Abstract
Loss of function of the tumor suppressor protein BRCA1 is responsible for a high percentage of familial and also sporadic breast cancers. Early work identified a stimulatory transcriptional coactivator function for the BRCA1 protein, and more recently, BRCA1 has been implicated in transcriptional repression, although few examples of repressed genes have been characterized. We recently used an in vitro transcription assay to identify a biochemical mechanism that explained the BRCA1 stimulatory activity. In this study, we identified an ubiquitin-dependent mechanism by which BRCA1 inhibits transcription. BRCA1 ubiquitinates the transcriptional preinitiation complex, preventing stable association of TFIIE and TFIIH, and thus blocks the initiation of mRNA synthesis. What is striking about this mechanism of regulation by BRCA1 is that the ubiquitination of the preinitiation complex is not targeting proteins for degradation by the proteasome, nor are ubiquitin receptors modifying the activity, but rather the ubiquitin moiety itself interferes with the assembly of basal transcription factors at the promoter. Using RNAi to knockdown expression of the endogenous BRCA1 protein, we assessed the level of repression dependent on BRCA1 in the cell, and we found that BRCA1 is at least as significant a transcriptional repressor as it is an activator. These results define a biochemical mechanism by which the BRCA1 enzymatic activity regulates a key cellular process.
Keywords: RNA polymerase II, TFIIE, transcription
BRCA1 is the breast and ovarian cancer specific tumor suppressor (1). Loss of BRCA1 can occur either by mutation of both alleles of the gene in the tumor cell (≈4% of all breast cancer cases) or by epigenetic down-regulation of the gene by methylation of its promoter (≈14% of sporadic breast cancer cases and up to 30% of ovarian cancer cases) (2, 3).
How BRCA1 protein exerts its tumor suppressor function remains unresolved, but it has been found to regulate a number of processes including transcription, repair of DNA damage, cell cycle checkpoints, and centrosome dynamics (4–6). The biochemical mechanism(s) by which BRCA1 regulates these diverse processes is unknown. The BRCA1 protein has the enzymatic activity of an E3 ubiquitin ligase when bound as a heterodimer to BARD1 (7, 8), and it is likely that the ubiquitin ligase activity is critical for BRCA1/BARD1 regulation of transcription and other processes. In this paper, the BRCA1/BARD1 heterodimer will be simply referred to as “BRCA1.”
Previously, we found that BRCA1 strongly stimulated transcription by stabilizing the preinitiation complex (PIC) on the core promoter (9). This activity was observed in either the presence or absence of BARD1 and was independent of ubiquitination function. Our results suggested that BRCA1 enhanced the stability of the PIC on promoter elements relative to bulk DNA (9). Because BRCA1 can ubiquitinate phosphorylated RNA polymerase II (RNAPII) both in vitro and in vivo (10, 11), we wondered whether the E3 ubiquitin ligase activity of BRCA1 might alter its stimulatory effect on transcription. We find in these experiments that the E3 ubiquitin ligase activity of BRCA1 strongly inhibits transcription by blocking PIC assembly.
Results
Ubiquitin-Dependent Repression of Transcription.
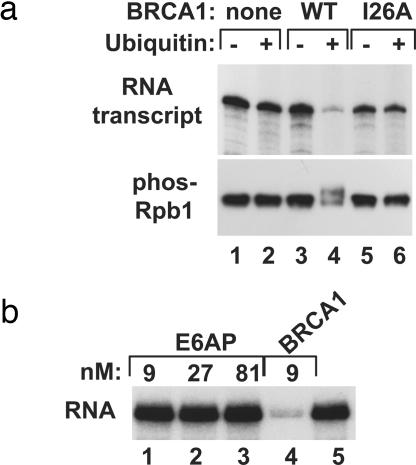
We tested the effects of BRCA1 E3 ubiquitin ligase activity in transcription reactions containing purified transcription and ubiquitination factors [TATA binding factor (TBP), TFIIB, RNAPII, TFIIF, TFIIE, TFIIH, E1, and E2 (UbcH5c)]. In the absence of the BRCA1/BARD1 heterodimer (BRCA1), the addition of ubiquitin had a negligible effect on RNA synthesis, and no ubiquitination of RNAPII was observed. However, when BRCA1 was included in the reaction, addition of ubiquitin repressed transcription nearly completely (Fig. 1a Upper, lanes 3 and 4). Transcriptional repression correlated with ubiquitination of the phosphorylated large subunit of RNAPII (Rpb1; Fig. 1a Lower, lanes 3 and 4). Importantly, BRCA1 exerts the repressive effect directly through its E3 activity. A mutant protein, BRCA1(I26A), which cannot bind the E2 enzyme (12), failed to ubiquitinate phospho-Rpb1 and did not repress transcription (lanes 5 and 6). These results link the enzymatic activity of BRCA1 to a previously unrecognized form of transcriptional repression.
Fig. 1.
The BRCA1 ubiquitin ligase ubiquitinates phosphorylated RNAPII and represses transcription. (a) Purified transcription/ubiquitination reactions containing TBP, TFIIB, RNAPII, TFIIF, TFIIE, and TFIIH and the ubiquitination factors E1 and E2 were assembled without BRCA1 (lanes 1 and 2), with full-length BRCA1 (lanes 3 and 4), or with the mutant BRCA1 protein (I26A) that is defective as an E3 ubiquitin ligase (lanes 5 and 6). Ubiquitin was included in reactions analyzed in lanes 2, 4, and 6. RNA products were analyzed (Upper). Immunoblots were probed with antibody against phosphorylated Rpb1 (Lower). (b) Transcription/ubiquitination reactions were assembled containing all factors except the E3 ubiquitin ligase, and RNA transcripts were analyzed. E6AP (lanes 1–3), BRCA1 (lane 4), or no E3 ubiquitin ligase (lane 5) were included in reactions at the indicated concentrations.
The transcriptional repression depends on the inclusion in reactions of each ubiquitination factor. In otherwise complete reactions, we omitted one ubiquitination factor per reaction, and we found that repression of transcription required all of the factors [supporting information (SI) Fig. 6], and the level of repression correlated with BRCA1 concentration (SI Fig. 7).
Repression of Transcription Is Specific to BRCA1.
The E3 ubiquitin ligase activity of BRCA1 exerted a strong repressive effect on RNA synthesis. Although TFIIH is another RING domain class of E3 ubiquitin ligase present in the transcription reaction (13), repression in this assay depends on the inclusion of BRCA1. To test further whether repression in this assay might be a general property of ubiquitin ligases, we assayed the E6AP E3 ubiquitin ligase for inhibition of transcription. E6AP is a HECT domain E3 that can use the same E2 enzyme (UbcH5c) in vitro as BRCA1 (14–16). Polyubiquitin chain formation assays confirmed that our preparation of E6AP was functional, and the activity observed in this nonspecific assay was similar to that of BRCA1 on a molar basis (data not shown). Unlike BRCA1, E6AP addition had no effect on transcription, even when added at 9-fold molar excess relative to BRCA1 (Fig. 1b). This result indicates that repression of transcription is specific to BRCA1 and not an effect of E3 ligases in general.
Repression of Transcription by BRCA1 Occurs During Initiation.
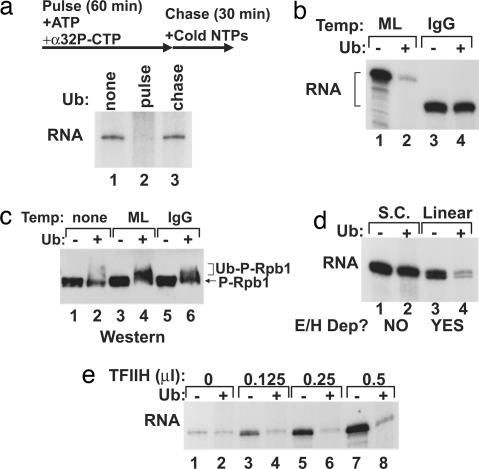
We tested whether the repressive activity of BRCA1 affects a specific stage of transcription by use of a pulse–chase approach. Transcription/ubiquitination reactions containing all factors but ubiquitin were assembled without UTP. In the absence of UTP, transcription initiates but stalls within four nucleotides, producing nascent, radiolabeled transcripts. Transcriptional elongation continues when a cold chase mix of complete nucleotides is added to the reaction (Fig. 2a). New initiations that occur during the chase phase are unlabeled, and thus only the transcripts initiated during the pulse phase are detectable. When ubiquitin was omitted from the reaction, normal RNA synthesis occurred, demonstrating that the pulse–chase design does not inhibit transcription (Fig. 2a, lane 1). If ubiquitin was added before the pulse (initiation), complete repression of transcription resulted (Fig. 2a, lane 2). By contrast, if ubiquitin was added before the chase (elongation), no repression occurred (Fig. 2a, lane 3). Therefore, BRCA1 must act during the initiation phase to repress transcription. This is consistent with our prior observation that BRCA1 preferentially associates with and ubiquitinates the hyperphosphorylated form of Rpb1 associated with transcriptional initiation (10).
Fig. 2.
