Abstract
A newly emerging family of phosphatases that are members of the haloacid dehalogenase superfamily contains the catalytic motif DXDX(T/V). A member of this DXDX(T/V) phosphatase family known as Dullard was recently shown to be a potential regulator of neural tube development in Xenopus [Satow R, Chan TC, Asashima M (2002) Biochem Biophys Res Commun 295:85–91]. Herein, we demonstrate that human Dullard and the yeast protein Nem1p perform similar functions in mammalian cells and yeast cells, respectively. In addition to similarity in primary sequence, Dullard and Nem1p possess similar domains and show similar substrate preferences, and both localize to the nuclear envelope. Additionally, we show that human Dullard can rescue the aberrant nuclear envelope morphology of nem1Δ yeast cells, functionally replacing Nem1p. Finally, Nem1p, has been shown to deposphorylate the yeast phosphatidic acid phosphatase Smp2p [Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S (2005) EMBO J 24:1931–1941], and we show that Dullard dephosphorylates the mammalian phospatidic acid phosphatase, lipin. Therefore, we propose that Dullard participates in a unique phosphatase cascade regulating nuclear membrane biogenesis, and that this cascade is conserved from yeast to mammals.
Keywords: C-terminal domain phosphatase, Dullard, lipin, nuclear envelope biogenesis, phosphatidic acid phosphatase
Reversible phosphorylation is governed by a large number of cellular protein kinases and protein phosphatases (1). The protein phosphatases can be classified into at least three major families based on amino acid sequences at their active sites (1). The three families include the protein serine/threonine phosphatases with the active site sequence: DXH(X)nDXXD(X)nNH(D/E) (2), the protein tyrosine phosphatases possessing the active site sequence C(X)5R (1), and a newly emerging protein phosphatase family containing the active site sequence DXDX(T/V). This latter family of phosphatases belongs to the haloacid dehalogenase (HAD) superfamily (3–6).
One of the best-studied DXDX(T/V) phosphatases dephosphorylates the phosphorylated C terminus of RNA polymerase II (7–9). This latter phosphatase is named TFIIF-interacting C-terminal domain (CTD) phosphatase 1 (FCP1). Using the human sequence of FCP1, it is possible to identify a small subfamily of the DXDX(T/V) phosphatases that include small CTD phosphatase 1 (SCP1), SCP2, SCP3, translocase of inner mitochondrial membrane 50 (TIMM50), HSPC129 (CTD small phosphatase like 2, CTDSPL2), ubiquitin-like domain containing CTD phosphatase (UBLCP), and Dullard. The biological functions of these phosphatases are just beginning to be understood. SCP1 was recently shown to function as a transcriptional corepressor inhibiting neuronal gene transcription in nonneuronal cells (10, 11). SCP2 is amplified in human sarcomas (12), whereas SCP3 was recently reported to be a tumor suppressor gene deleted in a lung carcinoma cell line (13). UBLCP possesses an ubiquitin-like domain (UBQ), but its function is unknown (14). TIMM50 localizes to mitochondria and is part of the molecular machinery for mitochondrial protein import (15, 16). Finally, Dullard was shown to be important in neural tube development in Xenopus (17), and recently it was also reported to inhibit bone morphogenic protein (BMP) receptor activation (18).
Some of the DXDX(T/V) phosphatases are conserved throughout evolution. Saccharomyces cerevisiae clearly has a protein that resembles human FCP1 (7). In addition, the yeast genome has four additional phosphatases that contain the DXDX(T/V) motif and also share sequence similarity to Fcp1p. Two of these phosphatases, Psr1p (plasma membrane sodium response protein 1) and Psr2p, are required for the activation of the sodium stress response in yeast, and these phosphatases dephosphorylate stress-regulated transcription factors (19, 20). Yeast also has Timm50p, which, like human TIMM50, plays a role in transporting proteins into the mitochondria (15, 16). Finally, a DXDX(T/V) protein known as Nem1p (nuclear envelope morphology protein 1) forms a complex with Spo7p (Sporulation-specific protein 7), and this complex is involved in the formation of a spherical nucleus (21). One target of the Nem1p-Spo7p complex is the nuclear membrane-associated phosphatidic acid phosphatase, Smp2p (22, 23). The yeast data suggest there is a critical signaling cascade involving the Nem1p/Spo7p protein complex and Smp2p, and that this pathway plays a critical role in nuclear membrane biogenesis (24–26). It is interesting to note that the phosphatidic acid phosphatase, Smp2p, is also a member of the superfamily of HAD proteins that contains a DXDX(T/V) phosphatase catalytic sequence (23).
