviral effect of Rev M10 in patient cells in vitro. CD4+ cells were activated and retrovirally transduced with pLJ-Rev M10 or pLJ-ΔRev M10 as described in Materials and Methods. After transduction and selection in G418 for 7 days, cells were challenged with HIVLAI at a multiplicity of infection of 0.05. After infection, RT activity was assayed in triplicate from culture supernatants at the indicated times.
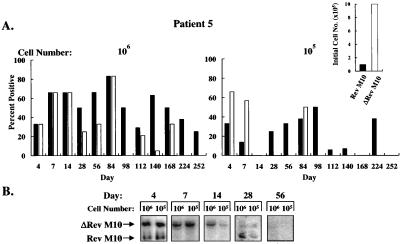
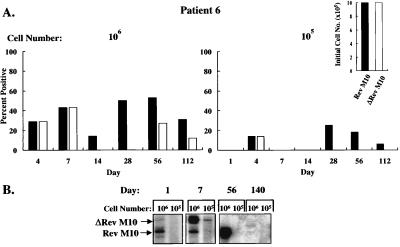
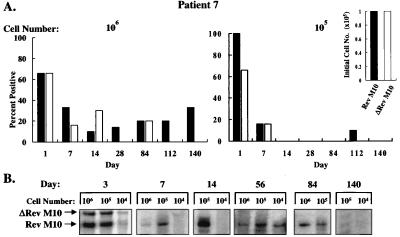
To analyze whether such inhibition might confer protective effects in vivo, these autologous gene-modified cell populations were mixed and infused into each patient, and the presence of the retroviral DNA in the cells was quantitated by limiting dilution PCR with a common pair of primers (Fig. 1B). With this method, retroviral vector DNA was readily detected, confirming that reverse transcription occurred in vector-transduced cells after infection by the retroviral vectors. Gene transfer efficiencies ranged from 1% to 10% in vitro, and G418 selection was not used for cells that subsequently were reinfused into patients. PBMC were isolated from patients at various times after reinfusion, and different cell numbers were used as the source of template DNA for PCR. Vector DNA was detected when 106 and 105 cells were analyzed but not lower cell numbers, as expected based on the dilution of these cells into a large pool of T cells in vivo. In all three patients, the DNA form of both vectors was readily detected at earlier time points (Figs. 3A, 4A, and 5A). With time, the Rev M10 retroviral vector DNA was detected either at higher frequencies or exclusively in comparison to the control vector. In patient 5, both vectors were detected at comparable frequencies up to day 84 when 106 cells were analyzed (Fig. 3A, Left). After this time period, Rev M10 was detected at higher frequencies, and by day 224, only the Rev M10 vector DNA was detected (Fig. 3A). As expected, selective persistence of Rev M10-transduced cells was more apparent when fewer cells, 105, were analyzed (Fig. 3A, Right). No signal was detected in patient 5 after day 252. Interestingly, this patient initially was infused with one order of magnitude more cells transduced with the control vector than the Rev M10-transduced cells (Fig. 3A, Inset), caused by an unintentional failure to account for the higher titer of the control retroviral vector preparation. Subsequently, in patients 6 and 7, the number of cells transduced with each vector were equalized before reinfusion (Figs. 4A and 5A, Insets). The pattern of DNA persistence in these two patients was similar to patient 5. In these patients, the frequency of each vector was similar on the day of infusion. With time, the control vector still could be detected, though at lower levels than Rev M10 in patient 6 (Fig. 4A, Left, days 14, 28, 56, and 112), and when 105 cells were analyzed, only the Rev M10 retroviral vector was detected (Fig. 4A, Right). Similarly, only the Rev M10 vector was detected at later times in patient 7 (Fig. 5A, Right, days 112 and 140). Some variation was detected in signals at higher cell concentrations. This variation may have been caused by several factors, including differences in cell trafficking, variation in the viability of cells isolated from the patients, or the variability in the time required to preserve these cells for additional analyses. In general, the lower cell number (105) was considered a more sensitive indicator of the selective persistence of each cell type, because at lower dilutions, the less frequent cell could be titrated out, and only the predominant cell would be detected.
