Abstract
Recognition of lysine-type peptidoglycan by peptidoglycan recognition protein (PGRP)-SA provokes the activation of the Toll and prophenoloxidase pathways. Here we reveal that a soluble fragment of lysine-type peptidoglycan, a long glycan chain with short stem peptides, is a potent activator of the Drosophila Toll pathway and the prophenoloxidase activation cascade in the beetle Tenebrio molitor. Using this peptidoglycan fragment, we present biochemical evidence that clustering of PGRP-SA molecules on the peptidoglycan is required for the activation of the prophenoloxidase cascade. We subsequently highlight that the lysozyme-mediated partial digestion of highly cross-linked lysine-type peptidoglycan dramatically increases the binding of PGRP-SA, presumably by inducing clustering of PGRP-SA, which then recruits the Gram-negative bacteria-binding protein 1 homologue and a modular serine protease containing low-density lipoprotein and complement control protein domains. The crucial role of lysozyme in the prophenoloxidase activation cascade is further confirmed in vivo by using a lysozyme inhibitor. Taken together, we propose a model whereby lysozyme presents a processed form of lysine-type peptidoglycan for clustering of PGRP-SA that recruits Gram-negative bacteria-binding protein 1 and the modular serine protease, which leads to the activation of both the Toll and prophenoloxidase pathways.
Keywords: innate immunity, pattern, prophenoloxidase, Toll
Insects rely entirely on innate immunity for defense against microbial pathogens. These defenses include both the inducible expression of antimicrobial peptides and the activation of prophenoloxidase cascades in hemolymph (insect blood) (1, 2). For example, Lys-type peptidoglycan (PG) from Gram-positive bacteria is recognized by the PG recognition protein (PGRP)-SA or PGRP-SD and thereby activates the Toll signaling pathway and the transcription of antimicrobial peptide genes (3, 4). This recognition signal is amplified in hemolymph by a proteolytic cascade similar to the vertebrate complement system. Drosophila genetic studies have shown that the Gram-negative bacteria-binding protein 1 (GNBP1), together with PGRP-SA, is also required to activate the Toll pathway in response to Gram-positive bacterial infection (5, 6). Diaminopimelic acid-type PG, usually found in Gram-negative bacteria, induces innate immune responses in Drosophila by activating the immune deficiency pathway (7, 8). A naturally occurring monomeric fragment of diaminopimelic acid-type PG, known as tracheal cytotoxin, specifically activates the immune deficiency pathway by inducing heterodimerization of its recognition receptors, PGRP-LCa and PGRP-LCx (9, 10). In sharp contrast, the Toll signaling pathway is not induced by muropeptide, a monomeric fragment of Lys-type PG composed of N-acetylglucosamine and N-acetylmuramic acid linked with a short peptide chain as a stem (11), although this muropeptide is the minimum binding unit for PGRP-SA (12–15).
The prophenoloxidase activation cascade, which leads to melanization of invading microbes, is another major innate immune defense mechanism in invertebrates that is triggered by PG and fungal β-1,3-glucan in the hemolymph (16, 17). We previously identified the Tenebrio PGRP-SA in the beetle Tenebrio molitor that exhibited the highest sequence homology with Drosophila PGRP-SA (18). We also reported that Tenebrio PGRP-SA is essential for the PG-dependent prophenoloxidase pathway activation and that a synthetic muropeptide dimer, linked by a β-1,4-glycosidic bond between the sugars, functions as a competitive inhibitor of soluble polymeric Lys-type PG in the activation of the prophenoloxidase system (18). However, it has not been known whether GNBP1 or its homologue is involved in the PG-dependent prophenoloxidase activation system.
Although insects encode several lysozymes that digest PG, these enzymes are likely to be involved in the immune response, but their roles have not yet been clearly delineated (19). Lysozyme present in the hemolymph may function directly as an antibacterial, or it could be involved in the processing and/or elimination of PG to modulate the immune response (20). However, highly cross-linked or modified Lys-type PG would be resistant to a complete digestion by lysozyme, resulting in a partially digested PG in bacteria (21). Here we focus on the biochemical and immunological properties of this partially digested PG. With multiple approaches, using the in vivo Drosophila Toll pathway, the in vitro prophenoloxidase activation system, and recombinant PGRP-SA proteins, we show that Lys-type PG triggers PGRP-SA clusters leading to the activation of the Toll and prophenoloxidase pathways by recruiting GNBP1 and a unique modular serine protease.
Results
A Linearized Lys-Type PG Activates both the Toll and Prophenoloxidase Pathways.