Herein, we demonstrate that the mammalian protein Dullard and the yeast protein Nem1p are orthologous protein phosphatases. This orthologous relationship is based on the evolutionary relationship between Dullard and Nem1p and the similarities among their transmembrane topologies, substrate preferences, and subcellular localizations. Most importantly, Dullard serves as a functional equivalent of Nem1p in yeast. In addition, our results suggest that Dullard dephosphorylates the mammalian equivalent of Smp2p, a recently identified protein known as lipin (24). Finally, we demonstrate that Dullard also regulates nuclear membrane biogenesis, suggesting that the essential elements of this phosphatase signaling cascade are conserved from yeast to humans.
Results
Phylogeny of CTD Phosphatases.
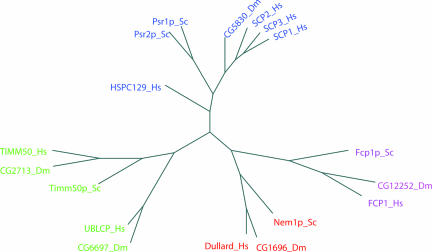
We analyzed the CTD phosphatases of Homo sapiens, Drosophila melanogaster, and S. cerevisiae to determine their phylogenetic relationships (Fig. 1). The first closely related group of proteins includes human SCP1, -2, and -3, HSPC129, and yeast Psr1p and -2p, along with CG5830 from D. melanogaster. A second group is divided into two subclasses, the TIMM50 and the UBLCP subclasses. Human UBLCP and fly CG6697 are distinct from the TIMM50 branch and appear to be restricted to higher eukaryotes. The third group is divided into the FCP1 and the Dullard subclasses. Both FCP1 and Dullard family members appear to be conserved in yeast, fly, and human.
Fig. 1.
A phylogeny of CTD phosphatases. Human Dullard (H. sapiens, Hs), fruit fly CG1696 (D. melanogaster, Dm), and yeast Nem1p (S. cerevisiae, Sc) are in red. GenBank accession numbers are provided in SI Methods.
Dullard Has Similar Domains to Nem1p.
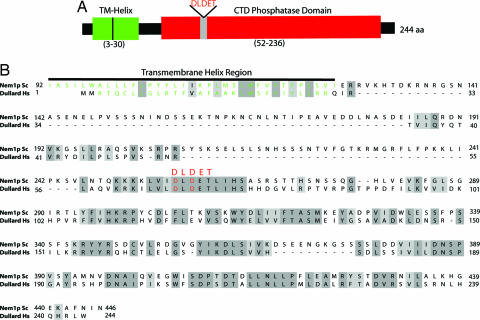
Sequence alignment of Dullard and Nem1p show they share ≈50% amino acid sequence similarity, however, their N termini are quite divergent (Fig. 2B). To determine whether the N termini of Nem1p and Dullard contain similar properties, we investigated both by bioinformatic analyses. The N terminus of Nem1p is predicted to contain a transmembrane helix motif of ≈40 residues with two potential membrane-spanning regions (Fig. 2B). The presence of these two membrane-spanning domains has been addressed experimentally for Nem1p, and this domain directs Nem1p to the nuclear envelope and endoplasmic reticulum (21). A similar analysis predicted that the N terminus of Dullard also possesses two membrane-spanning regions (Fig. 2B). Thus, despite primary sequence divergence, both Nem1p and Dullard contain predicted N-terminal transmembrane regions and a C-terminal DLDET phosphatase domain.
Fig. 2.
The CTD phosphatase domain and transmembrane helix (TM-Helix) of Dullard. (A) The representation of the domain organization of Dullard. TM-Helix, transmembrane-helix. The DLDET catalytic signature motif is in red. (B) The sequence alignment of Dullard and Nem1p. The residues in the transmembrane region in Dullard and Nem1p are in green.
To confirm that the N terminus of Dullard possesses elements to direct its localization, we generated an N-terminal truncation removing the first 45 residues of the protein (Δ1–45), which deletes the entire predicted membrane-spanning region. An EGFP fusion construct of Dullard was expressed in HeLa cells. Fluorescence microscopy demonstrated that Dullard localized to the perinuclear region [supporting information (SI) Fig. 6A]. A similar localization was observed by using other epitope tags (V5, FLAG, and myc; data not shown; ref. 27). This localization was abolished by the Δ1–45 truncation (SI Fig. 6B). Additionally, fusing the first 45 amino acids of Dullard to EGFP-targeted EGFP to the perinuclear region (SI Fig. 6C), supporting the prediction that Dullard possesses membrane-spanning regions in its N terminus. Therefore, both Nem1p and Dullard appear to be targeted to the nuclear envelope and endoplasmic reticulum by a similar membrane-spanning domain (Fig. 2B).
Dullard Is a Protein Ser/Thr Phosphatase.
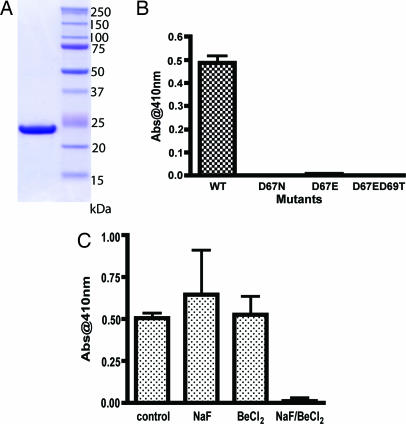
The N-terminal transmembrane region (Fig. 2B, residues 3–30) of Dullard is very hydrophobic and renders recombinant Dullard insoluble. Therefore, to test the enzymatic properties of Dullard, we constructed a C-terminal-His tagged protein lacking the first 45 amino acids of the protein. We produced the protein in Escherichia coli and purified it to near homogeneity (Fig. 3A). Dullard (46–244)-His6 was used for all biochemical characterizations. To determine whether Dullard acts as a protein phosphatase, we performed a kinetic analysis by using the artificial substrate, p-nitrophenyl phosphate (p-NPP). A pH/rate profile for Dullard, as measured by p-NPP hydrolysis, revealed that Dullard has a pH optimum at pH 5.5 (data not shown). Therefore, assays were performed at this pH. Recombinant Dullard exhibited a Km of 18 mM and kcat of 1 s−1 by using p-NPP as a substrate.
Fig. 3.
The activity of recombinant Dullard (46–244)-His6. (A) The purification of Dullard (46–244)-His6. Ten micrograms of protein was loaded onto gel and stained with Coomasie blue. (B) The activity against pNPP of wild-type and mutants (D67N, D67E, and D67ED69T) of recombinant human Dullard (46–244)-His6. The reactions were performed with 500 ng of each protein, and the absorbance at 410 nm was measured after termination of reaction with 0.25 M NaOH. The error bar is the standard deviation of three measurements. (C) The activity against pNPP of wild-type of recombinant human Dullard (46–244)-His6 with different inhibitors. The reactions were performed with 500 ng of wild-type recombinant protein and 1 mM NaF, 0.1 mM BeCl2, or a mixture of 1 mM NaF and 0.1 mM BeCl2. The error bar is the standard deviation of three measurements.
Because Dullard shares the DLDET motif with other CTD phosphatases (Fig. 2A), we examined the effect of mutating the predicted catalytic aspartate (DLDET) to asparagine (D67N) or to glutamate (D67E). As expected, mutation of the aspartate residue to either glutamate or asparagine abolished phosphatase activity (Fig. 3B). Additionally, we mutated the second conserved aspartic acid residue in the DLDET motif to threonine as well as producing the D67E mutation (D67ED69T) to see whether the EXTX motif, which is found in TIMM50, has catalytic activity against p-NPP. The D67ED69T mutant also lacked any enzymatic activity against p-NPP (Fig. 3B).
To explore the possibility that Dullard dephosphorylation proceeds by a pentacovalent phosphorus intermediate like other members of the HAD superfamily, Dullard activity was examined in the presence of several inhibitors (Fig. 3C). Like other HAD family proteins (28, 29), Dullard is inhibited by BeF3, which is known to form a pentacovalent intermediate; Dullard is not inhibited by NaF or BeCl2. The BeF3− most likely mimics phosphate coordination within Dullard's active site and thus effectively inhibits the enzyme. These results suggest that Dullard shares a similar catalytic mechanism to other proteins in the HAD superfamily.
