Abstract
The solution structure of the hepta-alanine polypeptide Ac-X2A7O2-NH2 (XAO) has been a matter of controversy in the current literature. On one side of the argument is a claim that the peptide adopts a mostly polyproline II (PPII) structure, with a <20% population of β conformations at room temperature [Shi Z, Olson CA, Rose GA, Baldwin RL, Kallenbach NR (2002) Proc Natl Acad Sci USA 99:9190–9195], whereas the other side of the argument insists that the peptide exists as an ensemble of conformations, including multiple β-turn structures [Makowska J, Rodziewicz-Motowidlo S, Baginska K, Vila JA, Liwo A, Chmurzynski L, Scheraga HA (2006) Proc Natl Acad Sci USA 103:1744–1749]. We have used an excitonic coupling model to simulate the amide I band of the FTIR, vibrational circular dichroism, and isotropic and anisotropic Raman spectra of XAO, where, for each residue, the backbone dihedral angle φ was constrained by using the reported 3JCαHNH values and a modified Karplus relation. The best reproduction of the experimental data could only be achieved by assuming an ensemble of conformations, which contains various β-turn conformations (≈26%), in addition to β-strand (≈23%) and PPII (≈50%) conformations. PPII is the dominant conformation in segments not involved in turn formations. Most of the residues were found to sample the bridge region connecting the PPII and right-handed helix troughs in the Ramachandran plot, which is part of the very heterogeneous ensemble of conformations generally termed type IV β-turn.
Keywords: alanine propensity, amide I, unfolded peptides, vibrational spectroscopy
The unfolded state of peptides and proteins has been the subject of an increasing number of experimental and theoretical studies (1–9), owing to the discovery of naturally disordered, although biologically functioning, proteins and peptides (9), and the general relevance for a thorough understanding of the protein folding process. In this context, the possible existence of local residue structure would certainly affect the initial phase of the folding process (10). The existence of such locally ordered segments has first been proposed by Tiffany and Krimm (11) based on electronic circular dichroism (ECD) measurements on poly-l-proline, poly-l-lysine, and poly-l-glutamic acid. They concluded that charged polypeptides assume, at least locally, a rather ordered polyproline II (PPII) conformation, which is the structure adopted by trans-poly-l-proline. This notion was later confirmed by vibrational circular dichroism (VCD) studies on a variety of unfolded polypeptides and proteins (12, 13).
PPII is a rather regular structural motif in that it exhibits a perfect, left-handed threefold rotational symmetry (31-helix) for its canonical conformation, with (φ, ψ) = (−78°, 146°) (14). Woody and coworkers (15–17) have published a series of papers proving that PPII gives rise to a far UV-ECD spectrum, which many in the scientific community still interpret as indicative of random coil (15). Recently, they reported a very convincing quantitative agreement between the PPII content of βII proteins, derived from crystallographic data, and ECD spectra (16). The PPII signal was also observed in the spectrum of the unfolded state of many proteins subjected to denaturing detergents (17). Thermal denaturation, however, often yields a conformation that is reflected by a weak, nearly symmetric, couplet with a positive maximum between 180 and 210 nm and a negative minimum between 210 and 230 nm (11, 16, 18). ECD and Raman optical activity spectra of some unstructured proteins (tau protein, casein, stathmin, Bob1) suggest that their structure contains a substantial amount of PPII (19–21).
More recently, the interest of the protein/peptide folding community has focused on the unfolded state of alanine-based peptides, after theoretical and experimental results suggested that it cannot be described by the classical statistical coil model of Tanford (22) and Flory (23). Thus, alanine has emerged as a paradigm for the breakdown of the statistical coil model. Shi et al. (24), for instance, used NMR and ECD measurements to study the structure of Ac-X2(A)7O2-NH2 (XAO, X and O denote diaminobutyric acid and ornithine, respectively), and interpreted their results as indicating that the individual residues predominantly adopt a PPII conformation at room temperature. The results of Shi et al. have been corroborated by numerous experimental and theoretical studies on short peptides, which all revealed a substantial PPII propensity for alanine (2, 4–8). This notion is also in perfect agreement with distributions that Serrano (25) and Avbelj and Baldwin (26) inferred from coil libraries, but at variance with a less restricted library investigated by Dobson and associates (27). The results of molecular dynamics (MD) simulations are force field-dependent and generally do not reproduce a PPII propensity of alanine without force-field modifications (28). Zagrovic et al. (29) investigated XAO by small angle x-ray scattering (SAXS) experiments and obtained a radius of gyration of 7.4 Å. If one assumes the “random walk scaling” between radius of gyration and end-to-end distance, this value corresponds to an average end-to-end distance of 18.1 Å, which is significantly shorter than what one would expect for a pure PPII structure (radius of gyration = 13.1 Å, end-to-end distance = 32.04 Å). Zagrovic et al. (29) also performed several MD simulations with six variants of the Amber and Gromos force fields. The simulations reproduced neither the extended, PPII-dominated structure, nor the very short radius of gyration obtained from the SAXS data. Makowska et al. (30) combined ECD and NMR measurements of XAO (involving the measurement of 3JCαHNH coupling constants, NOEs, and chemical shifts) with MD simulations performed with the AMBER 99 force field. The results led them to conclude that PPII is one of many conformations sampled by alanine. Additionally, the peptide's residues were found to sample multiple β-turn structures, without leading to the formation of stable structures with interpeptide hydrogen bonding. This result is surprising because alanine is not known for its propensity for turn structures (31). Altogether, the results of Makowska et al. reemphasize the validity of the classical statistical coil model, although their Ramachandran plot looks somewhat different from the random coil distribution of alanine (27, 32, 33). Recently, Jun et al. (34) used electron spin resonance distance measurements to probe the distance of two spin labels attached to an alanine-based peptide in the folded and (thermally) unfolded states and found the respective value for the latter to be too short for a pure PPII state of the peptide. Analogously, Tucker et al. (35) used an intrinsic FRET pair to deduce that the supposedly disordered mastoparan X peptide actually exists in aqueous solution as an ensemble of compact conformations.
One might try to resolve the above-discussed conflicting results by proposing that the PPII propensity of alanine solely applies to very short peptides, i.e., tri- and tetra-peptides (6, 36, 37), whereas it becomes more disordered in longer ones. A clarification of this issue has a high bearing on the understanding of both the unfolded state of peptides and proteins, as well as the protein unfolding process, in view of the abundance of alanine in nature, and its high propensity for helix formation (38). We therefore decided to subject XAO, which remains at the center of the debate, to a thorough experimental investigation combining FTIR, polarized Raman, VCD, and ECD spectroscopy with results obtained earlier from SAXS and NMR studies.
Theory
The underlying theory of our spectral modeling has been described in detail (37, 39). Briefly, we assume that the interaction between N amide I modes of a (blocked) polypeptide containing N amino acid residues can be described in terms of a coupled oscillator model. The validity and applicability of the coupled oscillator model for describing the delocalized excited states in polypeptides has recently been demonstrated by a variety of theoretical studies (40, 41). For N oscillators, the corresponding Hamiltonian is written as:
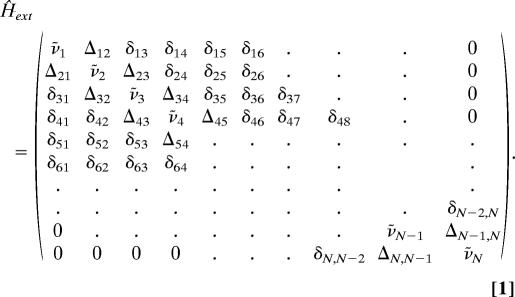 |
In our notation, we distinguish between nearest-neighbor coupling, Δj,j±1, and interactions between more distant residues, δj,j′ (j′ = j ± 2, j ± 3, j ± 4). The former involves through-bond and through-space coupling (42). Coupling parameter values were obtained from recent studies on tripeptides (5) and results of ab initio and density functional theory calculations (40–42), whereas δj, j′ were assumed to result from transition dipole coupling, which was explicitly calculated (43). The corresponding eigenfunctions of the Schrödinger equation can be written as a linear combination of local oscillator functions. As a consequence, the Raman tensor, and the transition dipole moment of an excitonic state can also be written as linear combinations of the Raman tensors and transition dipole moments of local amide I vibrations, respectively. To this end, the latter have to be transformed into a common coordinate system, which brings about an orientational dependence of IR and Raman scattering. The amide I band shapes can be calculated as a superposition of bands arising from the transitions into the N excitonic states. The Raman tensor of the ith excitonic state, α̂i, can be used to calculate the isotropic and anisotropic Raman intensities, which are proportional to the isotropic and anisotropic tensor invariants (44). The respective integrated IR absorptivity, εiIR, and the circular dichroism, Δεi, of the ith excitonic mode, can be calculated in units of M−1·cm−1 per residue. Finally, the Raman, IR, and VCD amide I profiles can be calculated as superpositions of Gaussian bands representing the excitonic modes (39, 45).
Results and Discussion
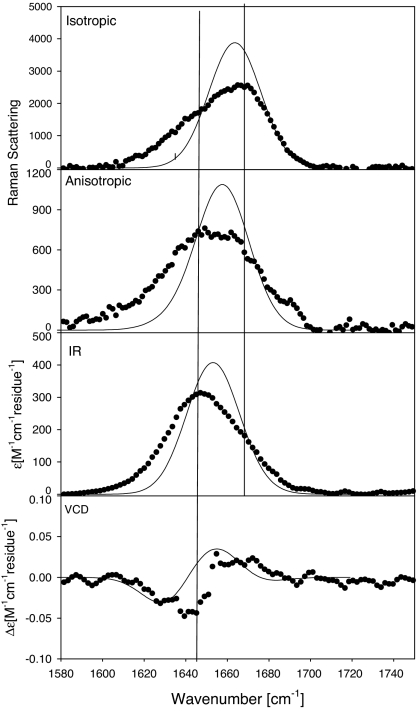
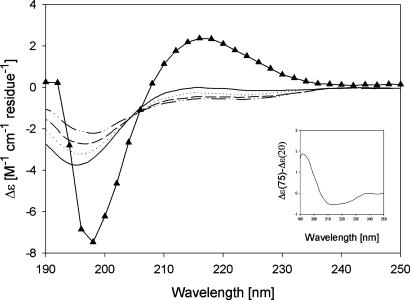
The amide I′ band profiles of the IR, isotropic Raman, anisotropic Raman, and VCD spectra of XAO in D2O (pD = 2.2) are shown in Fig. 1. We observed a clear noncoincidence between the isotropic and IR-band profiles, which suggests that the peptide predominantly samples the upper left quadrant of the Ramachandran plot. The anisotropic band is very broad and does not display the clearly discernable peak position in between the wavenumber positions of the IR and isotropic Raman bands, which has been shown to be diagnostic of PPII (39, 46, 47). The VCD spectrum displays the usual negative couplet of so-called unfolded peptides and proteins, but it is less pronounced than those observed for e.g., AAA, AAAA, AAKA and (AAKA)2 (5, 6, 37, 48, 49). The rotational strength per residue is substantially smaller than that observed for the amyloid peptide fragment Aβ1–28, which has been shown to contain a substantial PPII content (46, 47, 50), but similar to that of salmon calcitonin, which more closely resembles a statistical coil (47). All of these observations suggest that XAO has much less PPII content than suggested by Kallenbach and associates (24). This notion is corroborated by the ECD spectrum of XAO at 20°C, as shown in Fig. 2. As already argued by Vila et al. (51), the temperature-dependent spectra show the minimum at 195 nm, which is associated with PPII, but do not display the maximum at 216 nm, which is diagnostic of this structure. With respect to the minimum, the corresponding Δε value (per residue) is much smaller than that observed for AAAA (triangles in Fig. 2), and slightly smaller than that obtained for Aβ1–28 (46, 49).
Fig. 1.
Experimental isotropic and anisotropic Raman, FTIR, and VCD spectra of the amide I region of XAO, pD = 2.2 (●), and band profiles simulated by using the two-state model per residue (PPII/βs), in line with the results of Shi et al. (24).
Fig. 2.
Temperature-dependent ECD spectra of XAO, pD = 2.2 (black) at 20°C, 40°C, 60°C, and 80°C, where the arrows show increasing temperature, and tetraalanine, pD = 1 (triangles) at 20°C. (Inset) The difference spectrum Δε80° − Δε20°.
We performed a detailed analysis of the amide I′ band profiles based on a statistical model that assumes a blend of different conformations per residue. This approach is a substantial refinement of more crude models that were used previously to analyze the profiles of longer peptides (46, 47) and an extension of a recently applied simple statistical model (49). The model is based on the following representative conformations: (i) PPII with (φ, ψ) = (−68°, 150°), (ii) β-strand with (φ, ψ) = (−119°, 113°), and (iii) various β-turn conformations, i.e., type I′ [(φ, ψ)i = (60°, 30°), (φ, ψ)i+1 = (90°, 0°)], type II [(φ, ψ)i = (−60°, 100°), (φ, ψ)i+1 = (80°, −10°)], type III [(φ, ψ)i = (−60°, −30°), (φ, ψ)i+1 = (−60°, −30°)], type III′ [(φ, ψ)i = (60°, 30°), (φ, ψ)i+1 = (60°, 30°)], type V [(φ, ψ)i = (80°, −80°), (φ, ψ)i+1 = (−80°, 80°)], and a conformation representing the bridge region between the upper left quadrant and the helical region of the Ramachandran plot [(φ, ψ)i = (−20°, 20°), (φ, ψ)i+1 = (20°, 20°)], which emerged from the simulations of Makowska et al. (30) as part of the heterogeneous manifold of so-called type IV turns. The coordinates for PPII, β-strand, and 310 (type III) correspond to maxima of distributions inferred from coil libraries (26). The turn structures represent the structural manifolds sampled by MD simulations of Makowska et al. (ref. 30 and A. Liwo, personal communication).
The total intensity of an amide I′ band profile at a given wavenumber is written in terms of the above considered statistical ensemble:
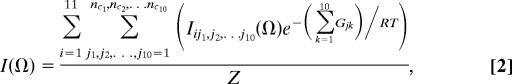 |
where Gjk is the Gibbs energy of the kth residue with conformation j. The subscript i labels the excitonic states of the amide I′ oscillators. Iij1,j2…, j10 is thus the intensity profile of the ith excitonic state associated with the configuration {j1, j2,…, j10} of the first 10 residues of the peptide. The conformation of the C-terminal residue does not affect the amide I′ band profiles. Hence, 11 excitonic states depend on the conformation of 10 residues. R is the gas constant, T the absolute temperature, Z the partition sum, and nc1, nc2, … nc10 are the numbers of conformations considered for the respective residues.
In our analysis we considered PPII (j = 1) and β-strand (j = 2) for all residues. For residues 3, 4, 7, and 8 (all alanines), we additionally considered type IV (j = 3), type III (j = 4) (type III′ for residue 3), type V (j = 5), type II (j = 6), and type I′ (j = 7). For residues 2 (X), 5, 6, and 9 (all Ala) we allowed type III′ and type IV turns as additional conformations. This blend represents, more or less, different clusters that emerged from the MD simulations of Makowska et al. (30). Their results suggest a limited sampling of conformations involving intrapeptide hydrogen bonding, hence this possibility was disregarded for the sake of simplicity. They did not obtain substantial fractions of right-handed helix-like conformations for alanine residues, but some coil libraries and many MD simulations indicate that alanine significantly samples helix-like conformations (51). We used the 3JCαHNH coupling constants reported by Makowska et al. (8) to restrict the choice of possible conformational mixtures by calculating:
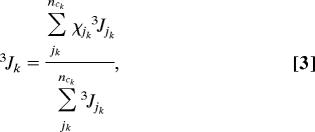 |
where 3Jk is the experimental coupling constant obtained for the kth residue (24), χjk is the mole fraction of the jth conformation of the kth residue, and 3Jjk is the respective coupling constant.
For the simulation of the band profiles, we used the nearest neighbor coupling Δi,i+1 (Eq. 1) constants, which Torii and Tasumi (42) obtained from ab initio calculations on a glycine dipeptide. These coupling constants have been shown to agree well with experimentally derived values for short peptides (5, 52). The non-nearest neighbor coupling constants δi,j were calculated by using the transition dipole-coupling formalism. We used the intrinsic amide I′ wavenumber of the central alanine residue of tetraalanine in water (37) as a reference for the PPII conformation of alanine. For the two charged residues, X and O, we estimated the respective intrinsic wavenumbers by using the wavenumber difference between K and A obtained from the anionic dipeptides AA and KA (50). The intrinsic wavenumbers for the other conformations considered in this study were obtained by considering their conformational dependence obtained from density functional theory studies on alanine dipeptides (53). The intrinsic dipole strengths of the individual amide I′ modes were obtained from the dipeptide study of Measey et al. (50). Here, we again used K as a model for X and O. Thus, we calculated the IR absorption and the VCD in absolute units. Finally, we used the earlier obtained (relative) Raman tensors for AA (for A) and KA (for X, O) to calculate the ratio of anisotropic and isotropic Raman scattering. Parameters used in the simulation can be found in Table 1.
Table 1.
Parameters used in the simulation, including dipole moments (μ), wavenumber positions (ν̃), and nearest-neighbor coupling constants (Δ)
| Parameter | Value |
|---|---|
| μAla | 2.7 × 10−19 esu cm |
| μLys | 3.0 × 10−19 esu cm |
| ν̃Ala | 1,658 cm−1 |
| ν̃Lys | 1,664 cm−1 |
| Δα | 12.0 cm−1 |
| Δ310 | 3.1 cm−1 |
| ΔPPII | 10.0 cm−1 |
| Δβs | 2.6 cm−1 |
| ΔTypeI | 7.0 cm−1, 2.0 cm−1 |
| ΔTypeII | −3.0 cm−1, −2.0 cm−1 |
| ΔTypeIII | 8.0 cm−1, 8.0 cm−1 |
| ΔTypeIV | −10.0 cm−1 |
| ΔTypeV | −1.0 cm−1, 20.0 cm−1 |
The two coupling constant values listed for the turn conformations, i.e. types I–V, are for residues i and i+1 of the turn, respectively.
We started the analysis by first testing the results of Shi et al. (24), which invoke a two-state model per residue, namely PPII and β-strand. In this case the measured 3JCαHNH coupling constants determine the respective molar fractions. The result of this simulation is depicted in Fig. 1. Apparently neither the asymmetry of the IR nor that of the isotropic Raman band profile is reproduced. The width of the anisotropic band profile is also not accounted for. Only the VCD signal is close to the experimentally obtained couplet. This simulation provides compelling evidence for the above formulated supposition that the two-state model of Shi et al. (24), which yields very high PPII fractions for the alanine residues (between 0.65 and 0.95), is not in agreement with our data. We calculated the respective average end-to-end distance by using:
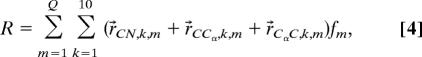 |
where m labels a given sequence of PPII and β-strand residues, and fm is the fraction of the sequence m. The total number of sequences for the two-state model is Q = 1,024. The vectors denote the length and orientations of the CN, NCα, and Cα C bonds with respect to a coordinate system defined in earlier papers (5, 39). Eq. 4 neglects the C-terminal residue for which we do not have any data. For the two-state model we obtained 30 Å, which far exceeds that of 18.1 Å, derived from the radius of gyration of 7.4 Å, as obtained from SAXS data (29).
In a second step, we performed a simulation that considered the entire above-mentioned conformational manifold, containing representatives of all of the local turn structures that emerged from the MD simulations of Makowska et al. (30). We used the result of these simulations to initially guess the mole fractions of the respective residue conformations and subsequently varied them to optimize the simulations with respect to the measured profiles. This process yielded a decent reproduction of the IR band profile, but the asymmetry of the isotropic band profile, the band profile of anisotropic scattering, and the observed noncoincidence between isotropic Raman scattering and IR absorption, all were not accounted for. Moreover, the VCD signal was nearly eliminated in all of these simulations. This result is per se important in that it indicates that a pronounced amide I′ VCD signal is inconsistent with a statistical coil sampling, in agreement with arguments by Dukor and Keiderling (12).
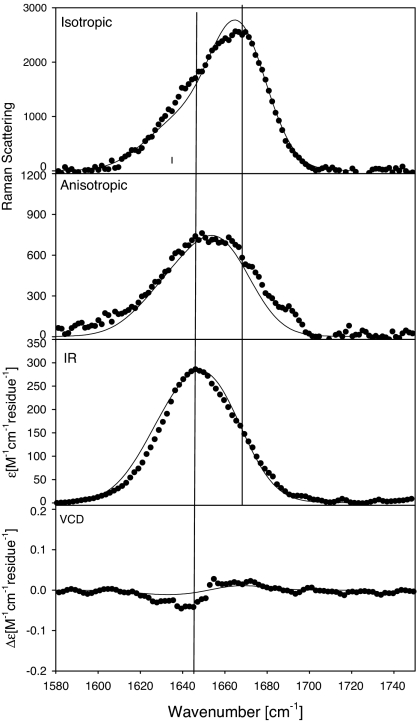
In the third and final step, we somewhat reduced the number of sampled conformations for all residues. Table 2 lists the (representative) conformations and the respective optimized mole fractions considered for this simulation. With respect to the turn structures, we confined ourselves to β-turn type III′ for residues 2 and 3 (XA), type III for residue 4 (A), type IV for residues 2–9 (XA7O), and type V for residues 2 and 3 (XA). PPII and β-strand conformations were considered for all residues. Altogether, we thus considered 142,884 different peptide conformations. Fig. 3 shows the best set of profiles obtained from simulations based on this structural model. The two Raman profiles are nearly perfectly reproduced. The simulated IR-band profile is slightly broader than the experimental one and the negative part of the VCD couplet is somewhat underestimated. Nevertheless, the agreement between experiment and simulation is more than satisfactory, particularly in view of the fact that the structural model is still crude, because it considers only representative conformations rather than distributions.
Table 2.
Molar fractions of different residue conformations used to simulate the amide I' band profiles
| Residue | PPII | β-strand | β-turn, type III | β-turn,type III' | β-turn, type IV | β-turn, type V | β-turn, type II | β-turn, type I |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.64 | 0.35 | ||||||
| 2 | 0.46 | 0.29 | 0.25 | |||||
| 3 | 0.21 | 0.09 | 0.20 | 0.25 | 0.15 | 0.05 | 0.05 | |
| 4 | 0.03 | 0.32 | 0.15 | 0.25 | 0.15 | 0.05 | 0.05 | |
| 5 | 0.68 | 0.12 | 0.2 | |||||
| 6 | 0.66 | 0.14 | 0.2 | |||||
| 7 | 0.57 | 0.18 | 0.25 | |||||
| 8 | 0.46 | 0.29 | 0.25 | |||||
| 9 | 0.77 | 0.12 | 0.11 | |||||
| 10 | 0.55 | 0.35 |
Fig. 3.
Experimental isotropic and anisotropic Raman, FTIR, and VCD spectra of the amide I' region of XAO, pD = 2.2 (●) and the band profiles simulated by using the conformations listed in Table 2.
We used Eq. 4 to calculate the end-to-end distance associated with the considered conformational blend and obtained a value of 19.1 Å. This finding is in excellent agreement with the values derived from the SAXS experiment (29) and underscores the suitability of our analysis.
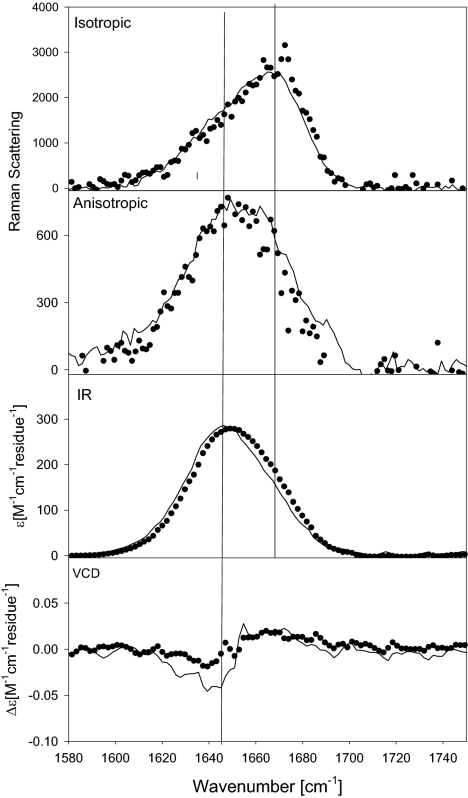
The temperature dependence of the ECD spectra (Fig. 2) and the 3JCαHNH coupling constants, as observed by Shi et al. (24), has been interpreted as indicating a larger fraction of β-strand at higher temperatures. Fig. 4 compares the IR, Raman, and VCD band profiles measured at 25°C (solid line) and 65°C (dots). To compare these profiles, we have to consider a wavenumber upshift of amide I′ of 4 cm−1 for this temperature interval, which is caused by anharmonic coupling to a low-frequency mode involving the hydrogen bonds to water molecules (54), which was accounted for by downshifting the profiles measured at 65°C by this amount. The remaining difference between the experimental profiles at 25°C and 65°C should result from changes of the equilibrium between different conformations. Apparently, the first moments of the experimentally observed anisotropic Raman and IR band profiles, and to a lesser extent also that of the isotropic Raman band profile, are at lower wavenumbers than those of the respective solid line profiles in Fig. 4. For IR, the high-temperature band profile is more asymmetric. The noncoincidence between the peak positions of the IR and isotropic Raman profiles is reduced from 20 cm−1 (25°C) to 14 cm−1 (65°C). The VCD couplet is reduced at high temperatures. All of these observations are consistent with the notion that a β-strand conformation is more populated at higher temperatures. This conformation gives rise to a reduced nearest-neighbor coupling, which leads to the observed reduction of the IR/isotropic Raman noncoincidence and the VCD signal. The reduced wavenumber shift at high temperature is consistent with the results of density functional theory calculations, which predict that amide I′ exhibits a slightly lower wavenumber in the β-strand conformation than in PPII (41).
Fig. 4.
Comparison of experimental isotropic and anisotropic Raman, FTIR, and VCD amide I' profiles of XAO, pD = 2.2 at 25°C (solid line) and 65°C (dots). For comparison, the spectra at 65°C were downshifted by 4 cm−1.
The result of their MD simulations led Makowska et al. (30) to the conclusion that PPII is only one of many possible conformations sampled by alanine residues, and that the notion of a PPII propensity for alanine has to be rejected. This finding is at variance with results obtained from experimental investigations on short peptides (6, 36, 37), which are clearly indicative of a PPII propensity for alanine, in accordance with recent MD simulations of Gnanakaran and Garcia (28, 55). On the other hand, it was clear that the radius of gyration value obtained from SAXS experiments was inconsistent with a predominant PPII population for all alanine residues of the XAO peptide. Our results strongly indicate that the truth lies in between the extremes invoked in the studies of Shi et al. (24) and Makowska et al. (30). The blend used for the simulation depicted in Fig. 3 still exhibits a substantial PPII fraction for residues 1 and 5–10. As a matter of fact, this is the only way to explain the significant noncoincidence between IR and isotropic Raman peak positions, which reflect the average nearest-neighbor coupling. This noncoincidence can only be achieved with a substantive fraction of PPII-like conformations (48, 56). However, our results also agree to a significant extent with Makowska et al., in that the consideration of turn-like conformations is necessary to explain our experimental results (30). We invoked the results of those authors in assuming that type III′ and V turn structures predominantly involve residues 2, 3, and 4, but we would have obtained similar results if we had selected another pair of residues. Makowska et al. obtained substantial type IV β-like conformations for residues 3–10. In principle, this is a rather heterogeneous class of conformations, containing all turn-like structures that are different from types I, II, III, and V, or their stereo-isomers. From the center of this region, we selected a representative conformation, i.e., (φ, ψ)i = (−20°, 20°), (φ, ψ)i+1 = (−20°, 20°), which allows negative nearest-neighbor coupling between amide I oscillators. The consideration of negative coupling is absolutely necessary for reproducing the asymmetry of the isotropic Raman band.
Taken together, our results suggest that longer alanine peptides can be more heterogeneous than one would expect based on the analysis of short peptide fragments. We recently reinvestigated trialanine and tetraalanine and found a substantial population of turn structures to be inconsistent with our spectroscopic data (49). The reason for the local turn propensity of alanine in XAO still needs to be explored. That the properties of XAO might reflect some general properties of unfolded alanine-based peptides is underscored by a recent, very elegant ESR study on the unfolded state of P2HG3WP(A4K)2CA4KA by Jun et al. (34), which also indicates a much smaller end-to-end distance than expected for an extended molecule. The results of the present study might indicate that alanine shows a turn propensity in the vicinity of charged residues (X). The possibility of turn formations as initiation of protein folding has been discussed in detail by Wright, Dyson, and Lerner (57), in view of their NMR studies on unfolded peptides and proteins. However, the undeniable occurrence of turn structures cannot obscure the fact that alanine still exhibits a substantial PPII propensity in longer peptides, which is in full accordance with findings derived from the coil libraries of Serrano (25) and Avbelji and Baldwin (26), but at variance with the more “disordered” library reported by Dobson and coworkers (27).
Materials and Methods
Material.
Ac-(Daba)2-(Ala)7-(Orn)2-NH2 was obtained as a gift from the laboratory of V. S. Pandé (Stanford University, Stanford, CA). To remove residual TFA, which absorbs in the vicinity of the amide I region, the peptide was dialyzed in a 1-ml Spectra/Por CE Float-A-lyzer dialysis bag, with a molecular weight cut-off of 500, and lyophilized overnight. For Raman, FTIR, and VCD experiments, the peptide was dissolved at a concentration of 25.1 mg/ml in acidified D2O. The pD of the resulting peptide solution was 2.2. For ECD measurements, the peptide solution was diluted 10-fold, with acidified D2O.
Methods.
The set-up for the IR, Raman, VCD and ECD measurements has been described in detail (49). Briefly, the polarized Raman spectra were obtained with the 442-nm (32 mW) excitation from a HeCd laser (model IK 4601R-E; Kimmon Electric, Englewood, CO). The spectra were recorded with a RM 100 confocal Raman microscope (Renishaw, Hoffman Estates, IL) as described. The x-polarized spectrum was measured seven times, and the y-polarized spectrum was measured 14 times. All spectra were averaged for each polarization direction to eliminate some of the noise. The reference spectra were appropriately subtracted from the sample spectra. High-temperature Raman measurements were carried out with an LTS-350 temperature-controlled microscope slide holder from Linkam (Surrey, U.K.).
The FTIR and VCD spectra were recorded with a Chiral IR Fourier Transform VCD spectrometer from Bio Tools (Jupiter, FL). The sample was placed into a cell with a pathlength of 42.5 μm. The spectral resolution was 4 cm−1 for both spectra. The VCD and IR were both collected as one measurement for a combined total time of 720 min (648 min for VCD, 72 min for IR). To eliminate any background and solvent contributions to the IR spectrum, the cell was first filled with the reference solvent, i.e., acidified D2O and was automatically subtracted by the Chiral IR software. The IR and VCD spectra were also obtained at 65°C, using a temperature-controlled cell.
The temperature-dependent UV ECD spectra in the wavelength range of 190 to 240 nm of XAO were measured with a J-810 spectrapolarimeter (Jasco, Easton, MD) (58) in a 0.1-mm quartz cell with 0.05-nm resolution and a scan speed of 500 nm/min at the Drexel University Medical School. We performed the measurements in D2O rather than in H2O to allow a direct comparison with structural data obtained by vibrational spectroscopies. For each measurement, the sample was allowed to equilibrate for 5 min at the adjusted temperature before acquisition. The spectra were obtained by averaging 10 scans and collected as ellipticity as a function of wavelength and converted to molar absorptivities per residue as described (52).
Acknowledgments
We thank Prof. Adam Liwo (University of Gdansk, Gdansk, Poland) for providing detailed results of the MD calculations reported in ref. 30, Dr. Bojan Zagrovic for many stimulating discussions and his critical reading of the manuscript, and Prof. Vijay S. Pande for XAO peptide. The project was in part supported by National Science Foundation Grant MCB-0318749 (to R.S.S.).
Abbreviations
- PPII
polyproline II
- ECD
electronic circular dichroism
- VCD
vibrational circular dichroism
- MD
molecular dynamics
- SAXS
small angle x-ray scattering.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Shi Z, Chen K, Liu Z, Kallenbach NR. Chem Rev. 2006;106:1877–1897. doi: 10.1021/cr040433a. [DOI] [PubMed] [Google Scholar]
- 2.Eker F, Xao C, Nafie L, Schweitzer-Stenner R. J Am Chem Soc. 2002;124:14330–14341. doi: 10.1021/ja027381w. [DOI] [PubMed] [Google Scholar]
- 3.Jha AK, Colubri A, Zaman MH, Koide S, Sosnick TR, Freed KF. Biochemistry. 2005;44:9691–9702. doi: 10.1021/bi0474822. [DOI] [PubMed] [Google Scholar]
- 4.Tran HT, Wang X, Pappu RV. Biochemistry. 2005;44:11369–11380. doi: 10.1021/bi050196l. [DOI] [PubMed] [Google Scholar]
- 5.Woutersen S, Hamm P. J Phys Chem B. 2000;104:11316–11320. [Google Scholar]
- 6.Eker F, Griebenow K, Cao X, Nafie L, Schweitzer-Stenner R. Proc Natl Acad Sci USA. 2004;101:10054–10059. doi: 10.1073/pnas.0402623101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chellgren BW, Creamer TP. Biochemistry. 2004;43:5864–5869. doi: 10.1021/bi049922v. [DOI] [PubMed] [Google Scholar]
- 8.Shi Z, Chen K, Liu Z, Ng A, Bracken WC, Kallenbach NR. Proc Natl Acad Sci USA. 2005;102:17964–17968. doi: 10.1073/pnas.0507124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. J Mol Graphics Modelling. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 10.Creighton TE. Biophys Chem. 1988;31:155–162. doi: 10.1016/0301-4622(88)80021-8. [DOI] [PubMed] [Google Scholar]
- 11.Tiffany ML, Krimm S. Biopolymers. 1968;6:1767–1770. doi: 10.1002/bip.1968.360061212. [DOI] [PubMed] [Google Scholar]
- 12.Dukor R, Keiderling T. Biopolymers. 1991;31:1747–1761. doi: 10.1002/bip.360311409. [DOI] [PubMed] [Google Scholar]
- 13.Keiderling TA, Xu Q. Adv Protein Chem. 2002;62:111–161. doi: 10.1016/s0065-3233(02)62007-8. [DOI] [PubMed] [Google Scholar]
- 14.Cowan PM, McGavin S. Nature. 1955;176:501–503. doi: 10.1038/1761062a0. [DOI] [PubMed] [Google Scholar]
- 15.Gokce I, Woody RW, Anderluh G, Lakey JH. J Am Chem Soc. 2005;127:9700–9701. doi: 10.1021/ja052632x. [DOI] [PubMed] [Google Scholar]
- 16.Sreerama N, Woody RW. Protein Sci. 2003;12:384–388. doi: 10.1110/ps.0235003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Z, Woody RW, Kallenbach NR. Adv Protein Chem. 2002;62:163–240. doi: 10.1016/s0065-3233(02)62008-x. [DOI] [PubMed] [Google Scholar]
- 18.Woody RW. Adv Biophys Chem. 1992;2:37–79. [Google Scholar]
- 19.Chang JF, Phillips K, Lundback T, Gstaiger M, Ladbury JE, Luisi B. J Mol Biol. 1999;288:941–952. doi: 10.1006/jmbi.1999.2711. [DOI] [PubMed] [Google Scholar]
- 20.Blanch EW, Morozowa-Roche LA, Cochran DAE, Doig AJ, Hecht L, Barron LD. J Mol Biol. 2000;301:553–563. doi: 10.1006/jmbi.2000.3981. [DOI] [PubMed] [Google Scholar]
- 21.Syme CD, Blanch EW, Holt C, Ross J, Goedert M, Hecht L, Barron LD. Eur J Biochem. 2002;269:148–156. doi: 10.1046/j.0014-2956.2001.02633.x. [DOI] [PubMed] [Google Scholar]
- 22.Tanford C. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 23.Flory PJ. Statistical Mechanics of Chain Molecules. New York: Wiley; 1969. [Google Scholar]
- 24.Shi Z, Olson CA, Rose GA, Baldwin RL, Kallenbach NR. Proc Natl Acad Sci USA. 2002;99:9190–9195. doi: 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serrano L. J Mol Biol. 1995;254:322–333. doi: 10.1006/jmbi.1995.0619. [DOI] [PubMed] [Google Scholar]
- 26.Avbelj F, Baldwin RL. Proc Natl Acad Sci USA. 2003;100:5742–5747. doi: 10.1073/pnas.1031522100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiebig KM, Schwalbe H, Buck M, Smith LJ, Dobson CM. J Phys Chem. 1996;100:2661–2666. [Google Scholar]
- 28.Gnanakaran S, Garcia AE. Proteins. 2005;59:773–782. doi: 10.1002/prot.20439. [DOI] [PubMed] [Google Scholar]
- 29.Zagrovic B, Lipfert J, Sorin EJ, Millett IS, van Gunsteren WF, Doniach S, Pande VS. Proc Natl Acad Sci USA. 2005;102:11698–11703. doi: 10.1073/pnas.0409693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makowska J, Rodziewicz-Motowidlo S, Baginska K, Vila JA, Liwo A, Chmurzynski L, Scheraga HA. Proc Natl Acad Sci USA. 2006;103:1744–1749. doi: 10.1073/pnas.0510549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou PY, Fasman GD. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 32.Ramachandran GN, Ramakrishnan C, Sasisekharan V. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 33.Brant DA, Flory PJ. J Am Chem Soc. 1965;87:2791–2800. [Google Scholar]
- 34.Jun S, Becker JS, Yonkunas M, Coalson R, Saxena S. Biochemistry. 2006;45:11666–11673. doi: 10.1021/bi061195b. [DOI] [PubMed] [Google Scholar]
- 35.Tucker MJ, Oyola R, Gai F. J Phys Chem B. 2005;109:4788–4795. doi: 10.1021/jp044347q. [DOI] [PubMed] [Google Scholar]
- 36.Eker F, Griebenow K, Cao X, Nafie L, Schweitzer-Stenner R. Biochemistry. 2004;43:613–621. doi: 10.1021/bi035740+. [DOI] [PubMed] [Google Scholar]
- 37.Schweitzer-Stenner R, