Abstract
ITK (IL-2-inducible T cell kinase), a Tec family protein tyrosine kinase (PTK), is one of three PTKs required for T cell antigen receptor (TCR)-induced activation of phospholipase C-γ1 (PLC-γ1). Like Src and Abl family PTKs, ITK adopts an inactive, “closed” conformation, and its conversion to the active conformation is not well understood, nor have its direct substrates been identified. In a side-by-side comparison of ITK and ZAP-70 (ζ chain-associated protein kinase of 70 kDa), ITK efficiently phosphorylated Y783 and Y775 of PLC-γ1, two phosphorylation sites that are critical for its activation, whereas ZAP-70 did not. SLP-76 (SH2-domain-containing leukocyte protein of 76 kDa), an adaptor required for TCR-induced activation of PLC-γ1, was required for the phosphorylation of both PLC-γ1 sites in intact cells. Furthermore, this event depended on the N-terminal tyrosines of SLP-76. Likewise, SLP-76, particularly its N-terminal tyrosines, was required for TCR-induced tyrosine phosphorylation and activation of ITK but was not required for the phosphorylation or activation of ZAP-70. Both ZAP-70 and ITK phosphorylated SLP-76 in vitro; thus, both PTKs are potential regulators of SLP-76, but only ITK is regulated by SLP-76. Upon TCR stimulation, a small fraction of ITK bound to SLP-76. This fraction, however, encompassed most of the catalytically active ITK. Catalytic activity was lost upon mild elution of ITK from the SLP-76-nucleated complex but was restored upon reconstitution of the complex. We propose that SLP-76 is required for ITK activation; furthermore, an ongoing physical interaction between SLP-76 and ITK is required to maintain ITK in an active conformation.
Keywords: adaptor protein, kinase regulation, phospholipase C-γ, signal transduction
A key event in antigen receptor signaling pathways is the tyrosine phosphorylation and activation of phospholipase C-γ (PLC-γ), triggering calcium flux and subsequent gene activation. Antigen-receptor-mediated activation of PLC-γ enzymes depends on Src-, Syk-, and Tec-family protein tyrosine kinases (PTKs) (1, 2). In the case of the T cell receptor antigen (TCR) signaling pathway, these roles are filled by Lck, ZAP-70 (ζ chain-associated protein kinase of 70 kDa), and ITK (IL-2-inducible T cell kinase), respectively. In addition, PLC-γ activation depends on the adapter proteins LAT (linker for activation of T cells) and SLP-76 (SH2-domain-containing leukocyte protein of 76 kDa), which are expressed in T cells, mast cells, platelets, and neutrophils; or the SLP-76 analog BLNK (B cell linker protein), which is expressed in B cells. These adaptors are substrates of the Syk family tyrosine kinases and are required to couple these kinases to the phosphorylation and activation of PLC-γ (3, 4). Thus, SLP-76-deficient Jurkat T cells (J14) exhibit severely compromised TCR-induced PLC-γ1 activation, which is restored upon reconstitution with WT SLP-76 (5).
The phosphorylation sites that are necessary and sufficient for TCR-induced activation of PLC-γ1 are Y783 and the recently described Y775 (6). Phosphorylation of the other known sites, Y472, Y771, and Y1254, is dispensable for the activation of the enzyme in this signaling pathway (6). One point that remains unclear is the identity of the PTK that directly phosphorylates PLC-γ1 in the context of the TCR signaling pathway; both ZAP-70 and ITK have been suggested to fulfill this role (2).
SLP-76 lacks intrinsic catalytic activity and acts as a scaffold, recruiting other proteins for correct localization during molecular signal transduction. SLP-76 contains three domains (3, 4): an N-terminal acidic domain, which includes three TCR-inducible tyrosine phosphorylation sites, Y113, Y128, and Y145; a central proline-rich domain; and a C-terminal SH2 domain. The N-terminal phosphorylation sites and central proline-rich domain are required for PLC-γ1 activation (7–9). Through these domains, SLP-76 interacts with a large number of signaling proteins, including Nck, Vav, Gads, and the Tec family kinase ITK (3, 4).
Tec family PTKs have been implicated in antigen-receptor signaling in many hematopoietic cell types. Three members of this family [ITK, RLK (resting lymphocyte kinase), and Tec] are expressed in T cells (10). Among these, ITK is most strongly implicated in the regulation of PLC-γ1, because the tyrosine phosphorylation and activation of PLC-γ1 are diminished in T cells from ITK-deficient mice (11). ITK comprises an N-terminal pleckstrin homology domain, a proline-rich Tec homology domain, an SH3 domain, an SH2 domain, and a C-terminal catalytic domain (10). It also contains two tyrosine phosphorylation sites that are thought to play a role in its activation (12): Y511, residing in the activation loop of the kinase domain; and Y180, residing in the SH3 domain.
In the resting state, ITK folds into an autoinhibited conformation, stabilized by an intramolecular interaction between the SH3 domain and the proline-rich region (13). After TCR stimulation, ITK undergoes a series of tyrosine phosphorylation events and biochemical interactions that result in the release of its kinase domain and its shift from an inactive to an active state. ITK activation depends on Lck (14, 15), ZAP-70 (16), and LAT (16, 17). Lck directly phosphorylates ITK at Y511, whereas ZAP-70 plays an indirect role in ITK activation. Finally, a possible role for ITK in regulating its own activity is shown by its ability to autophosphorylate at Y180 upon T cell stimulation (12).