Next, we used a phosphopeptide array of 360 phosphopeptides to explore Dullard's substrate preference. Recombinant Dullard hydrolyzed 73 of 174 phosphoserine peptides. Surprisingly, recombinant Dullard hydrolyzed only 1 of 57 phosphothreonine peptides and did not hydrolyze any of the 129 phosphotyrosine peptides. These data demonstrate that Dullard has a marked preference for dephosphorylating phosphoserine- or phosphothreonine-containing peptides, and that recombinant Dullard does not hydrolyze a variety of phosphotyrosine-containing peptides.
To explore the optimal phosphopeptide sequence motif necessary for recognition by Dullard, we followed the rates of phosphopeptide hydrolysis. Seven of the 360 phosphopeptides showed >75% hydrolysis by Dullard at early time points in the reaction. The best phosphopeptide substrates are shown in Table 1. These peptides contain basic residues in their N-terminal positions (relative to the sites of phosphorylation), and some of the peptides also have a proline residue following the phosphorylated residue. It is interesting to note that Nem1p displays a modest preference for the sequence motifs: RXX(pS) and (pS/pT)P (22), suggesting that Dullard and Nem1p are likely to have common biological substrates.
Table 1.
Results from a phosphopeptide array assay
The sequences having >75% of phosphate hydrolysis were selected. The prolines are in green, basic residues are in blue, and phosphorylated serines are in red.
Localization of Endogenous Dullard.
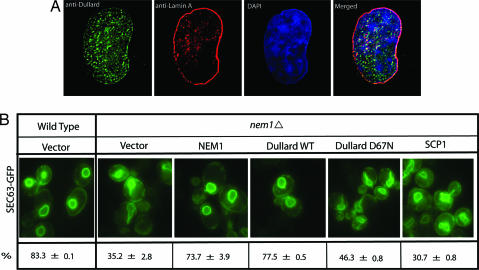
To determine the localization of endogenous Dullard, we generated a polyclonal antibody to recombinant Dullard and probed several mammalian cell lines (HEK 293, COS-7, and HeLa) with this antibody. This analysis showed that Dullard localizes in a punctuate pattern around and inside the nucleus. To test the specificity of this antibody, we preincubated the antibody with recombinant Dullard and found all localization was specific (data not shown). Because Fig. 4 seems to show a limited amount of Dullard localized to the same location as nuclear lamin, we conducted 3D imaging of Dullard nuclear localization by using deconvolution algorithms (SI Movie 1). This imaging elegantly shows that Dullard is present in the nuclear envelope membrane just beneath the nuclear envelope marker lamin A (SI Movie 1).
Fig. 4.
Subcelluar localization of endogenous Dullard and yeast complementation assay. (A) Subcellular localization of Dullard. Endogenous Dullard was detected with a polyclonal antibody to recombinant Dullard by immunocytochemistry and observed by deconvolution fluorescence microscopy. (B) Yeast complementation assay. nem1Δ cells expressing Sec63p-GFP were transformed with constructs containing NEM1, Dullard wild type, Dullard-D67N, SCP1, or empty vector. Cells were grown at 30°C to early log phase and analyzed by fluorescence microscopy. The scores in three independent experiments were converted to average percentage ± standard deviation.
Dullard Functionally Replaces Nem1p.
Deletion of NEM1 in yeast results in an aberrant nuclear membrane morphology (21). To investigate the functional conservation between Dullard and Nem1p, we expressed Dullard in yeast nem1Δ cells and scored the morphology of the nuclear envelope as normal or aberrant. Both Dullard and Nem1p rescued the aberrant nuclear envelope phenotype (Fig. 4B); however, Dullard-D67N, which is catalytically inactive, did not rescue the morphological phenotype (Fig. 4B). Likewise, SCP1, another member of the CTD phosphatase family, did not rescue the phenotype (Fig. 4B). These results demonstrate that Dullard can functionally replace Nem1p.
Dullard Dephosphorylates Lipin.
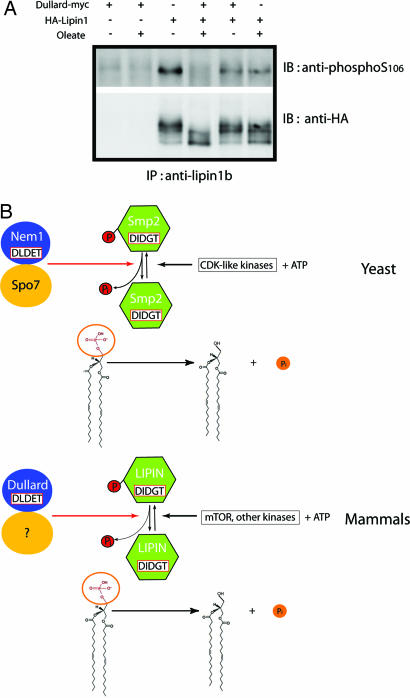
Nem1p, along with its interacting partner Spo7p, dephosphorylates the DXDX(T/V) phosphatidic acid phosphatase, Smp2p (22). Because Dullard can functionally replace Nem1p in nem1Δ cells, we hypothesized that Dullard could function to dephosphorylate the mammalian equivalent of Smp2p. Recent studies have suggested that a small family of proteins known as lipins (lipins 1, 2, and 3) shows sequence homology to Smp2p. Lipin 1 is also a DXDX(T/V) containing phosphatase which can be phosphorylated at multiple (pSer/pThr)Pro sequences (24). The apparent similarities in the yeast and mammalian cell signaling cascades prompted us to conduct in vitro dephosphorylation assays of lipin by using recombinant Dullard. We were surprised to observe that recombinant Dullard did not dephosphorylate lipin in vitro (data not shown). We speculated that Dullard might require a mammalian protein corresponding to the yeast counterpart of Spo7p. Therefore, we examined whether Dullard dephosphorylates lipin 1 in vivo in HeLa cells, HEK293A cells, and BHK cells. Dullard significantly dephosphorylates mouse lipin 1b only in BHK cells (Fig. 5A). This is most clearly seen by using the antibody prepared against the phosphorylation site Ser-106, because lipin can be dephosphorylated at multiple sites (24). Oleic acid is a known stimulant that induces dephosphorylation of lipin 1 (24). As expected, lipin 1 was dephosphorylated to a higher degree in cells expressing Dullard upon treatment with oleic acid. Additionally, there is no effect on the in vivo dephosphorylation of lipin 1 with the active site mutant (D67N) of Dullard (data not shown). Therefore, these results suggest that Dullard is responsible for the in vivo dephosphorylation of lipin 1 in BHK cells, which is enhanced by oleic acid. Our results suggest that Dullard dephosphorylates lipin in BHK cells but not in HeLa cells or HEK293A cells. Although we do not know the reason for this selective dephosphorylation of lipin in BHK cells, it is possible that HeLa cells and HEK293A cells do not have enough of the mammalian equivalent of the yeast protein Spo7p. Lack of the equivalent would render Dullard inactive toward lipin 1.
Fig. 5.
Dullard dephosphorylates lipin 1. (A) In vivo dephosphorylation of lipin by Dullard with oleic acid treatment in BHK cells. Cells were cotransfected with HA-lipin and Dullard-myc. Cell extracts were immunoprecipitated with antibody to lipin and then blotted with antibodies to phospho-S106 or HA tag. (B) Schematic of proposed relationship between Nem1p complex and Dullard complex.
Discussion
We have shown that Dullard is a protein Ser/Thr phosphatase localized to the nuclear envelope. Previous reports demonstrate that Dullard is localized to mitochondria (30), but we see no evidence for this in any of our localization studies. It appears that the NH2-terminal 45 residues of Dullard are capable of targeting EGFP to the nuclear membrane and endoplasmic reticulum. We also demonstrate that Dullard can functionally replace Nem1p in yeast and is a likely ortholog of yeast Nem1p. Finally, we show that Dullard can dephosphorylate lipin 1, suggesting that the biological role of Dullard is similar to that of yeast Nem1p in regulating nuclear membrane biogenesis.
A genetic interaction study with the nucleoporin Nup84p in yeast identified the two proteins, Nem1p and Spo7p (21). Subsequently, it was demonstrated that Nem1p and Spo7p form a protein complex, and that the two proteins have a role in nuclear envelope morphogenesis by regulating Smp2p (22). Smp2p is phosphorylated by cell cycle-related kinases and dephosphorylated by the complex of Nem1p and Spo7p (22). Dephosphorylated Smp2p then dephosphorylates phosphatidic acid and produces diacylglycerol in yeast (23, 25). The diacylglycerol is used in lipid synthesis, and dephosphorylation of phosphatidic acid induces translocation of the lipid synthesis regulator, Overproducer of inositol protein 1 (Opi1p) from the endoplasmic reticulum to the nucleus (25, 26). Nuclear Opi1p attenuates the transcription of lipid synthesis-related genes (25). Therefore, dephosphorylation of Smp2p by the Nem1p/Spo7p complex is an important regulatory mechanism of nuclear membrane biogenesis during cell division in yeast.