Achromobacter β-lytic protease is a lysostaphin-like enzyme that hydrolyzes the peptide bonds in the penta-Gly bridge present in Staphylococcus aureus PG (22). Using β-lytic protease, we solubilized S. aureus Lys-type PG by cleaving the penta-Gly bridge between the stem peptides, generating a linearized PG containing a long glycan chain with stem peptides (Fig. 1A Upper). Because this linearized PG contains many copies of muropeptide units, it should have multiple binding sites for PGRP-SA. Using the linearized PG, we confirmed that both the recombinant Drosophila PGRP-SA and Tenebrio PGRP-SA proteins bind to the linearized PG using size-exclusion chromatography [supporting information (SI) Fig. 7 B and C]. However, a mixture of Tenebrio PGRP-SA and the synthetic muropeptide dimer linked between the sugars (Fig. 1A Lower), which was previously used (18), showed the same elution profile as Tenebrio PGRP-SA alone, suggesting that the synthetic muropeptide dimer can bind to only one molecule of PGRP-SA (SI Fig. 7D).
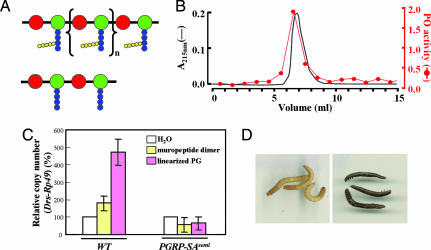
Fig. 1.
Linearized PG activates the Drosophila Toll pathway and induces Tenebrio melanin syntheses in vivo. (A) Expected structures of the linearized PG (Upper) and synthetic muropeptide dimer (Lower). Red, green, blue, and yellow balls indicate N-acetyl-glucosamine, N-acetylmuramic acid, stem peptides, and Gly residues of S. aureus PG, respectively. (B) The UV absorbance profile (black line) of the linearized PG fractionated on a Toyopearl HW-55S size-exclusion column and the phenoloxidase activity (red line) of each linearized PG fraction were plotted simultaneously. (C) Induction of the drosomycin (Drs)-Rp49 reporter gene after the injection of water (white), synthetic muropeptide dimer (yellow), or the linearized PG (purple) into wild-type female adult flies and PGRP-SAseml mutant flies. Drs expression was measured in four flies collected 18 h after challenge and normalized to the value obtained after injection of water (set at 100%). Bars represent the mean ± SD of four independent experiments. (D) One hundred nanograms of the synthetic muropeptide dimer (Left) or the linearized PG (Right) was injected into Tenebrio larvae. Within 18 h, appearance of melanin pigment was examined.
Consistent with these binding activities, the linearized PG, but not the synthetic muropeptide dimer, displayed potent immunostimulatory activities. To address these points, we used two innate immune responses: the well characterized in vivo Drosophila Toll pathway to examine antibacterial peptide expression and the Tenebrio prophenoloxidase cascade system in the large beetle to perform biochemical studies in vitro. We found that the linearized PG consistently induced strong phenoloxidase activity when the fractionated linearized PG was added to Tenebrio hemolymph solution (Fig. 1B), indicating that linearized PG can activate PG-dependent prophenoloxidase cascade. Subsequently, we injected the linearized PG into wild-type and PGRP-SAseml mutant flies and then monitored expression of the drosomycin-encoding gene to test whether the linearized PG can activate the Drosophila Toll pathway in vivo (Fig. 1C). Indeed, the linearized PG-injected wild-type flies induced drosomycin expression normally, but PGRP-SAseml mutant flies were defective in the induction of the antimicrobial peptide, demonstrating that the linearized PG activates the Toll pathway in a PGRP-SA-dependent manner. In contrast, the synthetic muropeptide dimer induced only very weak drosomycin expression in the wild-type flies whereas the PGRP-SAseml mutant flies were unresponsive (Fig. 1C). Likewise, the linearized PG, but not the synthetic muropeptide dimer, strongly induced melanin synthesis when injected into the larvae of Tenebrio, most likely by activation of the prophenoloxidase system (Fig. 1D). These results suggest that the PG fragment containing multiple binding sites for PGRP-SA can induce the Toll and prophenoloxidase pathways. Recently, Ligoxygakis and his colleagues (11) suggested that PG should be processed to increase the number of reducing ends to activate the Toll signaling pathway. Because both the linearized PG and the synthetic muropeptide dimer contain one reducing end each but only the linearized PG is able to induce activation of both Toll and prophenoloxidase cascades, this may suggest that the reducing ends may not be important.
Clustering of PGRP-SA Is Needed for the Activation of the Prophe-noloxidase Cascade.
We initially attempted to determine the minimal concentration of the linearized PG for activation of the PG-dependent prophenoloxidase cascade. The phenoloxidase activity was measured by incubation of hemolymph with different amounts of the linearized PG. Unexpectedly, the phenoloxidase activity was severely inhibited to the baseline level at high concentrations of the linearized PG, showing a classic bell-shaped dose–response curve (Fig. 2A). Moreover, the concentration of the linearized PG where the maximum phenoloxidase activity was produced could be significantly shifted with a stronger phenoloxidase activity if exogenous Tenebrio PGRP-SA protein was added to the reaction mixture (Fig. 2A). This shift in optimal concentration of linearized PG by addition of Tenebrio PGRP-SA protein suggests that the molar ratio between PGRP-SA and PG is important in the activation of prophenoloxidase cascade. These observations imply that too much linearized PG acts as a competitive inhibitor by sequestering PGRP-SA molecules, impairing the initial activating complex composed of clustered PGRP-SA molecules bound to one linearized PG molecule (see Fig. 2A Insets). Similar observations were reported in β-1,3-glucan recognition of the horseshoe crab factor G and in LPS recognition of the crayfish prophenoloxidase system (23, 24).
Fig. 2.
Clustered Tenebrio PGRP-SA is required for prophenoloxidase activation. (A) Lys-type PG-dependent phenoloxidase activity was measured with 10 nM Tenebrio PGRP-SA (0.2 μg·ml−1) and different amounts of the linearized PG (squares). The bell-shaped dose–response curve was shifted to the right by the addition of Tenebrio PGRP-SA to 120 nM (2.5 μg·ml−1), and a maximal point was observed at 100 ng of the linearized PG (circles). Insets indicate the putative initial complexed structures between Tenebrio PGRP-SA and the linearized PG. R and PG indicate Tenebrio PGRP-SA and linearized Lys-type PG, respectively. (B) In vitro reconstitution experiments were performed by using Tenebrio PGRP-SA or Drosophila PGRP-SA with the Tenebrio PGRP-SA-deficient hemolymph solution in the presence of the linearized PG (columns 2 and 3, respectively). Tenebrio PGRP-SA and Drosophila PGRP-SA were coincubated in the presence of the linearized PG (column 4).
We further investigated the initial activation step for Lys-type PG recognition using recombinant Drosophila PGRP-SA that is able to bind the linearized PG with a similar affinity as Tenebrio PGRP-SA (SI Fig. 7C), but the Drosophila PGRP-SA cannot induce activation of the Tenebrio prophenoloxidase cascade (Fig. 2B, column 3). The phenoloxidase activity induced by the linearized PG was severely inhibited by the addition of Drosophila PGRP-SA to the hemolymph solution even in the presence of exogenous Tenebrio PGRP-SA (Fig. 2B, column 4). This implies that the initial activating complex in the prophenoloxidase cascade is abolished by replacement of a part of Tenebrio PGRP-SA molecules by Drosophila PGRP-SA on the linearized PG. These results strongly suggest again that clustering of Tenebrio PGRP-SA on Lys-type PG is required for the initial activation of prophenoloxidase cascade.
Lys-Type PG Fragments That Accommodate at Least Two Tenebrio PGRP-SA Molecules Activate the Prophenoloxidase System.
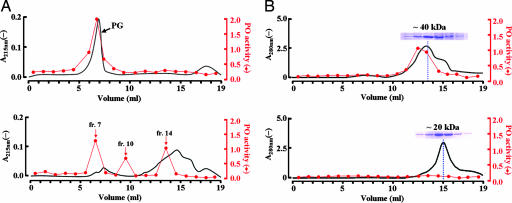
To determine how many molecules of Tenebrio PGRP-SA constitute the initial activating complex for the prophenoloxidase system, the various lengths of sugar chains of PG were generated by partial digestion of the linearized PG with lysozyme and were then fractionated according to their length on a size-exclusion column (Fig. 3A Upper and Lower). Three fractions (the 7th, 10th, and 14th fractions) showed the phenoloxidase activity when each fraction was incubated with the Tenebrio PGRP-SA-depleted hemolymph solution in the presence of the Tenebrio PGRP-SA protein and Ca2+. Of these, among the PG fragments in fraction 14 should be a smallest unit that is able to induce activation of the prophenoloxidase cascade. After adding an excess amount of Tenebrio PGRP-SA protein, we analyzed how many PGRP-SA molecules can bind to the PG fragment in fraction 14 by monitoring the apparent molecular mass on a size-exclusion column (Fig. 3B Upper). The apparent molecular mass of the complex between the PG fragments in the fraction and Tenebrio PGRP-SA was determined as ≈40 kDa, which indicates that the PG fragment binds to two molecules of PGRP-SA. When the PG fragment/Tenebrio PGRP-SA complex, isolated from this second sizing column, was incubated with the Tenebrio PGRP-SA-depleted hemolymph solution, it induced phenoloxidase activity even without adding Tenebrio PGRP-SA, clearly demonstrating that two molecules of PGRP-SA are sufficient to induce this activity (red line in Fig. 3B Upper). However, muropeptide monomer that was generated by a prolonged incubation of the linearized PG with lysozyme did not change the molecular mass of Tenebrio PGRP-SA protein on the size-exclusion column and did not activate the prophenoloxidase pathway (Fig. 3B Lower). The synthetic muropeptide dimer also showed the same pattern (data not shown). This observation indicates that the synthetic muropeptide dimer and the muropeptide monomer can bind to one PGRP-SA molecule without inducing phenoloxidase activity as shown in our previous study (18). It is noteworthy that the synthetic muropeptide dimer binds to only one PGRP-SA molecule although it contains two copies of muropeptide. We propose that this is a result of steric hindrance by the first bound PGRP-SA molecule on the muropeptide dimer because the two binding units on the synthetic muropeptide dimer are located too close in this dimeric molecule. However, another muropeptide dimer that is cross-linked by a penta-Gly bridge might be expected to accommodate two PGRP-SA molecules because the penta-Gly bridge provides sufficient space for binding of two PGRP-SA molecules. Consistently, the synthetic muropeptide dimer linked between the sugars was unable to activate the Toll and prophenoloxidase pathways as shown above, whereas the muropeptide dimer cross-linked by a penta-Gly bridge was reported to induce activation of the Toll pathway (11). Thus, it can be concluded that the PG fragment that accommodates two PGRP-SA molecules is the minimum unit that could induce the downstream events and result in activation of the Toll and prophenoloxidase pathways.
Fig. 3.
PG fragments that accommodate two Tenebrio PGRP-SA molecules activate the prophenoloxidase system. (A Upper) The linearized PG only. (A Lower) Partially digested linearized PG by lysozyme was fractionated by a Toyopearl HW-55S column equilibrated with 20 mM Tris·HCl (pH 8.0) containing 150 mM NaCl. The phenoloxidase activity profile of each fraction is shown in red. (B Upper) A mixture of Tenebrio PGRP-SA with fraction 14 was injected onto the same column. (B Lower) A mixture of Tenebrio PGRP-SA and the fully digested PG that was fully digested with lysozyme by incubation for 16 h at 37°C was injected onto the same column.
Lysozyme Presents a Processed Form of PG for PG Recognition Signals.
Most natural Gram-positive bacterial Lys-type PG is highly cross-linked between the glycan chains, which is different from that of the linearized PG. We anticipated that PGRP-SA might have limited access to natural Lys-type PGs because of the highly cross-linked structure of PG. Moreover, we previously observed that insoluble Lys-type PGs that were disrupted by sonication induced a strong phenoloxidase activity in vitro, whereas intact insoluble Lys-type PGs did not induce prophenoloxidase activation at a given time (18). To loosen the PG structure by an enzyme present in insect hemolymph, we chose lysozyme because it is able to hydrolyze almost all types of intact bacterial PG (25). We performed partial digestion of Lys-type PG from both S. aureus and Micrococcus luteus with lysozyme in vitro. Indeed, the partially digested Lys-type PGs induced a rapid and strong phenoloxidase activity in the Tenebrio hemolymph in vitro (data not shown). Moreover, when the partially digested insoluble Lys-type PGs were injected into Tenebrio larvae, stronger and faster melanin synthesis was observed in all of the injected larvae compared with intact insoluble Lys-type PGs (SI Fig. 8 B and C, respectively). However, when an inhibitor of lysozyme, N,N′,N″-triacetylchitotriose (26), was coinjected with the intact Lys-type PG, no melanin synthesis could be observed (SI Fig. 8D). In a control experiment, coinjection of lysozyme inhibitor and partially digested insoluble Lys-type PG induced melanin synthesis (SI Fig. 8E). These results strongly suggest that prior partial degradation of Lys-type PG by lysozyme is necessary for activation of the prophenoloxidase cascade.
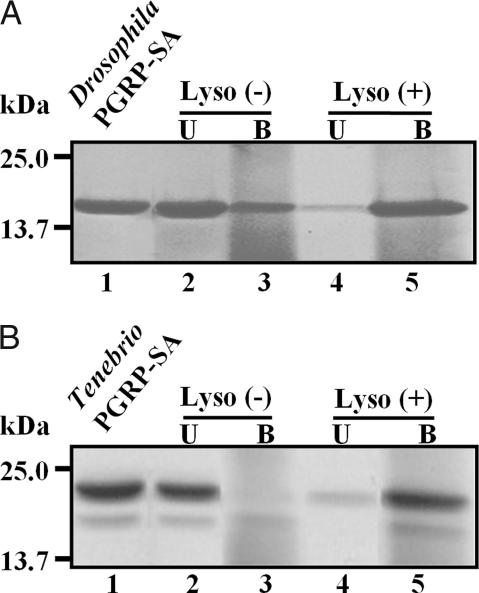
To ascertain the role of lysozyme in the recognition of Lys-type PG by PGRP-SAs in vitro, we examined the binding abilities of PGRP-SAs to the partially digested Lys-type PGs using Drosophila PGRP-SA and Tenebrio PGRP-SA. To our surprise, the partial digestion of Lys-type PG by lysozyme dramatically increased binding of both Drosophila PGRP-SA and Tenebrio PGRP-SA to PG (lane 5 in Fig. 4 A and B, respectively). The enhanced interaction between PGRP-SAs and PG should result in clustering of PGRP-SAs in PG, leading to the activation of the Toll and prophenoloxidase pathways. Our study presents the in vitro biochemical evidence that lysozyme plays a crucial role in enhancing the access of Drosophila PGRP-SA or Tenebrio PGRP-SA to insoluble Lys-type PG in the Toll and prophenoloxidase pathways, although the possibility that other proteins showing lysozyme-like activity process PGs for PGRP-SA binding in vivo cannot be excluded.
Fig. 4.
The ability of PGRP-SAs to bind to partially digested insoluble PG. (A) Ability of Drosophila PGRP-SA to bind to the partially digested insoluble Lys-type PG. Lyso (−) and Lyso (+) indicate intact PG without and with lysozyme (Lyso) treatment, respectively. Lane 1, Drosophila PGRP-SA only; lanes 2 and 4, the amounts of unbound (U) Drosophila PGRP-SA when Lyso (−) or Lyso (+) PG was incubated with Drosophila PGRP-SA, respectively; lanes 3 and 5, the amounts of bound (B) Drosophila PGRP-SA on Lys (−) or Lys (+) PG, respectively. (B) Binding ability of Tenebrio PGRP-SA to intact or the partially digested insoluble PG was examined. The assignment of each lane is the same as in A.
During the preparation of this article, a report appeared showing that Drosophila GNBP1 has lysozyme-like activity that hydrolyzes loosely cross-linked M. luteus Lys-type PG, but not highly cross-linked S. aureus Lys-type PG (27). Because the authors found that GNBP1 has the enzymatic activity, they proposed that Drosophila GNBP1 presents a processed form of PG for sensing by Drosophila PGRP-SA. Considering the limited lysozyme-like activity of GNBP1 and the fact that endogenous lysozymes in the insect hemolymph is active on highly cross-linked PGs, GNBP1 may have less importance for processing PG than the hemolymph lysozyme. However, the limited lysozyme-like activity of GNBP1 may play an important role in amplifying or scavenging the recognition signal.
PGRP-SA/PG Complex Recruits GNBP1 and Modular Serine Protease.
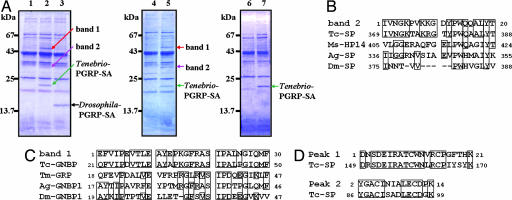
We next sought to identify the immediate downstream effector(s) that recognizes the clustered PGRP-SA molecules on partially digested Lys-type PGs. To this end, recombinant Tenebrio PGRP-SA was incubated with the partially digested S. aureus and M. luteus PGs and then added to the Tenebrio PGRP-SA-deficient hemolymph solution. Indeed, a 50-kDa protein (band 1 in Fig. 5A) and a 35-kDa protein (band 2 in Fig. 5A) were specifically enriched on the Tenebrio PGRP-SA-bound, partially digested Lys-type PGs (lanes 2 and 5 in Fig. 5A), but not on the Drosophila PGRP-SA/PG complexes (lane 3 in Fig. 5A), when analyzed by SDS/PAGE. It is notable that the Drosophila PGRP-SA bound to the partially digested PG did not interact with the two Tenebrio proteins (lane 3 in Fig. 5A). Also, Tenebrio PGRP-SA bound to synthetic muropeptide dimer coupled to Sepharose resin failed to recruit the proteins (lane 7 in Fig. 5A) under the same conditions, demonstrating that the two proteins are recruited as a result of clustered PGRP-SA molecules on Lys-type PG. We identified the two enriched proteins by using N-terminal amino acid sequencing (see SI Text). The 50-kDa protein (band 1 in Fig. 5A) was identified as a Tenebrio GNBP1, and the 35-kDa protein (band 2 in Fig. 5A) is a Tenebrio modular serine protease, but not containing a clip domain that is commonly found in the upstream proteases of the Toll and prophenoloxidase cascades (28) (Fig. 5 B–D). GNBP1 was reported to physically interact with PGRP-SA for the activation of the Toll pathway in Drosophila (5), but any strong binding of GNBP1 to PGRP-SA in vitro has not previously been observed. Our observation supports that clustered binding of PGRP-SA molecules on PG enhances the interaction of PGRP-SA with GNBP1 and that the GNBP1 homologue may also be involved in the prophenoloxidase pathway. Tenebrio modular serine protease contains low-density lipoprotein receptor A repeat domains, one complement control protein domain, and a serine protease domain. Manduca sexta hemolymph protease 14 (Ms-HP-14), containing a domain arrangement similar to that of the Tenebrio modular serine protease, was recently reported as an initiation enzyme of the prophenoloxidase activation system in M. sexta that binds curdlan, zymosan, and yeast and also interacts with PG (29, 30). In this study we present evidence that the modular serine protease is recruited to an initial activation complex consisting of the GNBP1 homologue, PGRP-SA, and PG.