BRCA1 E3 ubiquitin ligase represses initiation of transcription by blocking TFIIE and TFIIH function. (a) A pulse–chase protocol was used to separate the initiation and elongation phases. PICs in reactions containing the basal transcription factors, E1, E2, and BRCA1, were assembled during the pulse (UTP omitted) corresponding to the initiation phase. The addition of complete cold nucleotides in the chase allows elongation to proceed. Ubiquitin was omitted (lane 1) or added to otherwise complete transcription/ubiquitination reactions before the pulse (lane 2) or the chase (lane 3). RNA transcripts from each reaction are shown. (b) A comparison of the effect of the BRCA1 E3 ubiquitin ligase on transcription reactions from the adenoviral ML (lanes 1 and 2) and the IgG (lanes 3 and 4) promoters. Reactions were complete (basal transcription factors, E1, E2, and BRCA1) with the exception of ubiquitin, which was added to even-numbered lanes. RNA transcripts are shown. (c) In this immunoblot probed for phosphorylated Rpb1, the ubiquitination state of Rpb1 in the transcription/ubiquitination reaction was determined in the absence of template (lanes 1 and 2), with the ML template (lanes 3 and 4), and with the IgG template (lanes 5 and 6). Ubiquitin was added in even-numbered lanes. (d) BRCA1 E3 ubiquitin ligase repression of transcription was compared from negatively supercoiled (S.C.; lanes 1 and 2) and linear (lanes 3 and 4) IgG template. Ubiquitin was included in even-numbered lanes, and RNA transcripts are shown. Whether active transcription was dependent on the inclusion of TFIIE and TFIIH (E/H Dep?) is indicated for supercoiled and linear templates (17). (e) TFIIH was titrated into otherwise complete transcription/ubiquitination reactions: no TFIIH added (lanes 1 and 2), 0.125 μl (lanes 3 and 4), 0.25 μl (lanes 5 and 6), and 0.5 μl (lanes 7 and 8). Ubiquitin was included in even-numbered lanes, and resulting RNA transcripts are shown. Temp, template.
Promoter Specificity of Transcriptional Repression.
We tested several promoters in the BRCA1 repression assay. Similar to transcription from the adenoviral major late (ML) promoter, the adenoviral E4 promoter was repressed by BRCA1 ubiquitination activity (data not shown). In stark contrast, transcription from the IgG promoter was immune to repression by BRCA1 and ubiquitination factors (Fig. 2b). The ubiquitination of phospho-RNAPII was stimulated by the presence of a template (Fig. 2c, compare lane 2 to lanes 4 and 6), suggesting that PIC formation was important for BRCA1-dependent ubiquitination. Surprisingly, even though ubiquitination of RNAPII did not inhibit transcription from the IgG promoter, the phospho-RNAPII was ubiquitinated to a similar extent as with the ML promoter (Fig. 2c).
Because Rpb1 was ubiquitinated to a similar extent in transcription reactions from both the ML and IgG template, the resistance of the IgG promoter to transcriptional repression by BRCA1 must involve another factor. Repression by the BRCA1 ubiquitin ligase occurs during the initiation phase of the transcription reaction, so we examined initiation factor requirements. Transcription from most promoters used in the in vitro assay is highly stimulated by TFIIE and TFIIH. This reflects the requirement for promoter melting during the initiation phase. The IgG promoter, however, is active in the absence of TFIIE and TFIIH when the template is negatively supercoiled. When the same template is linearized, the negative superhelical tension is released, and TFIIE and TFIIH are then required for active transcription initiation (17). Although transcription from the supercoiled IgG template was resistant to repression, BRCA1 repressed transcription from a linear form of this plasmid (Fig. 2d). This finding indicates that ubiquitination of the transcription reaction by BRCA1 interferes with TFIIE and TFIIH function. We further tested this possibility by titrating TFIIH into transcription reactions with supercoiled ML plasmid template (Fig. 2e). Transcription from this template is weak in the absence of TFIIE and TFIIH but strongly stimulated by the addition of these basal factors. When TFIIH was omitted, transcription was minimal and the BRCA1 E3 ubiquitin ligase activity had no effect. Titration of TFIIH into the transcription reaction stimulated RNA synthesis in a dose-dependent manner in reactions lacking ubiquitin (Fig. 2e, odd lanes) but had no effect on reactions in which ubiquitin was included (even lanes). We conclude that the E3 ligase activity of BRCA1 blocks TFIIE and TFIIH function.
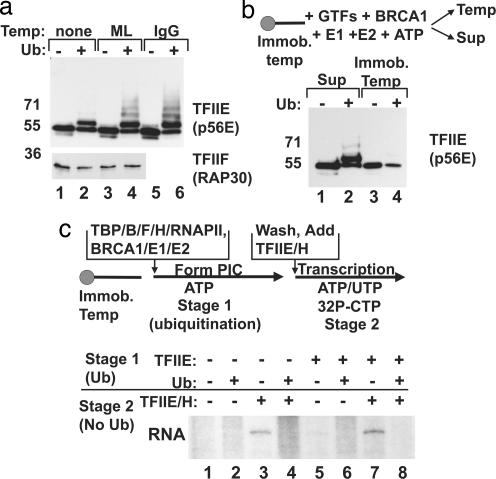
Although repression of transcription by BRCA1 is correlated with ubiquitination of phospho-Rpb1, the observation that TFIIE and TFIIH are the functional targets of the repression led us to investigate whether additional factors might be ubiquitinated. Western blot analysis of transcription reactions was limited by whether an available antibody could detect a given antigen at the concentration used in transcription reactions. Several basal factors were analyzed. We found that that the large subunit of TFIIE (p56E) is ubiquitinated by BRCA1 (Fig. 3a). In transcription reactions lacking template, more than one-half of the p56E was unmodified, and only monoubiquitination was apparent (Fig. 3a, lanes 1 and 2). The addition of either the ML or IgG template significantly enhanced ubiquitination levels, with multiubiquitination apparent (Fig. 3a, lanes 3–6). Thus, assembly of the transcription complex promotes ubiquitination of p56E, most likely by incorporating it into the PIC with BRCA1. Other factors, such as the RAP30 subunit of TFIIF (Fig. 3a, lower blot), were not targeted for ubiquitination, indicating that ubiquitination does not occur on all basal factors. Antibodies specific for TFIIH were insufficiently sensitive to determine whether any of the subunits of this factor were ubiquitinated in this reaction (data not shown).
Fig. 3.
BRCA1 ubiquitination of the PIC regulates the association of TFIIE. (a Upper) A Western blot of transcription reactions containing no template (lanes 1 and 2), ML template (lanes 3 and 4), and IgG template (lanes 5 and 6) was probed with TFIIE (p56E) antibody. (a Lower) A Western blot of transcription reactions containing no template (lanes 1 and 2) or ML template (lanes 3 and 4) was probed with TFIIF (RAP30)-specific antibody. (b) The fractionation of TFIIE (p56E) between the template (Temp) and supernatant (Sup) was analyzed by using an immobilized template (Immob. temp). PICs were assembled on bead-bound ML templates in the presence of the general transcription factors, BRCA1, E1, and E2 enzymes, and ATP. Reactions in lanes 2 and 4 also contained ubiquitin. The supernatant (lanes 1 and 2) was separated from the washed template (lanes 3 and 4), and the proteins were analyzed on protein gels. Western blots were probed with TFIIE (p56E) antibody. (c) A staged transcription protocol was used to test whether ubiquitination of a PIC lacking TFIIE could repress transcription. In Stage 1, transcription/ubiquitination reactions were assembled on an immobilized template in the presence of ATP only, permitting formation of the PIC and ubiquitination. The template was washed and incubated with complete nucleotides in Stage 2 plus TFIIE and TFIIH as indicated. Ubiquitin was included in even-numbered reactions in Stage 1. TFIIE was omitted from the Stage 1 reaction (lanes 1–4) or included (lanes 5–8). TFIIE and TFIIH were added back during Stage 2 (lanes 3, 4, 7, and 8), and transcription products were analyzed.
Ubiquitination of RNAPII Blocks TFIIE Association with the PIC.
During transcriptional initiation, TFIIE binds RNAPII and recruits TFIIH to the PIC (18). The finding that both Rpb1 and p56E are ubiquitinated in the transcription reaction led us to speculate that this modification might interfere with the stable association of TFIIE and RNAPII. Therefore we tested whether TFIIE binding to the transcription PIC was affected by BRCA1 E3 ligase activity. PICs were assembled on linear ML templates immobilized on agarose beads. In these reactions, ATP was the only nucleotide included and thus there could be no transcriptional elongation, but the RNAPII was phosphorylated, and ubiquitination was active. After incubation, the supernatant was collected, and the template was washed. The fractionation of p56E between the supernatant and template was monitored by Western blot analysis (Fig. 3b). The addition of ubiquitin to the PIC led to the appearance of ubiquitinated p56E in the supernatant (Fig. 3b, lane 2). In the bound fraction, the addition of ubiquitin reduced the amount of p56E associated with the template, and the remaining template-bound TFIIE was primarily unmodified (Fig. 3b, lane 4). Thus, ubiquitination of the PIC by BRCA1 interferes with TFIIE association, and this is likely the mechanism by which TFIIE/TFIIH function is blocked.
The dissociation of TFIIE from the transcription complex could occur because of ubiquitination of Rpb1, of p56E, or by some combination of the two. To distinguish the contribution of these modifications to transcriptional repression, we used a staged transcription assay with the immobilized template. In the first stage, PICs were assembled on linear template DNA with only ATP present. Ubiquitination factors were included in this reaction mixture. In the second stage, the templates were washed to remove all unbound factors, including the soluble ubiquitination factors. The templates were then incubated in a new transcription mix containing the complete nucleotide mixture so that elongation could occur (Fig. 3c). The important feature of this experimental design is that ubiquitination is only possible during Stage 1 (supporting controls are shown in SI Fig. 8). When TFIIE was omitted from the reaction entirely, no RNA synthesis occurred (Fig. 3c, lanes 1 and 2). Addition of TFIIE in Stage 2 rescued transcription (Fig. 3c, lane 3) but not if ubiquitin was present in Stage 1 (Fig. 3c, lane 4). Therefore, addition of unmodified TFIIE to ubiquitinated PICs cannot rescue transcription. The results were the same if TFIIE was added in both stages of transcription (Fig. 3c, lanes 5–8). We propose that the E3 ligase activity of BRCA1 targets a region adjacent to the binding interface of TFIIE and Rpb1, and thus both proteins are ubiquitinated. However, from a functional standpoint, our experiments suggest that RNAPII is the key ubiquitination substrate blocking initiation. In addition, the ubiquitination of phospho-Rpb1 in in vivo and in vitro systems, both in the absence of and also stimulated by DNA damage, has been well documented (10, 19, 20). By contrast, we have not detected TFIIE ubiquitination in cells (data not shown), suggesting that phospho-RNAPII ubiquitination is the critical modification for the regulation of transcription by the BRCA1 E3 ubiquitin ligase.