The mammalian ortholog of Smp2p appears to be lipin (22). Mutations in lipin result in lipodystrophy (31). Like Smp2p, lipin is a phosphatidic acid phosphatase and produces diacylglycerol in mammalian cells (24). Lipin also can be phosphorylated and dephosphorylated by cellular stimulants, such as insulin or oleic acid (24), and this regulation is connected to its function in adipocyte development (32–34). Lipin is phosphorylated on at least 19 residues (24) and can be phosphorylated by several kinases (34). Dephosphorylation of lipin results in its translocation to the nuclear envelope and endoplasmic reticulum membranes from the cytosol and generation of diacylglycerol (24). Our findings suggest that Dullard and lipin participate in a signaling cascade that regulates nuclear membrane biogenesis in a similar fashion to the Nem1p-Smp2p signaling module observed in yeast.
Interestingly, lipin and Smp2 have the same catalytic signature motif, DIDGT, and Nem1p and Dullard also have the same catalytic signature motif, DLDET. Both motifs originate from the HAD superfamily. This signaling cascade consists of two phosphatases with a DXDX(T/V) motif. Although kinase signaling cascades are extremely well known, there are few phosphatase signaling cascades that have been described. To illustrate the parallel nature of this signaling pathway, we have outlined the specific steps in both yeast and mammals (Fig. 5B). In yeast, Nem1p dephosphorylates Smp2p, and then dephosphorylated Smp2p dephosphorylates phosphatidic acid. In mammals, Dullard dephosphorylates lipin, and then dephosphorylated lipin dephosphorylates phosphatidic acid. The readout of either signaling cascades has a pronounced effect on regulating nuclear membrane biogenesis.
It was recently reported that Dullard can dephosphorylate the BMP receptor II, leading to down-regulation of the BMP pathway (18). Although this study may provide some useful insights into Dullard's biological function, the localizations of Dullard and the BMP receptor are expected to be in different cellular locations. It is interesting to note that Satow et al. (18) used an N-terminal tagged form of Dullard. Placing a tag on the N terminus might lead to mislocalization of the protein. In addition, overexpression of phosphatases in cultured cells can lead to nonspecific dephosphorylation. It would appear that additional work will be needed to clarify the role of Dullard in the BMP receptor signaling pathway.
Finally, it is interesting to note that inactivation of Dullard was originally shown to have a profound effect on brain development in Xenopus (17). Again, there does not appear to be a good explanation for this biological result. It is possible that altering nuclear membrane biogenesis could have a profound effect on brain development. Conversely, it is possible that Dullard may have multiple functions that in turn lead to the observed phenotype.
We have made considerable progress in elucidating the biological function of Dullard; however, there are many unanswered questions that remain to be addressed. It will be important to identify the mammalian equivalent of Spo7p (Fig. 5B) and to more fully characterize the biochemical interaction between Dullard and lipin 1. It is also critical to establish the role of dephosphorylated lipin in regulating the expression of genes involved in lipid synthesis in mammalian cells.
Materials and Methods
Cloning and expression constructs, protein expression and purification, phylogenetic analysis, generation of polyclonal antibodies to Dullard, and phosphatase assays are available as SI Methods.
Cell Culture, Transfection, and Immunocytochemistry (ICC).
HEK293A, HEK293T, COS-7, BHK, and HeLa cells were maintained at 37°C and 5% CO2 in DMEM (Invitrogen, San Diego, CA) containing 10% FBS. Cells were transfected by using Fugene 6 (Roche, Indianapolis, IN), following the manufacturer's recommended protocol. For ICC, cells were fixed with 3.7% formaldehyde and permeabilized with 0.3% Triton X-100. Cells were incubated with the above-mentioned anti-Dullard polyclonal antibody (1:1,000) and an anti-lamin A monoclonal antibody (Abcam, Cambridge, MA; 1:400) under conditions previously described (35), by using goat anti-rabbit Alexa-488 and goat anti-mouse Alexa-568 secondary antibodies (Invitrogen; 1:500 each). Cells were stained with DAPI to visualize DNA. Microscopy was performed with a Delta Vision Deconvolution Microscope System (Nikon, Melville, NY; TE-200 microscope) by using a ×100 objective and a step size of 0.1 μm. 3D perspectives of the captured images were generated with the Meshviewer program through the visualization services portal of the San Diego Supercomputer Center, University of California at San Diego.