Figure 3.
Prolonged survival and kinetics of Rev M10-transduced cells in patient 5. (A) After infusion of genetically modified CD4+ cells, peripheral blood samples were collected, PBMCs were isolated, 106 (Left) or 105 (Right) cells were lysed, and DNA analysis was performed for detection of Rev M10 or ΔRev M10 cells. Multiple limiting dilution and competitive DNA PCR analyses were performed at each time point. The bars represent the average of these analyses. (Inset) The relative percentage of the genetically modified cells in the PBMCs at the time of infusion, determined by DNA PCR. For the Rev M10 vector, 1% of 2.2 × 1010 cells were transduced, and for ΔRev M10, 10% of 2.2 × 1010 cells were genetically modified and infused. (B) A time course of RNA expression in patient 5 of cells stimulated with anti-CD3 and anti-CD28 antibodies by RT-PCR analysis is shown. Total RNA was extracted from 106 and 105 PBLs and used in the RT-PCR.
Figure 4.
Detection of pLJ Rev-M10 and pLJ-ΔRev M10 DNA or RNA in patient 6. (A) DNA PCR of 106 (Left) or 105 (Right) cells is shown with relative ratios at the time of infusion presented (Inset). For the Rev M10 vector, 2% of 4 × 109 cells were transduced and for ΔRev M10, 10% of 1 × 109 cells were gene-modified. Equal numbers of cells, genetically modified with each vector, were infused (8 × 108). (B) RT-PCR analysis of RNA from PBMCs stimulated with anti-CD3 and anti-CD28 is shown at the indicated times after reinfusion.
Figure 5.
Detection of pLJ-Rev M10 and pLJ-ΔRev M10 DNA or RNA in patient 7. (A) DNA PCR of 106 (Left) or 105 (Right) cells is shown with relative ratios at the time of infusion presented (Inset). For the Rev M10 vector, 1% of 9 × 109 cells were transduced and for ΔRev M10, 10% of 109 cells were gene-modified. Equal numbers of cells modified with each vector were reinfused (9 × 107). (B) RT-PCR analysis of RNA from PBMCs stimulated with anti-CD3 and anti-CD28 is shown at the indicated times after reinfusion.
Detection of DNA in transduced cells confirmed the presence of the relevant gene-modified cells at different times after gene transfer of autologous cells in these patients. To determine whether these cells could transcribe Rev M10, RT-PCR analysis was performed. Because these cells were largely quiescent, RNA was not readily detected unless cells were activated in vitro (data not shown; see also ref. 26). After stimulation with antibodies to CD3 and CD28 in vitro, which promotes the selective growth of CD4+ cells (31), RNA was detected for extended time periods, although not as long as those observed by using DNA PCR. By RT-PCR analysis, cells transduced with either vector were detected at the early time points (Figs. 3B, 4B, and 5B). At later time points, Rev M10 was preferentially detected. In patient 5, Rev M10 was detected up to day 28 but was undetectable at later times (Fig. 3B). In patient 6, only Rev M10 DNA was detected on day 56 (Fig. 4B), and Rev M10 RNA persisted as long as day 84 in patient 7, at which time no control vector RNA was detected (Fig. 5B). Thus, in all patients, the ability of genetically modified cells to synthesize vector mRNA after cell transfer in vivo was confirmed.
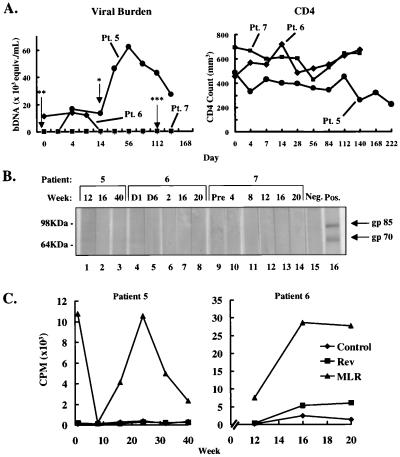
In addition to molecular genetic analyses, patients were evaluated for a variety of serum biochemical, hematological, and immune parameters. In all patients, no change in a variety of biochemical parameters were observed, including markers of major organ function or inflammation responses (data not shown). During the protocol, viral load was monitored by using the branched DNA protocol (Chiron). Though the patients displayed different levels at the time of entry and changes in viral load were observed when antiviral drug regimens were changed (Fig. 6A), no consistent changes were observed in association with the cell infusions. The increase in viral load observed on day 14 in patient 5 was related temporally to the discontinuation of AZT treatment. Because this patient had been on this medication for a short period of time, AZT likely exerted antiretroviral effects up to this time, and AZT withdrawal presumably allowed increased virus replication in vivo.
Figure 6.
Immune and virologic analyses. (A) Viral load and CD4 counts in experimental subjects before and after gene transfer. Viral load, detected by branched DNA analysis (Chiron), and CD4 counts, determined by FACS analysis, were monitored in the patients at specified times relative to infusion of gene-modified cells. At the time of infusion, patient 5, a 38-year-old male, was receiving AZT antiviral treatment, which was stopped on day 14 (∗, arrow); patient 6, a 39-year-old male, was maintained on AZT, which was begun ≈ 1 week before cell transfer (∗∗, arrow), followed by AZT and lamivudine (3TC), which was begun on day 112 (∗∗∗, arrow); and patient 7, a 48-year-old male was receiving AZT long term, dideoxyinosine (dDI), which was begun ≈1 month before the treatment, and acyclovir, which was begun ≈2 months before the treatment. (B) Western blot for the presence of Moloney murine leukemia virus (MoMuLV) reactive antibodies in the sera of the patients. Purified MoMuLV envelope glycoproteins gp70 and gp85 were separated by SDS electrophoresis and transferred to multiple nitrocellulose strips. Sera isolated from patient blood samples at intervals postinfusion were incubated with the protein-coated strips to detect the presence of gp70 and gp85 reactive antibodies. Neg, negative control sera obtained from an untreated donor. Pos, positive control sera. Pre, sera collected before infusion (pretreatment). (C) Lack of T cell response to Rev M10 after retroviral transduction. After infusion, PBLs were isolated from peripheral blood by Ficoll density gradient centrifugation and were seeded in flat-bottom 96-well plates in RPMI 1640 medium (0.2 × 106 cells/200 μl medium per well). The cells were cultured for 6 days in the absence or presence of the Rev protein (2 μg/ml) and analyzed by 3H-thymidine incorporation (48). Results obtained from two subjects, patients 5 and 6, are presented. There were not sufficient numbers of cells available from patient 7 to perform this analysis conclusively. In parallel, PBMCs derived from normal donors were irradiated (2,000 rads) and added to wells containing the PBMC of the HIV-seropositive patients in equal numbers, instead of the Rev protein, to serve as the positive control mixed lymphocyte reaction (MLR).
Immunologic analysis revealed that CD4 counts remained relatively stable throughout the study period (Fig. 6A). Another important issue regarding retroviral gene transfer is the potential immune response either to the vector or to the recombinant gene product. To detect immune responses to the murine retroviral vector, a Western blot analysis was performed by using patient sera from different times after cell transfer to detect reactivity to Moloney murine leukemia virus-derived gp70. This analysis revealed no serum antibodies reactive to gp70 by this method (Fig. 6B), indicating that the retroviral vector was not immunogenic as administered in this protocol. The potential of retroviral gene transfer of Rev M10 to induce immunity to Rev protein was determined by using a lymphocyte proliferation assay (32). This assay was chosen because it has proven to be the most sensitive and reliable indicator of T cell responses in mice immunized with Rev M10 (data not shown) and has been used to detect reactivity to Rev in HIV-infected individuals (33). In protocol subjects, however, minimal responses were detected to Rev, either before or after gene transfer, in contrast to their ability to respond to an allogeneic stimulus in the mixed lymphocyte proliferation assay (Fig. 6C).