A number of observations suggest that SLP-76 and ITK act in concert to mediate PLC-γ1 activation. Both are required to mediate TCR-induced activation of PLC-γ1 (5, 11); furthermore, both act downstream of ZAP-70, which is required for tyrosine phosphorylation of SLP-76 and for activation of ITK (16, 18, 19). Finally, ITK has been shown to bind to regions of SLP-76 that are required for PLC-γ1 activation (7, 20). Based on these findings, SLP-76 might act as a passive adaptor protein to facilitate the colocalization of ITK with PLC-γ1 or other substrates; alternatively, SLP-76 might play an active role in regulating the catalytic activity of ITK. Although attractive, this hypothesis has never been directly proven.
In this study, we present evidence that ITK, but not ZAP-70, is capable of phosphorylating PLC-γ1 at the two sites required for the activation of PLC-γ1. We further show that catalytically active ITK is found primarily in complex with SLP-76 and present evidence that SLP-76 constitutes both a positive regulator of ITK and a potential substrate. Finally, we demonstrate that a continued interaction with SLP-76 is required to maintain ITK in an active state.
Results
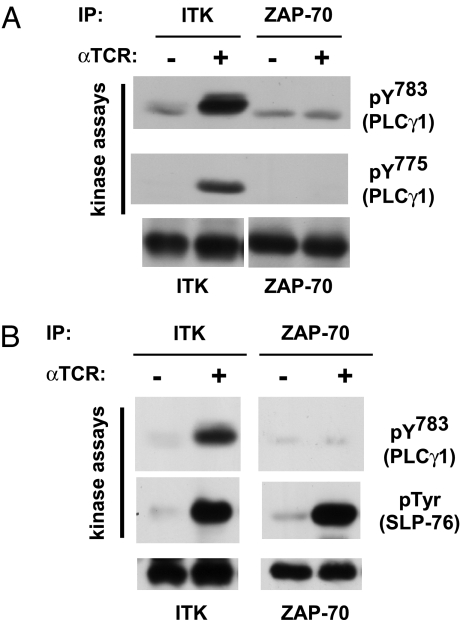
PLC-γ1 phosphorylation and subsequent activation depend on both ITK and ZAP-70 (1, 2). The PLC-γ1 phosphorylation sites required for its activation are Y783 and Y775 (6). To test which kinase most efficiently phosphorylated these sites, we isolated ITK and ZAP-70 by immunoprecipitation from stimulated and unstimulated cells and compared their ability to phosphorylate a recombinant PLC-γ1 fragment in vitro. PLC-γ1 phosphorylation was detected by using phospho-specific antibodies. The specificity of the antisera was confirmed by using an appropriate site-specific, phosphorylated blocking peptide (data not shown). ITK, isolated from TCR-stimulated cells, efficiently phosphorylated the PLC-γ1 fragment at Y783 and at Y775, whereas ZAP-70 did not (Fig. 1A). In contrast, both kinases exhibited similar catalytic activity when recombinant SLP-76 was used as a substrate (Fig. 1B), suggesting that both kinases were isolated in an active conformation but differ in their catalytic specificity. Additional immune-complex kinase assays revealed that Lck, Tec, and Txk/RLK, isolated from TCR-stimulated cells, failed to phosphorylate PLC-γ1 at Y783, suggesting that these kinases cannot phosphorylate PLC-γ1 at this activation site or that they were immunoprecipitated in an inactive conformation (data not shown). Together, these experiments demonstrate that catalytically active ZAP-70 is unable to directly phosphorylate the tyrosines critical for PLC-γ1 activation, whereas ITK efficiently phosphorylates both PLC-γ1 sites.
Fig. 1.
ITK, but not ZAP-70, efficiently phosphorylates both PLC-γ1 activation sites, Y783 and Y775, in vitro. J14 cells stably reconstituted with WT FLAG-tagged SLP-76 were mock-stimulated or stimulated for 1 min with anti-TCR and then lysed as described in Materials and Methods. (A) ITK or ZAP-70 immune complexes were prepared from the lysates of 5 × 106 cells, and their catalytic activity was assayed, using a GST–PLCγ1SH2-SH2-SH3 fusion protein as a substrate. Kinase reaction products (top and middle blots) and immune complex beads (bottom blot) were separated by SDS/PAGE, and phospho-specific antibodies, anti-PLC-γ1 pY783 or anti-PLC-γ1 pY775, were used to detect phosphorylation of the GST–PLC-γ1 fusion protein. The beads were probed by Western blotting with anti-ITK and anti-ZAP-70 antibodies, as indicated. The data are representative of at least two independent experiments. (B) Conditions were the same as in A, except that the kinase substrate was GST–PLCγ1SH2-SH2-SH3 (top blot) or His-tagged SLP-76 (middle blot). Anti-pTyr (4G10) was used to detect phosphorylation of His-tagged SLP-76. The data are representative of at least two independent experiments. IP, immunoprecipitation.
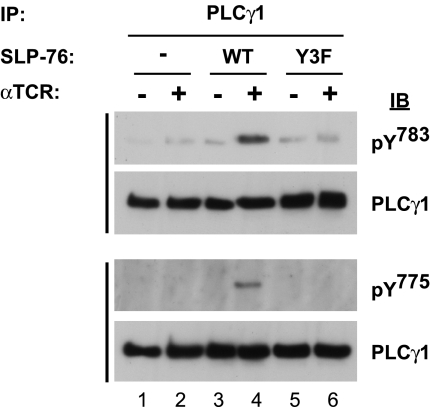
In intact cells, TCR-induced phosphorylation of PLC-γ1 at Y783 depends on SLP-76 (21), but the requirements for Y775 phosphorylation are not known. We addressed this question by using a SLP-76-deficient, Jurkat-derived T cell line (J14) as well as J14 cells that were stably reconstituted with WT or mutant FLAG-tagged SLP-76. SLP-76-expressing cells exhibited TCR-induced phosphorylation of PLC-γ1 at both Y783 and Y775 (Fig. 2, lanes 3 and 4), but neither site was phosphorylated in the absence of SLP-76 nor upon mutation of its N-terminal phosphorylation sites (Fig. 2). Thus, SLP-76 and, in particular, its N-terminal phosphorylation sites, are required to mediate TCR-induced phosphorylation of both PLC-γ1 activation sites in intact cells.