Fig. 5.
Tenebrio GNBP1 protein and modular serine protease are recruited to Tenebrio PGRP-SA-bound partially digested PG. (A) Proteins were extracted from the intact insoluble PG (lane 1), Tenebrio PGRP-SA-bound PG (lane 2), or Drosophila PGRP-SA-bound PG (lane 3) after incubation with Tenebrio PGRP-SA-deficient hemolymph solution and then analyzed by SDS/PAGE. M. luteus insoluble PG (lanes 4 and 5) and the synthetic muropeptide dimer-coupled resin (lanes 6 and 7) without and with Tenebrio PGRP-SA were treated as described in A. It is notable that the Drosophila PGRP-SA bound to the partially digested PG did not interact with the two Tenebrio proteins (lane 3). (B) N-terminal amino acid sequence comparison between band 2 and Tribolium castaneum serine protease (Tc-SP, XP_967486), M. sexta hemolymph protease 14 (Ms-HP14) (30), A. gambiae serine protease (Ag-SP, XP_321263), and D. melanogaster modular serine protease (Dm-SP, CG31217). (C) Comparison of the N-terminal sequences of band 1 and T. castaneum GNBP-like protein (Tc-GNBP, XP_969449), T. molitor glucan recognition protein (32) (Tm-GRP), Anopheles gambiae GNBP1 (Ag-GNBP1, AAR13751), and Drosophila melanogaster GNBP1 (Dm-GNBP1) (33). Boxes indicate residues identical to those in the sequence of band 1. (D) Sequence identities between two internal sequences (Peak 1 and Peak 2) of band 2 and low-density lipoprotein receptor A repeat domain sequence of Tc-SP.
Discussion
Gram-negative bacteria are innately resistant to lysozyme because the LPS layer of the outer membrane protects the inner PG layer. When Gram-negative bacteria invade insect hemolymph, the immune deficiency pathway is elicited by recognition of monomeric and/or polymeric diaminopimelic acid-type PG fragments that are released during cell growth and division. In contrast, when Gram-positive bacteria invade insect hemolymph, the hemolymph lysozyme is able to digest the exposed multi-PG layer of Gram-positive bacteria. If the PG layer of the bacteria is completely degraded by lysozyme, leading to lysis of the bacteria, the bacteria are not harmful to the insect anymore. Consistently, the monomeric end products of PG by lysozyme do not induce the immune responses of insects, although it is bound to its receptor molecule, PGRP-SA (Fig. 6B′). However, several Gram-positive bacteria produce a highly modified or cross-linked PG that is not completely digested by lysozyme (25), and they could survive in the insect hemolymph without other innate defense systems (Fig. 6C). Here we found that the partially digested PG with lysozyme generates a stronger and faster phenoloxidase activity in vivo and in vitro and recruits a larger number of PGRP-SA molecules in vitro compared with the intact PG. We have shown that clustered PGRP-SA plays a critical role in inducing the Toll and the prophenoloxidase pathways in vivo and/or in vitro comparing a linearized PG fragment to the natural or synthetic dimeric muropeptide. Finally, we found that the Lys-type PG/PGRP-SA complex recruits GNBP1 and modular serine protease in the beetle system. We propose that this Lys-type PG/PGRP-SA/GNBP1/modular serine protease complex is the initial activator that triggers serine protease cascades in the Toll and prophenoloxidase pathways, where lysozyme presents a processed form of PG for binding of PGRP-SA, GNBP1, and modular serine protease (Fig. 6). In conclusion, the present work provides biochemical data about how the Lys-type PG recognition signal is transferred downstream.
Fig. 6.
A model summarizing the molecular events in the initiation of the Toll and prophenoloxidase pathways. Although a few PGRP-SA (R) molecules bind to intact PG, it is not able to activate the immune responses (A). PG of Gram-positive bacteria is digested partially (B) or completely (B′) by lysozyme. Whereas the partially digested PG recruits more PGRP-SA molecules binding to the bacterial surface (B), the fully digested PG cannot recruit PGRP-SA on the bacterial surface leading to lysis of the bacterial cell (B′). The clustered PGRP-SA molecules recruit GNBP1 and a modular serine protease (SP) containing low-density lipoprotein receptor A repeat domains (LDL), resulting in the activation of the modular serine protease (C). Then the activated SP triggers the proteolytic cascade leading to activation of the Toll and prophenoloxidase (proPO) pathways that produce antimicrobial peptide (AMP) and melanin around the invading bacteria.