DISCUSSION
Recent improvements in antiretroviral drug treatments have resulted in remarkable reductions of viral loads in HIV-seropositive patients. Unless virus can be completely eliminated, however, the concern remains that low levels of viral replication will lead to continued depletion and damage to the immune system. In fact, the selective pressure of chemotherapy may generate drug-resistant strains of virus that retain the pathogenic virulence of wild-type strains (34). Additional therapeutic approaches to enhance immune function therefore are likely to be beneficial. Gene transfer approaches may complement antiretroviral therapy and facilitate reconstitution of the immune system, and such combinations potentially could provide greater protection against HIV replication and delay disease progression.
Among recombinant genes explored in gene transfer studies, Rev M10 has been shown by several laboratories to protect human T cells from productive HIV replication after HIV-1 challenge in vitro (1–3, 15, 25). Cells that express Rev M10 inhibit productive viral replication of both laboratory and clinical HIV isolates without affecting several normal T cell functions (4, 25). We also recently have demonstrated that transduction of human primary PBMCs with Rev M10 using gold microparticle-mediated DNA transfer can lead to stable Rev M10 gene expression in these cells (4). Despite promising results in preclinical and early clinical studies, it remains important to determine whether Rev M10 protects CD4+ T cells in vivo and to promote prolonged engraftment of functional lymphocytes resistant to HIV infection.
In the present study, survival of cells transduced using retroviral vectors was determined. The persistence of gene-modified cells was established by PCR of DNA extracted from PBMCs. After a single infusion of transduced cells, the Rev M10 retroviral vector DNA was detected for over 4 months in all three patients and was detected for 9 months in one patient. Rev M10-transduced cells thus have now been shown to prolong survival compared with ΔRev M10 negative controls using either nonviral or viral vectors. In comparison to the previous study with nonviral gene delivery, engraftment with retroviral vector transduction appeared more persistent. In addition, RNA expression from cells recovered from patients was detected for longer times. This finding may be caused, in part, by the enhanced efficiency of stable gene transfer. In addition, the cell activation signals used in vitro in this study are more effective in stimulating CD4 cell growth and less likely to induce apoptosis (31, 35), which may have provided a more effective method to propagate and activate cells to transcribe vector RNA. DNA was more readily detected than RNA by PCR, suggesting that the majority of cells introduced into the patient were quiescent. The DNA signal reflected the total number of cells that received the vector and/or had undergone cell division subsequent to this event, whereas the RNA expression was detected only in a subset of these cells that were activated to transcribe the gene in vitro. These data demonstrated that inducible transcription of Rev M10 mRNA was detectable for considerable time periods after reinfusion of gene-modified T cells.
A potential obstacle to the persistence of cells expressing foreign proteins in vivo is the development of host immune responses to recombinant proteins. Although Rev M10 RNA was induced in cells after extended periods postinfusion, it is likely that Rev M10 expression was not maintained at high levels constitutively in quiescent T cells. In addition, this study, as well as those reported previously (36), suggest that Rev is not highly antigenic. Multiple administrations of gene-modified cells may induce such immunity, as reported for a distinct gene product, a thymidine kinase/hygromycin-resistance fusion protein in a different T cell subset, CD8+ cells (37). Based on the prolonged persistence, as well as our inability to demonstrate active immunity in patients 5 and 6, it appeared that the protocol described here did not induce a significant immune response to Rev M10 in CD4+ cells.
Other studies have documented a lack of host immunity to gene-modified cells modified by retroviral vectors encoding selectable markers or other potentially therapeutic genes (38–42). In contrast, increased immune reactivity has been observed after infusion of gene-modified HIV-specific cytotoxic T lymphocytes in HIV-infected patients (37). Alternative viral vectors, such as adenovirus, have been limited by the immunogenicity of recombinant proteins eliciting a strong host immune response against genetically modified cells (43–46). Immune responses to fetal calf serum also have been described that can considerably diminish the half-life of cells grown in the presence of heterologous sera. Thus, although it cannot be assumed that such responses will be uniformly induced and limit the efficiency of gene therapy, it is important to monitor these potential responses and to develop strategies to modulate them if necessary.
From this study, the mode of gene delivery appears likely to influence the persistence of genetically modified cells. The survival time of retrovirally transduced Rev M10 T cells generally was longer than was observed with particle-mediated transduced cells reported in the previous trial (168 ± 74 vs. 23 ± 28 days; P = .0017 by Student’s t test). Because this difference was apparently not caused by the immune response, it is likely that the increased persistence of genetically modified cells using retroviral vectors reflected the ability of retroviruses to integrate more efficiently, perhaps in transcriptionally active regions (47), in contrast to the first-generation plasmid vectors used in the previous study (26).
This protocol was aimed primarily at assessing molecular markers of T cell survival after gene transfer and to gather data from pilot studies on the safety and toxicity of the transduction, expansion, and infusion procedures. No toxicity from these procedures was observed in these patients, and there were no major changes in viral load that correlated with the gene or adoptive cell transfer protocols. In addition, patient sera displayed no reactivity to gp70, the envelope protein of Moloney murine leukemia virus, indicating that the persistence of murine retrovirus was unlikely. As expected with the low number of genetically modified cells, no change in CD4 counts or viral load was observed in any patient associated with the cell and gene transfer procedure. Although the results showed enhanced persistence of Rev M10-transduced cells and toxicities that would limit further studies were not evident, additional studies will be required to define the potential clinical efficacy of gene transfer approaches. In addition, it will be important to develop strategies that use immune stimulation procedures to facilitate the engraftment of larger numbers of genetically protected cells in vivo. This combination of treatments, together with antiretroviral agents, may be required to reconstitute immune function needed to eliminate the virus in vivo. The data reported here suggest that T cell-based gene transfer protocols may facilitate efforts to develop such combinational approaches to the treatment of AIDS.
Acknowledgments
We thank the patients who participated in this study, as well as Dr. Paul Watkins and the staff of the General Clinical Research Center for generous assistance. We also thank Ms. Donna Gschwend and Ms. Nancy Barrett for manuscript preparation, and we are also grateful to Drs. Gary Tarpley and Dean Griffith (director of general medicine) of the Upjohn Company, Kalamazoo, MI, for kindly providing delavirdine. We also would like to thank Microbiological Associates (Gaithersburg, MD) for performing GP70 Western blots. This work was supported by National Institutes of Health Grants AI36606, AI36207, and RR00042.
ABBREVIATIONS
- RT
reverse transcriptase
- PBL
peripheral blood lymphocyte
- PBMC
peripheral blood mononuclear cell
- AZT
azidothymidine
References
- 1.Malim M H, Freimuth W W, Liu J, Boyle T J, Lylerly H K, Cullen B R, Nabel G J. J Exp Med. 1992;176:1197–1201. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bevec D, Dobrovnik M, Hauber J, Bohnlein E. Proc Natl Acad Sci USA. 1992;89:9870–9874. doi: 10.1073/pnas.89.20.9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahner I, Zhou C, Yu X, Hao Q, Guatelli J C, Kohn D B. J Virol. 1993;67:3199–3207. doi: 10.1128/jvi.67.6.3199-3207.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woffendin C, Yang Z, Udaykumar, Xu L, Yang N, Sheehy M J, Nabel G J. Proc Natl Acad Sci USA. 1994;91:11581–11585. doi: 10.1073/pnas.91.24.11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullenger B A, Gallardo H F, Ungers G E, Gilboa E. Cell. 1990;63:601–608. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- 6.Sarver N, Cantin E M, Chang P S, Zaia J A, Ladne P A, Stephens D A, Rossi J J. Science. 1990;247:1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- 7.Ojwang J O, Hampel A, Looney D J, Wong-Staal F, Rappaport J. Proc Natl Acad Sci USA. 1992;89:10802–10806. doi: 10.1073/pnas.89.22.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu M, Ojwang J, Yamada O, Hampel A, Rappaport J, Looney D, Wong-Staal F. Proc Natl Acad Sci USA. 1993;90:6340–6344. doi: 10.1073/pnas.90.13.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu M, Poeschla E, Wong-Staal F. Gene Ther. 1994;1:13–26. [PubMed] [Google Scholar]
- 10.Chatterjee S, Johnson P R, Wong K K., Jr Science. 1992;258:1485–1488. doi: 10.1126/science.1359646. [DOI] [PubMed] [Google Scholar]
- 11.Trono D, Feinberg M, Baltimore D. Cell. 1989;59:113–120. doi: 10.1016/0092-8674(89)90874-x. [DOI] [PubMed] [Google Scholar]
- 12.Pearson L, Garcia J, Wu F, Modesti N, Nelson J, Gaynor R. Proc Natl Acad Sci USA. 1990;87:5079–5083. doi: 10.1073/pnas.87.13.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marasco W A, Haseltine W A, Chen S Y. Proc Natl Acad Sci USA. 1993;90:7889–7893. doi: 10.1073/pnas.90.16.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan L, Bagasra O, Laughlin M A, Oakes J W, Pomerantz R J. Proc Natl Acad Sci USA. 1994;91:5075–5079. doi: 10.1073/pnas.91.11.5075. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Liu J, Woffendin C, Yang Z, Nabel G J. Gene Ther. 1994;1:32–37. [PubMed] [Google Scholar]
- 16.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammarskjold M L, Heimer J, Hammarskjold B, Sangwan I, Albert L, Rekosh D. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malim M H, Hauber J, Le S Y, Maizel J V, Cullen B R. Nature (London) 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 19.Luo Y, Yu H, Peterlin B M. J Virol. 1994;68:3850–3856. doi: 10.1128/jvi.68.6.3850-3856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruhl M, Himmelspach M, Bahr G M, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington G K, Probst H, Bevec D, Hauber J. J Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogerd H P, Fridell R A, Madore S, Cullen B R. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 22.Malim M H, Cullen B R. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 23.Malim M H, McCarn D F, Tiley L S, Cullen B R. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malim M H, Bohnlein S, Hauber J, Cullen B R. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 25.Fox B A, Woffendin C, Yang Z Y, San H, Ranga U, Gordon D, Osterholzer J, Nabel G J. Hum Gene Ther. 1995;6:997–1004. doi: 10.1089/hum.1995.6.8-997. [DOI] [PubMed] [Google Scholar]
- 26.Woffendin C, Ranga U, Yang Z, Xu L, Nabel G J. Proc Natl Acad Sci USA. 1996;93:2889–2894. doi: 10.1073/pnas.93.7.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nabel G J, Fox B A, Post L, Thompson C B, Woffendin C. Hum Gene Ther. 1994;5:79–92. doi: 10.1089/hum.1994.5.1-79. [DOI] [PubMed] [Google Scholar]
- 28.Woffendin C, Ranga U, Nabel G J. In: Current Protocols in Human Genetics. Dracopoli N C, Haines J L, Korf B R, Moir D T, Morton C C, Seidman C E, Seidman J G, Smith D R, editors. New York: Wiley; 1996. p. 13.6. [Google Scholar]
- 29.Korman A J, Frantz J D, Strominger J L, Mulligan R C. Proc Natl Acad Sci USA. 1987;84:2150–2154. doi: 10.1073/pnas.84.8.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potts B J. In: Mini Reverse Transcriptase Assay. Aldovini A, Walker B D, editors. New York: Stockton; 1990. pp. 103–106. [Google Scholar]
- 31.Levine B L, Mosca J D, Riley J L, Carroll R G, Vahey M T, Jagodzinski L L, Wagner K F, Mayers D L, Burke D S, Weislow O S, St. Louis D C, June C H, Louis D C. Science. 1996;272:1939–1943. doi: 10.1126/science.272.5270.1939. [DOI] [PubMed] [Google Scholar]
- 32.Orosz C G, Horstemeyer B, Zinn N E, Bishop D K. Transplantation. 1989;47:189–194. doi: 10.1097/00007890-198901000-00039. [DOI] [PubMed] [Google Scholar]
- 33.Blazevic V, Ranki A, Krohn K J E. AIDS Res Hum Retroviruses. 1995;11:1335–1342. doi: 10.1089/aid.1995.11.1335. [DOI] [PubMed] [Google Scholar]
- 34.Emery S, Lane H C. AIDS. 1996;10:159–163. doi: 10.1097/00002030-199601001-00022. [DOI] [PubMed] [Google Scholar]
- 35.Boise L H, Minn A J, Noel P J, June C H, Accavitti M A, Lindsten T, Thompson C B. Immunity. 1995;3:87–98. doi: 10.1016/1074-7613(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 36.Reiss P, De Ronde A, Lange J M, De Wolf F, Dekker J, Danner S A, Debouck C, Goudsmit J. AIDS Res Hum Retroviruses. 1989;5:621–628. doi: 10.1089/aid.1989.5.621. [DOI] [PubMed] [Google Scholar]
- 37.Riddell S R, Elliott M, Lewinsohn D A, Gilbert M J, Wilson L, Manley S A, Lupton S D, Overell R W, Reynolds T C, Corey L, Greenberg P D. Nat Med. 1996;2:216–223. doi: 10.1038/nm0296-216. [DOI] [PubMed] [Google Scholar]
- 38.Dai Y, Roman M, Naviaux R K, Verma I M. Proc Natl Acad Sci USA. 1992;89:10892–10895. doi: 10.1073/pnas.89.22.10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Culver K W, Morgan R A, Osborne W R, Lee R T, Lenschow D, Able C, Cornetta K, Anderson W F, Blaese R M. Hum Gene Ther. 1990;1:399–410. doi: 10.1089/hum.1990.1.4-399. [DOI] [PubMed] [Google Scholar]
- 40.Rosenberg S A, Aebersold P, Cornetta K, Kasid A, Morgan R A, Moen R, Karson E M, Lotze M T, Yang J C, Topalian S L, Merino M J, Culver K, Miller A D, Blaese R M, Anderson W F. N Engl J Med. 1990;323:570–578. doi: 10.1056/NEJM199008303230904. [DOI] [PubMed] [Google Scholar]
- 41.Brenner M K, Rill D R, Holladay M S, Heslop H E, Moen R C, Buschle M, Krance R A, Santana V M, Anderson W F, Ihle J N. Lancet. 1993;342:1134–1137. doi: 10.1016/0140-6736(93)92122-a. [DOI] [PubMed] [Google Scholar]
- 42.Kohn D B, Weinberg K I, Nolta J A, Heiss L N, Lenarsky C, et al. Nat Med. 1995;1:1017–1023. doi: 10.1038/nm1095-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Nunes F A, Berencsi K, Gonczol E, Engelhardt J F, Wilson J M. Nat Genet. 1994;7:362–369. doi: 10.1038/ng0794-362. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y, Li Q, Ertl H C, Wilson J M. J Virol. 1995;69:2004–2015. doi: 10.1128/jvi.69.4.2004-2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brody S L, Metzger M, Danel C, Rosenfeld M A, Crystal R G. Hum Gene Ther. 1994;5:821–836. doi: 10.1089/hum.1994.5.7-821. [DOI] [PubMed] [Google Scholar]
- 46.Crystal R G, McElvaney N G, Rosenfeld M A, Chu C S, Mastrangeli A, Hay J G, Brody S L, Jaffe H A, Eissa N T, Danel C. Nat Genet. 1994;8:42–51. doi: 10.1038/ng0994-42. [DOI] [PubMed] [Google Scholar]
- 47.Varmus H. Science. 1988;240:1427–1435. doi: 10.1126/science.3287617. [DOI] [PubMed] [Google Scholar]
- 48.Bishop D K, Orosz C. Transplantation. 1989;47:671–677. [PubMed] [Google Scholar]