Yeast Complementation Assay.
All yeast strains used in this study are derivatives of BY4741 (EUROFAN II), genotype MATa;his3D1;leu2D0;met15D0;ura3D0. The Sec63-GFP construct was kindly provided by Charles N. Cole (Dartmouth Medical School, Dartmouth, MA). To monitor nuclear membrane morphology, nem1Δ cells were transformed with SEC63-GFP. nem1Δ cells expressing Sec63p-GFP were then transformed with a construct containing NEM1, Dullard, Dullard-D67N, SCP1, or empty vector. Cells containing both constructs were grown in the appropriate selective medium at 30°C to early log phase and analyzed by fluorescence microscopy. At least 200 cells in three independent experiments were scored as having or lacking wild-type nuclear membrane morphology.
In Vivo Dephosphorylation and Coimmunoprecipitation.
Ten-centimeter plates of BHK cells were transfected with 10 μg of plasmid, 8 μg of Dullard-myc/2 μg of pcDNA3, 8 μg of pcDNA3/2 μg of HA-lipin, or 8 μg of Dullard-myc/2 μg of HA-lipin for 48 h. After 24 h, transfected cells were serum-starved in DMEM/0.1% BSA overnight before addition of 0.33 mM oleate (conjugated to 3% BSA) for 1 h. Cells were lysed in 1 ml of immunoprecipitation buffer (50 mM NaF/1 mM EDTA/1 mM EGTA/10 mM sodium phosphate/50 mM β-glycerophosphate, pH 7.4/0.1% Triton X-100) supplemented with 1 mM phenylmethylsulfonylfluoride/10 μg/ml leupeptin/10 μg/ml aprotinin/10 μg/ml pepstatin/0.5 μM microcystin. Cell lysate was incubated for 3 h with 2 μg of antilipin antibodies prebound to 15 μl of protein A-agarose. After thorough washing, the immune complexes were resuspended in SDS load buffer, eluted, and resolved on SDS/PAGE gel. Proteins were detected with antibodies to phospho-S106 and HA tag (Roche). The preparation of antibodies to phospho-S106 of mouse lipin 1b and lipin was reported previously (24).
Supplementary Material
Acknowledgments
This manuscript is dedicated to the memory of Dr. John C. Lawrence, Jr. We thank members of the Dixon laboratory for insightful discussion and critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) Training Grant T32 HD 007203-23 (to M.S.G.), NIH Grant 18024, funds from the Walther Cancer Institute (to J.E.D.), and NIH Grants DK52753 and DK28312 (to J.C.L.). Y.K. was supported by Training Fellowship from the California Institute of Regenerative Medicine (CIRM).
Abbreviations
- CTD
C-terminal domain
- HAD
haloacid dehalogenase
- p-NPP
p-nitrophenyl phosphate
- FCP1
TFIIF-interacting CTD phosphatase 1
- SCP
small CTD phosphatase
- TIMM50
translocase of inner mitochondrial membrane 50
- UBLCP
ubiquitin-like domain containing CTD phosphatase
- BMP
bone morphogenetic protein.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702099104/DC1.