Acute Silencing of BRCA1 Reveals a Large Number of Repressed Genes.
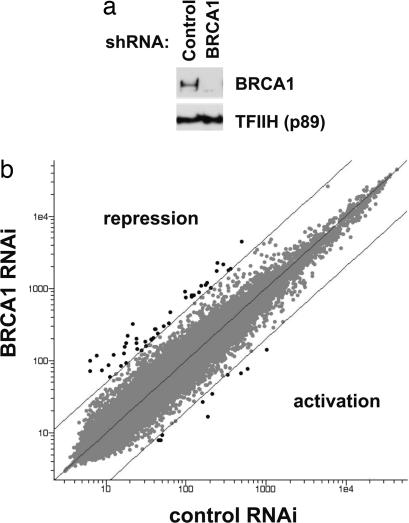
The effects of BRCA1 on gene expression have largely been studied by overexpression of the BRCA1 protein in cells already expressing BRCA1 (for example, refs. 21 and 22). In these studies, exogenous expression of BRCA1 stimulated a large number of genes and repressed few genes. We found that after acutely silencing BRCA1 expression in HeLa cells using RNA interference, loss of BRCA1 resulted in higher expression of a large number of genes, indicating that BRCA1 repressed those targets (Fig. 4). Among the genes altered 2-fold or more, BRCA1 repressed ≈700 genes and stimulated ≈600 genes. Using a more stringent criterion of 5-fold effects, BRCA1 repressed 33 genes and stimulated eight. The effects of BRCA1 suppression on a number of these genes were confirmed by RT-PCR (SI Fig. 9). Although it is possible that many of the repressed genes were indirect targets of depletion of BRCA1, we suggest that the mechanism of ubiquitin-dependent repression of transcription identified in this study is an important component of the function of BRCA1 in the cell.
Fig. 4.
RNAi knockdown of BRCA1 reveals a significant transcriptional repressor function. (a) HeLa cells were transfected with a plasmid expressing shRNA specific for GFP (control) or for BRCA1. Cells were harvested 72 h after transfection, and Western blots were probed with antibodies against BRCA1 (Upper) and the p89 subunit of TFIIH (loading control; Lower). (b) RNA isolated from the same cells (a) was used in microarray analysis. Genes with 5-fold or greater altered expression are highlighted in black. Those above the diagonal marking 5-fold effects were derepressed in the cells with knocked down BRCA1 expression.
Discussion
In this study we found that BRCA1 represses transcription by preventing full assembly of the PIC (Fig. 5). BRCA1, in the absence of ubiquitin, binds to the PIC and even stimulates the level of transcription (9). Recruitment of the charged E2 to this promoter site would result in the ubiquitination of RNAPII, TFIIE, and also BRCA1. We have found that preubiquitinated BRCA1 protein does not disrupt the PIC (A.A.H., unpublished observations). Rather, the ubiquitination of the PIC, probably via phospho-Rpb1, results in the destabilization of TFIIE and TFIIH in the complex and the concomitant inactivation of transcription. The ubiquitinated RNAPII could still function in transcription on synthetic templates such as the IgG promoter under conditions in which TFIIE and TFIIH are not required. This is a previously unrecognized mode of transcriptional repression, whereby ubiquitination sterically blocks protein–protein association. Ubiquitination has been shown to affect a target protein by inducing binding to the proteasome (23, 24) or to other ubiquitin receptors (25). For the reaction described in this study, regulation by ubiquitin requires neither of these pathways but instead regulates the assembly of a multiprotein complex. Repression of transcription was observed here in the absence of proteasome or other ubiquitin receptors.
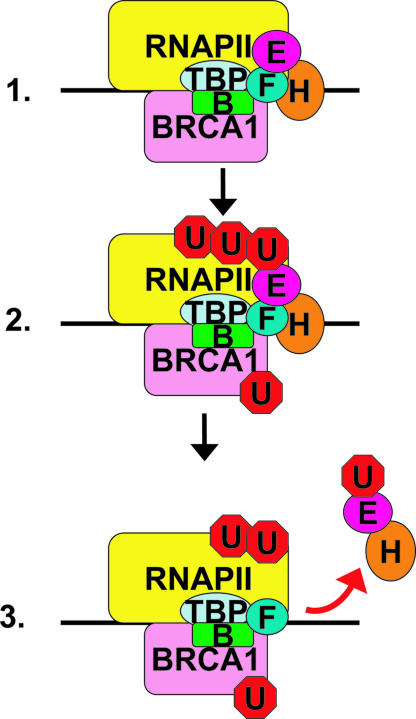
Fig. 5.
Model diagram of BRCA1 repression of transcription: BRCA1 binds initiation competent PIC (1.), BRCA1 ubiquitinates itself, RNAPII, and TFIIE (2.), and TFIIE and TFIIH elute from the ubiquitinated PIC (3.).
Does this mechanism, detected in vitro, operate in the cell? We argue that the answer is yes. Ubiquitinated phospho-RNAPII, although strongly stimulated by DNA damage, has also been detected in the undamaged cell (10, 19), and the overexpression of BRCA1 raises the level of RNAPII ubiquitination independent of DNA damage (10). These observations are consistent with a small percentage of promoters being repressed by BRCA1 ubiquitination of RNAPII. Whether the ubiquitinated RNAPII in the cell is subsequently degraded, because the proteasome would be present in such a setting, is unknown. BRCA1 has also been found to ubiquitinate RNAPII at 3′ processing sites of genes associated with the process of polyadenylation. In this latter case, the RNAPII is targeted for degradation (11). Clearly, BRCA1 interacts with the transcription apparatus, but it is uncertain whether the two identified mechanisms of BRCA1 regulation of transcription are mutually exclusive.
In combination with our previous study (9), these results demonstrate that BRCA1 regulates formation of the PIC, acting as a repressor or activator depending on the context. In the purified transcription assay, we controlled the switch between activation and repression by addition of the ubiquitination factors. This may parallel the situation in the cell. Recent work indicates that the association of BRCA1 with its E2 enzyme is a regulated process (26), and we suggest that this could determine the activity of BRCA1 at a promoter. A second issue raised by our results is promoter specificity. In the in vitro system, protein concentrations are such that BRCA1 interacts with RNAPII directly. In the cell, however, we expect that protein partners of BRCA1 confer gene specificity. Sequence-specific factors, such as ZBRK1, c-Myc, and ERα, all recruit BRCA1 to genes for repression (27–33). In support of the concept that the promoter specificity of BRCA1 repression is due to specific DNA-binding factors, we located putative ZBRK1 binding sites in 19 of the 33 genes most repressed in our microarray study, but no identifiable ZBRK1 binding sites were observed in the genes stimulated by BRCA1 (data not shown). One function of BRCA1 in these repression complexes may be to recruit other repressors, such as CtIP (27), but the results shown herein using a defined transcription assay reveal that BRCA1 also has the capacity to directly regulate basal transcription factor function at the promoter.
How might the transcriptional repression of BRCA1 contribute to its function as a tissue-specific tumor suppressor? Several of the most significantly repressed targets identified in the initial microarray analysis are implicated in breast development or breast cancer (SI Fig. 9). Of particular interest are amphiregulin (AREG) and early growth response-1 (EGR-1). AREG is a ligand for the epidermal growth factor receptor (EGFR) that is essential for postnatal breast development (34) and overexpression of AREG has been noted in primary breast cancers (35). EGR-1 is a transcription factor with a variety of gene targets involved in angiogenesis, including EGFR (36, 37). Interestingly, high EGFR expression in breast cancers is correlated with loss of BRCA1 as well as poor clinical outcomes (38, 39). Based on the examples of repression of AREG and EGR-1 by BRCA1, we propose that transcriptional repression by BRCA1 may contribute to its tissue-specific tumor suppression.
Methods
Transcription Factors.
The transcription factors used in these assays were purified by using established techniques (40–42).
Ubiquitination Factors.
Full-length BRCA1/BARD1 were purified from baculovirus-infected insect cells as described (43). When indicated, the mutant BRCA1 containing isoleucine-26 to alanine substitution was purified by using the same methods as the wild type and was used in place of the wild-type protein. E1 and E2 (UbcH5c) were expressed in bacteria and purified (43). Bovine ubiquitin was purchased from Sigma (St. Louis, MO).
Plasmid Templates.
G-less cassette templates were based on the p(C2AT)19 vector (44) and have been described (45). The linearized IgG template was prepared by digestion with XmnI and subsequent gel purification. The immobilized ML template was prepared by excising the template from its plasmid with HindIII/XmnI digest. The fragment was gel purified, and the 5′ overhang produced by HindIII digestion was filled in with Biotin-14-dATP (Invitrogen, Carlsbad, CA) using the Klenow fragment of DNA Polymerase I. The excised DNA template was immobilized on streptavidin M-280 beads according to the manufacturer's directions (Dynal, Great Neck, NY).
Transcription/Ubiquitination Assay.
Transcription assays were based on reactions described (9). Reactions contained 20 mM Hepes-NaOH (pH 7.9), 20% glycerol, 1 mM EDTA, 60 mM KCl, 0.1 mM each ATP and UTP, 0.05 mM 3′-O-methyl-GTP, 0.003 mM CTP, 1 mM DTT, 0.15 mg/ml BSA, 2 mM MgCl2, 0.003 mM ZnSO4, 1.2 mg/ml plasmid template (1 nM), 10 μCi (1 Ci = 37 GBq) of [α-32P]CTP (800 Ci/mmol; PerkinElmer, Boston, MA), and transcription factors. Unless otherwise noted, the amounts of each factor used per 25-μl reaction were as follows: 8 ng of yeast TATA box-binding protein (16 nM), 60 ng of TFIIB (60 nM), 100 ng of calf thymus RNAPII, 100 ng of TFIIF (40 nM), 4 ng of TFIIE (1.8 nM), and 0.5 μl of TFIIH fraction. Ubiquitination factors were included in the reaction mixture or added to individual reactions before incubation: BRCA1 (9 nM; 50 ng of BRCA1 and 24 ng of BARD1), 6× His-E1 ubiquitin ligase (40 nM), 0.75 μg of 6× His-UbcH5c (2 μM), and 2 μg of ubiquitin (12 μM).
Reactions were assembled on ice and then incubated at 30°C for 90–120 min. Reactions were terminated by addition of 200 μl of transcription stop mix (7 M urea, 0.5% SDS, 2 mM EDTA, 0.1 M LiCl, 0.35 M NH4OAc), extracted in phenol/chloroform, precipitated in ethanol, and resolved on 6% polyacrylamide gels containing 8.3 M urea. Gels were dried and exposed to film with an intensifying screen. PhosphorImager analysis was performed by using a Molecular Dynamics (Sunnyvale, CA) PhosphorImager and ImageQuant software.
Preparation of cDNA for Microarray and TaqMan Analysis.
HeLa cells were cotransfected (Lipofectamine; Invitrogen) with 5 μg of shRNA expression plasmid (46) and 20 ng of pBabe-puro. BRCA1 shRNA (gccacaggaccccaagaatgag) was targeted to the 3′ untranslated region. The control shRNA was targeted against a mutant GFP construct (gggccatggcacgtacggcaag). Puromycin selection (3 μg/ml) was applied 24 h after transfection, and cells were harvested after 72 h. RNA was prepared with Tri Reagent (Molecular Research, Cincinnati, OH) and further purified over RNeasy columns (Qiagen, Valencia, CA). Microarray analysis was performed on separate samples at the Harvard Biopolymers Facility on the Affymetrix (Santa Clara, CA) HG-U133_plus_2 chip. For TaqMan assays, 10 ng of cDNA template was used in standard reactions.
Supplementary Material
Acknowledgments
We thank Karen Daniels and Matthew Spaits for technical assistance and R. Baer (Columbia University, New York, NY) for E6AP expression plasmid. This work was supported by a predoctoral fellowship from the Department of Defense Breast Cancer Research Program (to A.A.H.) and National Institutes of Health Grant CA90281 (to J.D.P.).
Abbreviations
- RNAPII
RNA polymerase II
- PIC
preinitiation complex
- ML
major late.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610481104/DC1.
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Rahman N, Stratton MR. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 3.Turner N, Tutt A, Ashworth A. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 4.Starita LM, Parvin JD. Curr Opin Cell Biol. 2003;15:345–350. doi: 10.1016/s0955-0674(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 5.Venkitaraman AR. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 6.Parvin JD, Sankaran S. Cell Cycle. 2006;5:1946–1950. doi: 10.4161/cc.5.17.3208. [DOI] [PubMed] [Google Scholar]
- 7.Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 8.Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 9.Horwitz AA, Sankaran S, Parvin JD. J Biol Chem. 2006;281:8317–8320. doi: 10.1074/jbc.C500475200. [DOI] [PubMed] [Google Scholar]
- 10.Starita LM, Horwitz AA, Keogh MC, Ishioka C, Parvin JD, Chiba N. J Biol Chem. 2005;280:24498–24505. doi: 10.1074/jbc.M414020200. [DOI] [PubMed] [Google Scholar]
- 11.Kleiman FE, Wu-Baer F, Fonseca D, Kaneko S, Baer R, Manley JL. Genes Dev. 2005;19:1227–1237. doi: 10.1101/gad.1309505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D, III, Fukuda M, Ohta T, Klevit R. Proc Natl Acad Sci USA. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagi Y, Masuda CA, Chang WH, Komori H, Wang D, Hunter T, Joazeiro CA, Kornberg RD. Mol Cell. 2005;18:237–243. doi: 10.1016/j.molcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Wu-Baer F, Lagrazon K, Yuan W, Baer R. J Biol Chem. 2003;278:34743–34746. doi: 10.1074/jbc.C300249200. [DOI] [PubMed] [Google Scholar]
- 15.Scheffner M, Huibregtse JM, Howley PM. Proc Natl Acad Sci USA. 1994;91:8797–8801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 17.Parvin JD, Sharp PA. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 18.Maxon ME, Goodrich JA, Tjian R. Genes Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 19.Ratner JN, Balasubramanian B, Corden J, Warren SL, Bregman DB. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 20.Lee KB, Wang D, Lippard SJ, Sharp PA. Proc Natl Acad Sci USA. 2002;99:4239–4244. doi: 10.1073/pnas.072068399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD, Haber DA. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 22.MacLachlan TK, Somasundaram K, Sgagias M, Shifman Y, Muschel RJ, Cowan KH, El-Deiry WS. J Biol Chem. 2000;275:2777–2785. doi: 10.1074/jbc.275.4.2777. [DOI] [PubMed] [Google Scholar]
- 23.Coux O, Tanaka K, Goldberg AL. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 24.Voges D, Zwickl P, Baumeister W. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 25.Katzmann DJ, Odorizzi G, Emr SD. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 26.Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. EMBO J. 2006;25:2178–2188. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuta S, Wang JM, Wei S, Jeng YM, Jiang X, Gu B, Chen PL, Lee EY, Lee WH. Cancer Cell. 2006;10:13–24. doi: 10.1016/j.ccr.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Zheng L, Pan H, Li S, Flesken-Nikitin A, Chen PL, Boyer TG, Lee WH. Mol Cell. 2000;6:757–768. doi: 10.1016/s1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]
- 29.Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos MR, Pestell RG, Yuan F, Auborn KJ, Goldberg ID, Rosen EM. Science. 1999;284:1354–1356. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 30.Zheng L, Annab LA, Afshari CA, Lee WH, Boyer TG. Proc Natl Acad Sci USA. 2001;98:9587–9592. doi: 10.1073/pnas.171174298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Zhang H, Kajino K, Greene MI. Oncogene. 1998;17:1939–1948. doi: 10.1038/sj.onc.1202403. [DOI] [PubMed] [Google Scholar]
- 32.Xiong J, Fan S, Meng Q, Schramm L, Wang C, Bouzahza B, Zhou J, Zafonte B, Goldberg ID, Haddad BR, et al. Mol Cell Biol. 2003;23:8668–8690. doi: 10.1128/MCB.23.23.8668-8690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy RD, Gorski JJ, Quinn JE, Stewart GE, James CR, Moore S, Mulligan K, Emberley ED, Lioe TF, Morrison PJ, et al. Cancer Res. 2005;65:10265–10272. doi: 10.1158/0008-5472.CAN-05-1841. [DOI] [PubMed] [Google Scholar]
- 34.Sternlicht MD, Sunnarborg SW, Kouros-Mehr H, Yu Y, Lee DC, Werb Z. Development (Cambridge, UK) 2005;132:3923–3933. doi: 10.1242/dev.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desruisseau S, Palmari J, Giusti C, Romain S, Martin PM, Berthois Y. Int J Cancer. 2004;111:733–740. doi: 10.1002/ijc.20312. [DOI] [PubMed] [Google Scholar]
- 36.Fahmy RG, Dass CR, Sun LQ, Chesterman CN, Khachigian LM. Nat Med. 2003;9:1026–1032. doi: 10.1038/nm905. [DOI] [PubMed] [Google Scholar]
- 37.Nishi H, Nishi KH, Johnson AC. Cancer Res. 2002;62:827–834. [PubMed] [Google Scholar]
- 38.Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, Bishop T, Benitez J, Rivas C, Bignon YJ, et al. Clin Cancer Res. 2005;11:5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 39.Sainsbury JR, Farndon JR, Sherbet GV, Harris AL. Lancet. 1985;1:364–366. doi: 10.1016/s0140-6736(85)91385-6. [DOI] [PubMed] [Google Scholar]
- 40.Haile DT, Parvin JD. J Biol Chem. 1999;274:2113–2117. doi: 10.1074/jbc.274.4.2113. [DOI] [PubMed] [Google Scholar]
- 41.Mondal N, Parvin JD. Nature. 2001;413:435–438. doi: 10.1038/35096590. [DOI] [PubMed] [Google Scholar]
- 42.Schlegel BP, Green VJ, Ladias JA, Parvin JD. Proc Natl Acad Sci USA. 2000;97:3148–3153. doi: 10.1073/pnas.070452397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starita LM, Machida Y, Sankaran S, Elias JE, Griffin K, Schlegel BP, Gygi SP, Parvin JD. Mol Cell Biol. 2004;24:8457–8466. doi: 10.1128/MCB.24.19.8457-8466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawadogo M, Roeder RG. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 45.Parvin JD, Shykind BM, Meyers RE, Kim J, Sharp PA. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 46.Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC. Proc Natl Acad Sci USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.