Materials and Methods
Details of some materials and methods are available in the SI Text.
Fly Stock and drosomycin Expression.
OregonR flies were used as the wild-type strain. PGRP-SAseml is a line carrying the semmelweis mutation (Cys54Tyr) in Drosophila PGRP-SA (3). Ten nanoliters of water, the linearized PG (10 mg·ml−1), or synthetic muropeptide dimer (10 mg·ml−1) was injected into the thorax of the wild-type or PGRP-SAseml female adults (3–4 days old) by using a Nanoject apparatus (Drummond, Broomall, PA). After injection, the flies were then incubated for 18 h at 25°C. drosomycin expression level was measured as previously described (31).
Binding Assay of PGRP-SAs to Partially Digested Insoluble PGs.
The binding assay was performed according to the previously reported method (18). Briefly, 10 μg of the purified Tenebrio PGRP-SA or Drosophila PGRP-SA was mixed with 40 μl of a 50% (vol/vol) suspension of the partially digested S. aureus or M. luteus PG (500 μg) in 50 mM Tris·HCl (pH 7.0) for 12 h at 4°C in a shaker. Unbound PGRP-SA proteins in the supernatant and bound PGRP-SAs in the pellet fraction were analyzed by SDS/PAGE.
Identification of Tenebrio GNBP1 and Associated Modular Serine Protease.
For this experiment, we prepared a Tenebrio PGRP-SA-deficient hemolymph solution according to the previously reported method by using the synthetic muropeptide dimer-coupled affinity column (18). This solution contains all of the essential components, except for Tenebrio PGRP-SA, necessary for the activation of the prophenoloxidase system by Lys-type PG. Tenebrio PGRP-SA (40 μg) was incubated with partially digested insoluble Lys-type PG (8 mg) in 200 μl of PBS for 12 h at 4°C. After incubation, Tenebrio PGRP-SA-bound insoluble Lys-type PG was recovered by centrifugation at 20,000 × g for 10 min at 4°C, washed three times first with 20 mM Tris·HCl (pH 8.0) and next with 50 mM Tris·HCl (pH 6.0). The recovered Tenebrio PGRP-SA-bound Lys-type PG was incubated with 2.5 ml of the Tenebrio PGRP-SA-deficient hemolymph solution (20 mg of total protein) for 3 h at 4°C. After removing the insoluble residue by centrifugation, the mixture was washed twice with 50 mM Tris·HCl (pH 6.0). The bound proteins on the insoluble Lys-type PG were extracted with 100 μl of 2× SDS/PAGE loading buffer and then separated by SDS/PAGE. The protein bands on the polyacrylamide gel were transferred onto a PVDF membrane, and then the N-terminal amino acid sequences of the 50-kDa (Tenebrio GNBP1) and 35-kDa (Tenebrio modular serine protease) proteins were determined on an automatic gas-phase amino acid sequencer (Applied Biosystems, Foster City, CA).
Supplementary Material
Acknowledgments
We thank Y. Fujimoto and K. Fukase (Osaka University, Osaka, Japan) for providing a synthetic muropeptide dimer and Dr. Neal Silverman for critical reading of the manuscript. This work was supported by the programs of the National Research Laboratory (M10400000028-04J0000-02), the Bio-discovery Research, and the Creative Research Initiatives (Center for Biomolecular Recognition) of the Ministry of Science Technology/Korea Science and Engineering Foundation. This work was also partially supported by the BK21 program and a Pusan National University postdoctor fellow grant (J.-W.P., 2006).
Abbreviations
- PG
peptidoglycan
- PGRP
PG recognition protein
- GNBP1
Gram-negative bacteria-binding protein 1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610924104/DC1.
References
- 1.Royet J, Reichhart JM, Hoffmann JA. Curr Opin Immunol. 2005;17:11–17. doi: 10.1016/j.coi.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Söderhäll K, Cerenius L. Curr Opin Immunol. 1998;10:23–28. doi: 10.1016/s0952-7915(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 3.Michel T, Reichhart JM, Hoffmann JA, Royet J. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 4.Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J. Nat Immunol. 2004;5:1175–1180. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- 5.Gobert V, Gottar M, Matskevich AA, Rutschmann S, Royet J, Belvin M, Hoffmann JA, Ferrandon D. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- 6.Pili-Floury S, Leulier F, Takahashi K, Saigo K, Samain E, Ueda R, Lemaitre B. J Biol Chem. 2004;279:12848–12853. doi: 10.1074/jbc.M313324200. [DOI] [PubMed] [Google Scholar]
- 7.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 8.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 9.Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J. Science. 2006;311:1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- 10.Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh BH. J Biol Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- 11.Filipe SR, Tomasz A, Ligoxygakis P. EMBO Rep. 2005;6:327–333. doi: 10.1038/sj.embor.7400371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang CI, Ihara K, Chelliah Y, Mengin-Lecreulx D, Wakatsuki S, Deisenhofer J. Proc Natl Acad Sci USA. 2005;102:10279–10284. doi: 10.1073/pnas.0504547102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan R, Roychowdhury A, Ember B, Kumar S, Boons GJ, Mariuzza RA. Proc Natl Acad Sci USA. 2004;101:17168–17173. doi: 10.1073/pnas.0407856101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim MS, Byun M, Oh BH. Nat Immunol. 2003;4:787–793. doi: 10.1038/ni952. [DOI] [PubMed] [Google Scholar]
- 15.Chang CI, Pili-Floury S, Herve M, Parquet C, Chelliah Y, Lemaitre B, Mengin-Lecreulx D, Deisenhofer J. PLoS Biol. 2004;2:E277. doi: 10.1371/journal.pbio.0020277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerenius L, Söderhäll K. Immunol Rev. 2004;198:116–126. doi: 10.1111/j.0105-2896.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 17.Kanost MR, Jiang H, Yu XQ. Immunol Rev. 2004;198:97–105. doi: 10.1111/j.0105-2896.2004.0121.x. [DOI] [PubMed] [Google Scholar]
- 18.Park JW, Je BR, Piao S, Inamura S, Fujimoto Y, Fukase K, Kusumoto S, Söderhäll K, Ha NC, Lee BL. J Biol Chem. 2006;281:7747–7755. doi: 10.1074/jbc.M510058200. [DOI] [PubMed] [Google Scholar]
- 19.Daffre S, Kylsten P, Samakovlis C, Hultmark D. Mol Gen Genet. 1994;242:152–162. doi: 10.1007/BF00391008. [DOI] [PubMed] [Google Scholar]
- 20.Gorman MJ, Kankanala P, Kanost MR. Insect Mol Biol. 2004;13:19–24. doi: 10.1111/j.1365-2583.2004.00454.x. [DOI] [PubMed] [Google Scholar]
- 21.Masschalck B, Michiels CW. Crit Rev Microbiol. 2003;29:191–214. doi: 10.1080/713610448. [DOI] [PubMed] [Google Scholar]
- 22.Li S, Norioka S, Sakiyama F. J Biochem (Tokyo) 1998;124:332–339. doi: 10.1093/oxfordjournals.jbchem.a022116. [DOI] [PubMed] [Google Scholar]
- 23.Muta T, Seki N, Takaki Y, Hashimoto R, Oda T, Iwanaga A, Tokunaga F, Iwanaga S. J Biol Chem. 1995;270:892–897. [PubMed] [Google Scholar]
- 24.Söderhäll K, Unestam T. Can J Microbiol. 1979;25:406–414. doi: 10.1139/m79-062. [DOI] [PubMed] [Google Scholar]
- 25.Shockman GD, Daneo-Moore L, Kariyama R, Massidda O. Microb Drug Resist. 1996;2:95–98. doi: 10.1089/mdr.1996.2.95. [DOI] [PubMed] [Google Scholar]
- 26.Fukamizo T, Ikeda Y, Ohkawa T, Goto S. Eur J Biochem. 1992;210:351–357. doi: 10.1111/j.1432-1033.1992.tb17428.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Weber AN, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. EMBO J. 2006;25:5005–5014. doi: 10.1038/sj.emboj.7601363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piao S, Song YL, Kim JH, Park SY, Park JW, Lee BL, Oh BH, Ha NC. EMBO J. 2005;24:4404–4414. doi: 10.1038/sj.emboj.7600891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji C, Wang Y, Guo X, Hartson S, Jiang H. J Biol Chem. 2004;279:34101–34106. doi: 10.1074/jbc.M404584200. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Jiang H. J Biol Chem. 2006;281:9271–9278. doi: 10.1074/jbc.M513797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Mengin-Lecreulx D, Lemaitre B. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 32.Zhang R, Cho HY, Kim HS, Ma YG, Osaki T, Kawabata S, Söderhäll K, Lee BL. J Biol Chem. 2003;278:42072–42079. doi: 10.1074/jbc.M307475200. [DOI] [PubMed] [Google Scholar]
- 33.Kim YS, Ryu JH, Han SJ, Choi KH, Nam KB, Jang IH, Lemaitre B, Brey PT, Lee WJ. J Biol Chem. 2000;275:32721–32727. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.