References
- 1.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg J, Huang HB, Kwon YG, Greengard P, Nairn AC, Kuriyan J. Nature. 1995;376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 3.Allen KN, Dunaway-Mariano D. Trends Biochem Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Rebay I, Silver SJ, Tootle TL. Trends Genet. 2005;21:163–171. doi: 10.1016/j.tig.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Gohla A, Birkenfeld J, Bokoch GM. Nat Cell Biol. 2005;7:21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- 6.Glasner ME, Gerlt JA, Babbitt PC. Curr Opin Chem Biol. 2006;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, Gill GN, Dixon JE, Noel JP. Mol Cell. 2006;24:759–770. doi: 10.1016/j.molcel.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin PS, Dahmus ME. Methods Enzymol. 2003;370:155–165. doi: 10.1016/S0076-6879(03)70013-5. [DOI] [PubMed] [Google Scholar]
- 9.Varon R, Gooding R, Steglich C, Marns L, Tang H, Angelicheva D, Yong KK, Ambrugger P, Reinhold A, Morar B, et al. Nat Genet. 2003;35:185–189. doi: 10.1038/ng1243. [DOI] [PubMed] [Google Scholar]
- 10.Yeo M, Lin PS, Dahmus ME, Gill GN. J Biol Chem. 2003;278:26078–26085. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- 11.Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- 12.Su YA, Lee MM, Hutter CM, Meltzer PS. Oncogene. 1997;15:1289–1294. doi: 10.1038/sj.onc.1201294. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa S, Kai M, Tamari M, Takei Y, Takeuchi K, Bandou H, Yamane Y, Ogawa M, Nakamura Y. DNA Res. 1997;4:35–43. doi: 10.1093/dnares/4.1.35. [DOI] [PubMed] [Google Scholar]
- 14.Zheng H, Ji C, Gu S, Shi B, Wang J, Xie Y, Mao Y. Biochem Biophys Res Commun. 2005;331:1401–1407. doi: 10.1016/j.bbrc.2005.04.065. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto H, Esaki M, Kanamori T, Tamura Y, Nishikawa S, Endo T. Cell. 2002;111:519–528. doi: 10.1016/s0092-8674(02)01053-x. [DOI] [PubMed] [Google Scholar]
- 16.Meinecke M, Wagner R, Kovermann P, Guiard B, Mick DU, Hutu DP, Voos W, Truscott KN, Chacinska A, Pfanner N, Rehling P. Science. 2006;312:1523–1526. doi: 10.1126/science.1127628. [DOI] [PubMed] [Google Scholar]
- 17.Satow R, Chan TC, Asashima M. Biochem Biophys Res Commun. 2002;295:85–91. doi: 10.1016/S0006-291X(02)00641-1. [DOI] [PubMed] [Google Scholar]
- 18.Satow R, Kurisaki A, Chan TC, Hamazaki TS, Asashima M. Dev Cell. 2006;11:763–774. doi: 10.1016/j.devcel.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Siniossoglou S, Hurt EC, Pelham HR. J Biol Chem. 2000;275:19352–19360. doi: 10.1074/jbc.M001314200. [DOI] [PubMed] [Google Scholar]
- 20.Kaida D, Yashiroda H, Toh-e A, Kikuchi Y. Genes Cells. 2002;7:543–552. doi: 10.1046/j.1365-2443.2002.00538.x. [DOI] [PubMed] [Google Scholar]
- 21.Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. EMBO J. 1998;17:6449–6464. doi: 10.1093/emboj/17.22.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han GS, Wu WI, Carman GM. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris TE, Huffman TA, Chi A, Shabanowitz J, Hunt DF, Kumar A, Lawrence JC., Jr J Biol Chem. 2007;282:277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- 25.Carman GM, Han GS. Trends Biochem Sci. 2006;31:694–699. doi: 10.1016/j.tibs.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Hara L, Han GS, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S. J Biol Chem. 2006;281:34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schirmer EC, Florens L, Guan T, Yates JR, III, Gerace L. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 28.Kamenski T, Heilmeier S, Meinhart A, Cramer P. Mol Cell. 2004;15:399–407. doi: 10.1016/j.molcel.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 29.Cho H, Wang W, Kim R, Yokota H, Damo S, Kim SH, Wemmer D, Kustu S, Yan D. Proc Natl Acad Sci USA. 2001;98:8525–8530. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Cheong N, Zhang Z, De Rose R, Deng Y, Farber SA, Fernandes-Alnemri T, Alnemri ES. J Biol Chem. 2004;279:24813–24825. doi: 10.1074/jbc.M402049200. [DOI] [PubMed] [Google Scholar]
- 31.Peterfy M, Phan J, Xu P, Reue K. Nat Genet. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 32.Fagone P, Sriburi R, Ward-Chapman C, Frank M, Wang J, Gunter C, Brewer JW, Jackowski S. J Biol Chem. 2007;282:7591–7605. doi: 10.1074/jbc.M608175200. [DOI] [PubMed] [Google Scholar]
- 33.Peterfy M, Phan J, Reue K. J Biol Chem. 2005;280:32883–32889. doi: 10.1074/jbc.M503885200. [DOI] [PubMed] [Google Scholar]
- 34.Huffman TA, Mothe-Satney I, Lawrence JC., Jr Proc Natl Acad Sci USA. 2002;99:1047–1052. doi: 10.1073/pnas.022634399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Copp J, Wiley S, Ward MW, van der Geer P. Am J Physiol. 2005;288:C403–C415. doi: 10.1152/ajpcell.00095.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.