Fig. 2.
SLP-76 and its N-terminal tyrosine phosphorylation sites are required for TCR-induced phosphorylation of PLC-γ1 at Y783 and Y775. J14-derived cell lines stably expressing the indicated FLAG-tagged alleles of SLP-76 were mock-stimulated or stimulated for 1 min with anti-TCR and lysed. Anti-PLCγ1 immune complexes prepared from lysates of 5 × 106 cells were separated by SDS/PAGE and probed by Western blotting with phospho-specific antibodies (anti-PLCγ1 pY783 or anti-PLCγ1 pY775), as indicated. Each blot was then stripped and reprobed with anti-PLCγ1. The results are representative of at least three independent experiments. IB, immunoblot; IP, immunoprecipitation.
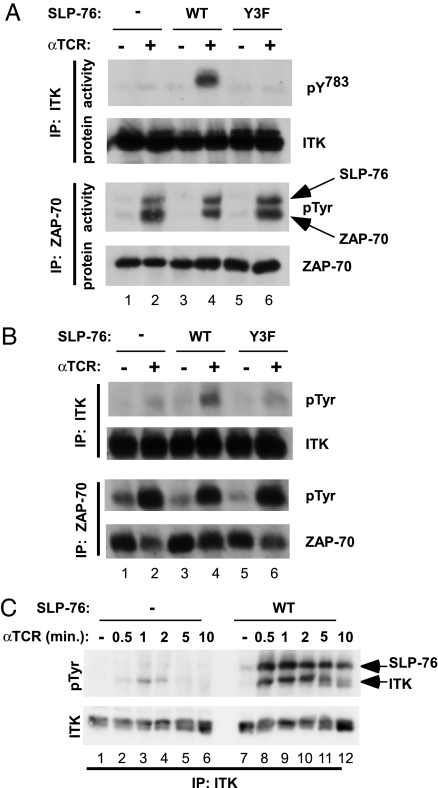
The above results suggested that SLP-76 may regulate PLC-γ1 phosphorylation by regulating the activity of ITK. The immune-complex kinase assay described above reflects an increase in the catalytic activity of ITK and ZAP-70, which occurs upon TCR stimulation. We used this assay to test whether SLP-76 is required for the TCR-induced activation of ITK or ZAP-70. Whereas TCR-induced activation of ZAP-70 was independent of SLP-76, this adaptor was required for the TCR-induced activation of ITK (Fig. 3A, lanes 1–4). Moreover, the N-terminal phosphorylation sites of SLP-76 were required for the TCR-induced activation of ITK but not for activation of ZAP-70 (Fig. 3A, lanes 5 and 6).
Fig. 3.
TCR-induced activation and tyrosine phosphorylation of ITK depend on SLP-76 and the N-terminal tyrosine phosphorylation sites of SLP-76. The indicated J14-derived cell lines were stimulated with anti-TCR for 1 min, and ITK or ZAP-70 was immunoprecipitated from the lysates, as indicated. (A) SLP-76 is required to activate ITK. Immune complexes were assayed for catalytic activity as in Fig. 1B, using GST–PLCγ1SH2-SH2-SH3 as a substrate for ITK (first blot) and His-tagged SLP-76 as a substrate for ZAP-70 (third blot). The electrophoretic mobility of the substrate (SLP-76) and enzyme (ZAP-70) are indicated by arrows on the right. In parallel, the immune complexes were probed for the presence of the relevant kinase (second and fourth blots). A representative result of at least three similar experiments is shown. (B) SLP-76 is required for TCR-induced phosphorylation of ITK. Immune complexes were probed with anti-phosphotyrosine and then stripped and reprobed with anti-ITK or anti-ZAP-70, respectively. (C) Time course of TCR-induced phosphorylation of ITK in the presence and absence of SLP-76. ITK was immunoprecipitated from the lysates of 10 × 106 cells, then probed with anti-phosphotyrosine, and then stripped and reprobed with anti-ITK. The bands corresponding to tyrosine-phosphorylated ITK and coimmunoprecipitating SLP-76 are indicated by arrows. This experiment was repeated three times with similar results. IP, immunoprecipitation.
Activation of tyrosine kinases is accompanied by their phosphorylation on tyrosine residues, raising the possibility that SLP-76 may influence the tyrosine phosphorylation of ITK. At 1 min after TCR stimulation, TCR-induced phosphorylation of ITK was substantially reduced in SLP-76-deficient cells, or upon mutation of the N-terminal tyrosine phosphorylation sites of SLP-76 (Fig. 3B, upper two blots). As a control for cell stimulation, we examined TCR-induced tyrosine phosphorylation of ZAP-70 which, as previously described (5), did not depend on SLP-76 (Fig. 3B, lower two blots). Thus, at 1 min after TCR stimulation, SLP-76 is required for both the tyrosine phosphorylation and activation of ITK.
In contrast, at 10 min after TCR stimulation, TCR-induced phosphorylation of ITK was reported previously to be independent of SLP-76 (5). To further probe this apparent contradiction, we examined the time course of TCR-induced ITK tyrosine phosphorylation in the presence and absence of SLP-76. As previously reported (16), TCR-induced ITK phosphorylation peaked at ≈1 min and was close to baseline levels by 10 min (Fig. 3C, lanes 7–12). Over the entire time course, ITK tyrosine phosphorylation was substantially reduced in SLP-76-deficient cells as compared with SLP-76-reconstituted cells (Fig. 3C). We therefore suggest that the previously reported results reflect the choice of an inappropriate time point for measuring TCR-induced ITK phosphorylation.
Taken together, the above results demonstrate that SLP-76 is required for the tyrosine phosphorylation and activation of ITK, which, in turn, is uniquely capable of phosphorylating PLC-γ1 at Y783 and Y775. Indeed, ITK and SLP-76 have been shown to associate upon TCR stimulation (22), raising the possibility that SLP-76 may activate ITK by directly binding to it and altering its conformation. This model has been previously proposed (20) but never demonstrated. We therefore examined the role that this association may play in the SLP-76-dependent activation of ITK.
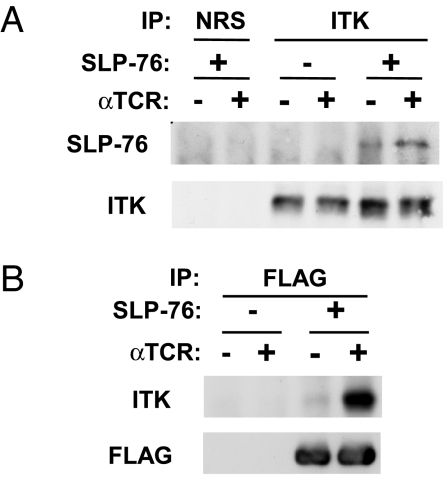
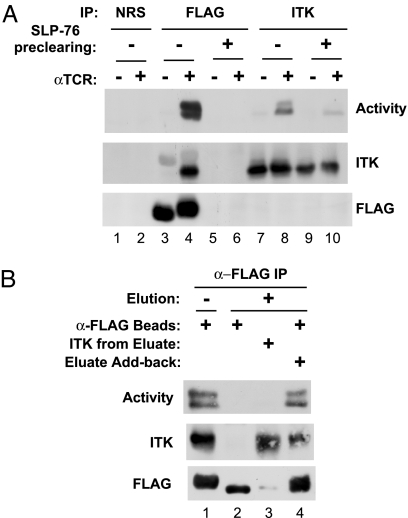
We first confirmed the previously reported TCR-inducible association of ITK with SLP-76 (22). Upon immunoprecipitation of ITK, we detected a coprecipitating, tyrosine-phosphorylated band with electrophoretic mobility identical to that of SLP-76 (Fig. 3C, lanes 8–12). Indeed, when anti-ITK immune complexes were probed with anti-SLP-76, we detected a weak, inducible association of ITK with SLP-76 (Fig. 4A). As a control for specificity, no SLP-76-reactive band was detected in ITK immunoprecipitates from SLP-76-deficient cells or in immunoprecipitates prepared by using preimmune serum (normal rabbit serum) (Fig. 4A). In a reciprocal experiment, we immunoprecipitated FLAG-tagged SLP-76 and clearly detected an inducible association with the ITK protein, an association that was not seen in SLP-76-deficient cells (Fig. 4B). To further substantiate the inducible association of SLP-76 with ITK, we tested whether ITK kinase activity could be detected in association with SLP-76. Indeed, a kinase activity, capable of phosphorylating Y783 of PLC-γ1, was observed in anti-FLAG immunoprecipitates from TCR-stimulated cells but was not found in normal rabbit serum immunoprecipitates (Fig. 5A, lanes 1–4). This activity was abrogated by preclearing the lysates with anti-ITK before the anti-FLAG immunoprecipitation (23); thus, the SLP-76-associated kinase activity can be identified as ITK.
Fig. 4.
TCR-inducible association of SLP-76 and ITK. J14 cells or J14 cells that were stably reconstituted with FLAG-tagged WT SLP-76 were mock-stimulated or stimulated with anti-TCR and then lysed as in Fig. 1. (A) Anti-ITK immune complexes were prepared from the lysates of 6 × 106 cells and sequentially probed with anti-SLP-76 and then with anti-ITK. Normal rabbit serum (NRS) was used to control for nonspecific absorption to the beads. (B) Anti-FLAG immune complexes were prepared from the lysates of 15 × 106 cells and sequentially probed with anti-ITK and then with anti-FLAG. The data are representative of at least three independent experiments. IP, immunoprecipitation.
Fig. 5.
Catalytically active ITK is found in a complex with SLP-76. J14 cells stably reconstituted with FLAG-tagged WT SLP-76 were mock-stimulated or stimulated with anti-TCR and lysed as in Fig. 1. (A) SLP-76 was removed from the lysates by three rounds of immunoprecipitation with anti-FLAG (SLP-76 preclearing), or lysates were untreated (no preclearing). Subsequently, anti-FLAG immune complexes were prepared from the lysates of 15 × 106 cells, or anti-ITK immune complexes were prepared from the lysates of 5 × 106 cells. Normal rabbit serum (NRS) was used to control for nonspecific absorption to the beads (lanes 1 and 2). Immune complexes were assayed for kinase activity, using GST–PLCγ1SH2-SH2-SH3 as a substrate (top blot). After the kinase assay, the immunoprecipitation beads were analyzed separately for the presence of ITK (middle blot) and FLAG-tagged SLP-76 (bottom blot). (B) Anti-FLAG immune complexes prepared from the lysates of 20 × 106 TCR-stimulated cells were left alone (lane 1) or were eluted with kinase reaction buffer supplemented with 0.2 M LiCl and then washed with kinase reaction buffer (lane 2). Eluates were desalted by dialysis against kinase reaction buffer and reimmunoprecipitated with anti-ITK (lane 3) or added back to the eluted anti-FLAG beads (lane 4). All beads were then washed before measuring in vitro kinase activity (top blot). After the reaction, the immunoprecipitation beads were analyzed for the presence of ITK (middle blot) and FLAG-tagged SLP-76 (bottom blot). IP, immunoprecipitation.
We wondered whether catalytically active ITK is found primarily in complex with SLP-76 or in the much larger pool of ITK that is not associated with SLP-76. The specific activity of ITK, defined as its catalytic activity normalized to the amount of ITK protein, was much higher in the anti-FLAG immune complexes, as compared with the activity found in direct anti-ITK immunoprecipitates (Fig. 5A, compare lanes 4 and 8). To test whether catalytically active ITK is found exclusively in association with SLP-76, we removed SLP-76 from the lysates by three rounds of immunoprecipitation with anti-FLAG. After preclearing, no FLAG-tagged SLP-76 was detected in the precleared lysates (Fig. 5A, lanes 5 and 6). This procedure did not substantially reduce the overall level of ITK protein, but the remaining ITK was not catalytically active (Fig. 5A, lanes 7–10). Thus, most of the catalytically active ITK in the cell is found in association with SLP-76.
Because catalytically active ITK is found in association with SLP-76, we wondered whether it might lose its activity upon dissociation from the complex. To address this question, immunoprecipitates of FLAG-tagged SLP-76 prepared from TCR-stimulated cells were eluted with 200 mM LiCl, followed by washing the beads to remove the salt. This mild procedure removed ITK protein and catalytic activity from the anti-FLAG beads, whereas SLP-76 remained bound to the beads (Fig. 5B, lanes 1 and 2). After desalting the eluate by dialysis, ITK was reimmunoprecipitated from the eluate and was found to lack catalytic activity (Fig. 5B, lane 3). Activity was recovered by recombining the desalted eluate with the eluted, washed beads for 90 min at 4°C, followed by washing of the beads. Under these conditions, ITK protein and activity were observed to reassociate with the beads (Fig. 5B, lane 4). Interestingly, SLP-76 underwent a mobility shift, in only those lanes in which active ITK was present, which was consistent with the notion that SLP-76 itself represents a substrate of ITK. Taken together, this experiment provides strong evidence that an ongoing physical interaction with SLP-76 is required to maintain ITK in a catalytically active conformation.
Discussion
The regulation and function of ITK are the subject of much research and speculation. Like Src and Abl family PTKs, ITK folds into an autoinhibited conformation, but the mechanisms that activate the kinase, in the context of physiologic signaling pathways, are not well understood (13, 24, 25). Some have speculated that SLP-76 could activate ITK, by sequestering domains that are required for autoinhibitory interactions. To date, this speculation is based primarily on interactions between recombinant fragments of ITK and SLP-76-derived peptides (20). According to another model, the ITK SH2 domain forms a complex with cyclophilin A, a peptidyl-prolyl cis/trans isomerase, and this interaction inhibits ITK catalytic activity (26). Still others have suggested that an inducible interaction between ITK and tyrosine phosphorylated LAT may play a role in its activation (16, 17).
Of the five known PLC-γ1 phosphorylation sites, Y775 and Y783 are necessary and sufficient for its activation (6). SLP-76 is required for TCR-induced phosphorylation of PLC-γ1 at Y783 (21, 27). We now show that SLP-76, in particular its N-terminal tyrosine phosphorylation sites, also is required for the TCR-induced phosphorylation of the recently identified site Y775. We therefore set out to identify the kinase that directly phosphorylates these two sites and to investigate whether and how it is regulated by SLP-76.
The two kinases that are most directly implicated in TCR-induced PLC-γ1 activation are ITK and ZAP-70. Each kinase may influence PLC-γ1 activation by phosphorylating PLC-γ1 itself or by phosphorylating one or both of the adaptors LAT and SLP-76, which, in turn, are required for PLC-γ1 activation. To better identify their direct targets, we established an in vitro, immune-complex kinase assay, using physiologically relevant, recombinant substrates. Both ITK and ZAP-70 exhibited TCR-inducible activity in this assay system, which was reflected in their ability to phosphorylate recombinant SLP-76 when they were isolated from TCR-stimulated cells. More importantly, this side-by-side comparison revealed a striking difference in the substrate specificity of ITK and ZAP-70. Whereas ITK phosphorylated a recombinant PLC-γ1 fragment at both Y775 and Y783, ZAP-70 did not efficiently phosphorylate either site. Similarly, Bruton's tyrosine kinase, a Tec family PTK expressed in B cells, phosphorylates the corresponding sites on PLC-γ2 more efficiently than Syk does (28, 29). ITK-dependent phosphorylation of PLC-γ1 Y783 has not been conclusively demonstrated in intact cells (27); nonetheless, the above evidence suggests that Tec family kinases are uniquely competent to phosphorylate PLC-γ enzymes at the sites critical for their activation. In intact T cells, phosphorylation of these same sites depends on SLP-76. We now propose an explanation for this correlation by demonstrating that SLP-76 directly regulates the catalytic activity of ITK.
The TCR-inducible tyrosine phosphorylation and activation of ITK were abrogated in SLP-76-deficient T cells, clearly demonstrating that SLP-76 is an upstream regulator of ITK. This function of SLP-76 requires its N-terminal tyrosine phosphorylation sites, which are known to bind to the SH2 domain of ITK (20, 22) and are likely responsible for the inducible interaction of SLP-76 with ITK. Each of the three tyrosine phosphorylation sites of SLP-76 contributes to PLC-γ1 activation (30); their individual and combined contribution to ITK activation remains to be determined.
Several models may explain the SLP-76-dependent phosphorylation and activation of ITK. The ITK–SLP-76 interaction may unmask phosphorylation sites within the kinase required for its activation. Another possibility is that SLP-76 may bridge an interaction between Lck and ITK, thereby bringing about the transphosphorylation and activation of ITK. Indeed, Lck is known to transphosphorylate ITK at Y511, which is found within the activation loop of ITK (14, 15), and has also been shown to bind to SLP-76 (31, 32). Finally, the TCR-inducible interaction between the SH2 domain of ITK and the N-terminal tyrosine phosphorylation sites on SLP-76 may compete with interactions that maintain the autoinhibited conformation of ITK, as previously suggested (20). ITK activation, in turn, would lead to its increased tyrosine phosphorylation via increased autophosphorylation at Y180 (12). Of course, these possibilities are not mutually exclusive. In a similar fashion, Bruton's tyrosine kinase requires the SLP-76 analog B cell linker protein to mediate its B cell antigen receptor-induced transtyrosine phosphorylation and autophosphorylation (33).
All of the above models postulate that ITK activation takes place within a SLP-76 nucleated complex. To test this assumption, we confirmed a TCR-inducible interaction between full-length SLP-76 and ITK. The selective removal of SLP-76-associated ITK by preclearing experiments demonstrated that the relatively small proportion of ITK, found in complex with SLP-76, encompasses nearly all of the catalytically active ITK in the cell. This result was surprising because it suggests that, once phosphorylated and activated, ITK cannot dissociate from SLP-76 without losing its activity. Indeed, the catalytic activity of SLP-76-associated ITK was lost upon its mild elution from the SLP-76 complex and was restored upon reconstitution of the physical interaction between SLP-76 and ITK. Because this mild elution step should not affect the state of ITK phosphorylation, we suggest that SLP-76 exerts an ongoing effect on tyrosine-phosphorylated ITK, which is required to maintain ITK in an active state.
Several models may explain the ongoing requirement for SLP-76 association to maintain ITK activity. SLP-76 may act as a regulatory subunit, which prevents ITK from reassuming the “closed,” inactive conformation, by binding to ITK, as suggested in the “bidentate” model of ITK regulation (13); alternatively, SLP-76 may compete with the interaction of ITK with molecules that inhibit its catalytic activity, such as cyclophilin A (26). Another possibility is that SLP-76 and Vav may act in concert to maintain the active form of ITK. Indeed, Vav, which binds to the N-terminal tyrosines of SLP-76 (3), also is required for the TCR-induced phosphorylation and activation of ITK (34).
When comparing the catalytic activity of ITK and ZAP-70, we observed similar, robust phosphorylation of recombinant SLP-76 by both kinases. This result was unexpected, given the strong evidence that ZAP-70 phosphorylates SLP-76: a dramatic reduction in the TCR-induced phosphorylation of SLP-76, observed in ZAP-70-deficient cells (18), and the efficient ZAP-70-mediated tyrosine phosphorylation of SLP-76, observed when the two proteins are coexpressed in a heterologous system (19, 35). Indeed, ZAP-70 can phosphorylate SLP-76-derived peptides in vitro, with a Km in the 10–50 μM range (19). Nonetheless, our result raises the possibility that SLP-76 may be a physiologically relevant substrate of both ITK and ZAP-70. In further support of this idea, the kinase domain of ITK, expressed as a GST fusion protein, was shown to phosphorylate a GST fusion protein encompassing the N terminus of SLP-76, with a Km of 16 μM (36). In another study, SLP-76 was efficiently phosphorylated in COS cells, upon coexpression of another Tec family kinase, RLK (37). Our experiment demonstrates direct phosphorylation of full-length SLP-76, by full-length ITK, which was activated by a physiologically relevant signal. It also compares the potential of ITK and ZAP-70 to mediate SLP-76 phosphorylation. Given the different catalytic specificity of ITK and ZAP-70, as evidenced by their strikingly different abilities to phosphorylate a PLC-γ1 fragment, we speculate that these enzymes may target different sites on SLP-76, and, indeed, ITK is not required to phosphorylate Y145 of SLP-76 (30). Further work will be required to identify the site(s) phosphorylated by ITK and to determine their functional relevance in the context of intact cells.
The Src, Lck, and Abl PTKs can be activated by well studied, oncogenic mutations that disrupt the closed conformation of the kinase (25). Examples of adaptors that bind to and activate WT PTKs in the context of normal physiologic signaling pathways are few, and in this sense the SLP-76-ITK interaction may become an important paradigm for understanding PTK regulation.
Materials and Methods
Antibodies.
The monoclonal antibody C305 was used for anti-TCR stimulations (38). Monoclonal anti-FLAG epitope (M2) was from Sigma (St. Louis, MO). Polyclonal anti-PLC-γ1 was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phosphotyrosine (4G10) was from Upstate Cell Signaling Solutions (Lake Placid, NY). Phospho-specific anti-PLC-γ1 pY783 was from Biosource (Camarillo, CA). Phospho-specific anti-PLC-γ1 pY775 (6) was provided by Barbara L. Rellahan (Laboratory of Immunobiology, Division of Monoclonal Antibodies, Center for Drug Evaluation and Research, Food and Drug Administration, Bethesda, MD). Polyclonal anti-ITK (BL12) (39) was provided by Michael G. Tomlinson (University of Birmingham, Birminigham, U.K.) and Joseph Bolen (Millennium Pharmaceuticals, Inc., Cambridge, MA). Polyclonal anti-ZAP-70 (40) was provided by Dapeng Qian (Progenics Pharmaceuticals, Inc., Tarrytown, NY) and Arthur Weiss (University of California, San Francisco, CA). Polyclonal anti-SLP-76 has been described previously (21).
Cell Culture.
The SLP-76-deficient Jurkat-derived cell line J14, its WT SLP-76-reconstituted derivative, J14–76-11, and the SLP-76-Y3F-reconstituted derivative have been described previously (5, 7)
Recombinant Proteins.
A GST fusion protein encompassing residues 538–851 of PLC-γ1 (GST-PLCγ1SH2-SH2-SH3) (7) was purified and eluted from glutathione agarose (Sigma) by using standard procedures. BL21-CodonPlus-RIL bacteria (Stratagene, La Jolla, CA) were transformed with N-terminally His-tagged, full-length human SLP-76, cloned into pDEST17 (Invitrogen, Carlsbad, CA), and grown in M9 minimal medium (41) containing 50 μg/ml carbenicillin and 34 μg/ml chloramphenicol at 24°C to the mid-log phase. Expression was induced by adding 0.4 mM isopropyl-β-d-thiogalactopyranoside at 16°C for 24 h. Bacteria were collected by centrifugation; frozen at −70°C; thawed into PBS containing 1 mM DTT, 2 mM phenylmethanesulfonyl fluoride, 1 μg/ml pepstatin, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 5 μg/ml RNase, 5 μg/ml DNase, and 10 μg/ml lysozyme at 4°C; and disrupted by sonication with a probe sonicator at 4°C. The lysate was cleared by centrifugation at 9,000 × g for 12 min at 4°C, and the supernatant was incubated with Ni-NTA agarose beads (Qiagen, Valencia, CA). His-tagged SLP-76 was eluted from the beads by using 200 mM imidazole at pH 6.
Cell Lysis and Immunoprecipitation.
Jurkat cells were washed in PBS containing Ca2+ and Mg2+ (Dulbecco's PBS), preheated to 37°C for 10 min, and stimulated for 1 min with α-TCR (C305) or mock-stimulated, collected, and lysed at 108 cells/ml in cold lysis buffer (20 mM Hepes, pH 7.3/1% Triton X-100/150 mM NaCl/10% glycerol/10 mM NaF/1 mM Na3VO4/10 μg/ml aprotinin/2 mM EGTA/10 μg/ml leupeptin/2 mM phenylmethanesulfonyl fluoride). After 20 min at 4°C, lysates were centrifuged at 4°C in a microcentrifuge at 16,000 × g for 10 min. Supernatants were used directly for immunoprecipitation of kinases or were ultracentrifuged at 137,000 × g for 11 min at 4°C, before immunoprecipitation with anti-FLAG.
In Vitro Kinase Assay.
Anti-Flag or anti-ZAP-70 immunoprecipitates were assayed for kinase activity essentially as described (23), but with the modification of using 10 μM ATP and either 0.1–1 μg recombinant GST–PLCγ1SH2-SH2-SH3 or 1 μg of His-tagged, recombinant SLP-76 as a substrate. Reactions were terminated after 15 min by adding EDTA to a concentration of 12.5 mM. Supernatants and beads were analyzed separately by SDS/PAGE, followed by detection with ECL.
Acknowledgments
We thank Michael G. Tomlinson, Joe Bolen, Dapeng Qian, Arthur Weiss, and Barbara L. Rellahan for providing reagents; Ron Wange, Ami Aronheim, and Itzik Kehat for valuable technical insights and advice; and all members of the D.Y. laboratory for support and advice. This work was supported by The Rappaport Family Institute for Research in the Medical Sciences.
Abbreviations
- LAT
linker for activation of T cells
- PLC-γ1
phospholipase C-γ1
- PTK
protein tyrosine kinase
- TCR
T cell antigen receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Carpenter G, Ji Q. Exp Cell Res. 1999;253:15–24. doi: 10.1006/excr.1999.4671. [DOI] [PubMed] [Google Scholar]
- 2.Rhee SG. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koretzky GA, Abtahian F, Silverman MA. Nat Rev Immunol. 2006;6:67–78. doi: 10.1038/nri1750. [DOI] [PubMed] [Google Scholar]
- 4.Yablonski D, Weiss A. Adv Immunol. 2001;79:93–128. doi: 10.1016/s0065-2776(01)79003-7. [DOI] [PubMed] [Google Scholar]
- 5.Yablonski D, Kuhne MR, Kadlecek T, Weiss A. Science. 1998;281:413–416. doi: 10.1126/science.281.5375.413. [DOI] [PubMed] [Google Scholar]
- 6.Serrano CJ, Graham L, DeBell K, Rawat R, Veri MC, Bonvini E, Rellahan BL, Reischl IG. J Immunol. 2005;174:6233–6237. doi: 10.4049/jimmunol.174.10.6233. [DOI] [PubMed] [Google Scholar]
- 7.Yablonski D, Kadlecek T, Weiss A. Mol Cell Biol. 2001;21:4208–4218. doi: 10.1128/MCB.21.13.4208-4218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myung PS, Derimanov GS, Jordan MS, Punt JA, Liu Q-H, Judd BA, Meyers EE, Sigmund CD, Freedman BD, Koretzky GA. Immunity. 2001;15:1011–1026. doi: 10.1016/s1074-7613(01)00253-9. [DOI] [PubMed] [Google Scholar]
- 9.Kumar L, Pivniouk V, de la Fuente MA, Laouini D, Geha RS. Proc Natl Acad Sci USA. 2002;99:884–889. doi: 10.1073/pnas.022619199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartzberg PL, Finkelstein LD, Readinger JA. Nat Rev Immunol. 2005;5:284–295. doi: 10.1038/nri1591. [DOI] [PubMed] [Google Scholar]
- 11.Liu KQ, Bunnell SC, Gurniak CB, Berg LJ. J Exp Med. 1998;187:1721–1727. doi: 10.1084/jem.187.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilcox HM, Berg LJ. J Biol Chem. 2003;278:37112–37121. doi: 10.1074/jbc.M304811200. [DOI] [PubMed] [Google Scholar]
- 13.Andreotti AH, Bunnell SC, Feng S, Berg LJ, Schreiber SL. Nature. 1997;385:93–97. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- 14.Gibson S, August A, Kawakami Y, Kawakami T, Dupont B, Mills GB. J Immunol. 1996;156:2716–2722. [PubMed] [Google Scholar]
- 15.Heyeck SD, Wilcox HM, Bunnell SC, Berg LJ. J Biol Chem. 1997;272:25401–25408. doi: 10.1074/jbc.272.40.25401. [DOI] [PubMed] [Google Scholar]
- 16.Shan X, Wange RL. J Biol Chem. 1999;274:29323–29330. doi: 10.1074/jbc.274.41.29323. [DOI] [PubMed] [Google Scholar]
- 17.Ching KA, Grasis JA, Tailor P, Kawakami Y, Kawakami T, Tsoukas CD. J Immunol. 2000;165:256–262. doi: 10.4049/jimmunol.165.1.256. [DOI] [PubMed] [Google Scholar]
- 18.Williams BL, Schreiber KL, Zhang W, Wange RL, Samelson LE, Leibson PJ, Abraham RT. Mol Cell Biol. 1998;18:1388–1399. doi: 10.1128/mcb.18.3.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wardenburg JB, Fu C, Jackman JK, Flotow H, Wilkinson SE, Williams DH, Johnson R, Kong G, Chan AC, Findell PR. J Biol Chem. 1996;271:19641–19644. doi: 10.1074/jbc.271.33.19641. [DOI] [PubMed] [Google Scholar]
- 20.Bunnell SC, Diehn M, Yaffe MB, Findell PR, Cantley LC, Berg LJ. J Biol Chem. 2000;275:2219–2230. doi: 10.1074/jbc.275.3.2219. [DOI] [PubMed] [Google Scholar]
- 21.Gonen R, Beach D, Ainey C, Yablonski D. J Biol Chem. 2005;280:8364–8370. doi: 10.1074/jbc.M409437200. [DOI] [PubMed] [Google Scholar]
- 22.Su Y-W, Zhang Y, Schweikert J, Koretzky GA, Reth M, Wienands J. Eur J Immunol. 1999;29:3702–3711. doi: 10.1002/(SICI)1521-4141(199911)29:11<3702::AID-IMMU3702>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 23.Beach D, Gonen R, Bogin Y, Reischl IG, Yablonski D. J Biol Chem. 2007;282:2937–2946. doi: 10.1074/jbc.M606697200. [DOI] [PubMed] [Google Scholar]
- 24.Boggon TJ, Eck MJ. Oncogene. 2004;23:7918–7927. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- 25.Harrison SC. Cell. 2003;112:737–740. doi: 10.1016/s0092-8674(03)00196-x. [DOI] [PubMed] [Google Scholar]
- 26.Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Proc Natl Acad Sci USA. 2002;99:1899–1904. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houtman JC, Houghtling RA, Barda-Saad M, Toda Y, Samelson LE. J Immunol. 2005;175:2449–2458. doi: 10.4049/jimmunol.175.4.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez R, Matsuda M, Perisic O, Bravo J, Paul A, Jones NP, Light Y, Swann K, Williams RL, Katan M. J Biol Chem. 2001;276:47982–47992. doi: 10.1074/jbc.M107577200. [DOI] [PubMed] [Google Scholar]
- 29.Humphries LA, Dangelmaier C, Sommer K, Kipp K, Kato RM, Griffith N, Bakman I, Turk CW, Daniel JL, Rawlings DJ. J Biol Chem. 2004;279:37651–37661. doi: 10.1074/jbc.M311985200. [DOI] [PubMed] [Google Scholar]
- 30.Jordan MS, Sadler J, Austin JE, Finkelstein LD, Singer AL, Schwartzberg PL, Koretzky GA. J Immunol. 2006;176:2430–2438. doi: 10.4049/jimmunol.176.4.2430. [DOI] [PubMed] [Google Scholar]
- 31.Sanzenbacher R, Kabelitz D, Janssen O. J Immunol. 1999;163:3143–3152. [PubMed] [Google Scholar]
- 32.Kumar L, Feske S, Rao A, Geha RS. Proc Natl Acad Sci USA. 2005;102:19063–19068. doi: 10.1073/pnas.0509176102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baba Y, Hashimoto S, Matsushita M, Watanabe D, Kishimoto T, Kurosaki T, Tsukada S. Proc Natl Acad Sci USA. 2001;98:2582–2586. doi: 10.1073/pnas.051626198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds L, Smyth LA, Norton T, Freshney N, Downward J, Kioussis D, Tybulewicz VLJ. J Exp Med. 2002;195:1103–1114. doi: 10.1084/jem.20011663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raab M, da Silva AJ, Findell PR, Rudd CE. Immunity. 1997;6:155–164. doi: 10.1016/s1074-7613(00)80422-7. [DOI] [PubMed] [Google Scholar]
- 36.Lin TA, McIntyre KW, Das J, Liu C, O'Day KD, Penhallow B, Hung CY, Whitney GS, Shuster DJ, Yang X, et al. Biochemistry. 2004;43:11056–11062. doi: 10.1021/bi049428r. [DOI] [PubMed] [Google Scholar]
- 37.Schneider H, Guerette B, Guntermann C, Rudd CE. J Biol Chem. 2000;275:3835–3840. doi: 10.1074/jbc.275.6.3835. [DOI] [PubMed] [Google Scholar]
- 38.Weiss A, Stobo JD. J Exp Med. 1984;160:1284–1299. doi: 10.1084/jem.160.5.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomlinson MG, Kurosaki T, Berson AE, Fujii GH, Johnston JA, Bolen JB. J Biol Chem. 1999;274:13577–13585. doi: 10.1074/jbc.274.19.13577. [DOI] [PubMed] [Google Scholar]
- 40.Qian D, Lev S, van Oers NS, Dikic I, Schlessinger J, Weiss A. J Exp Med. 1997;185:1253–1259. doi: 10.1084/jem.185.7.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]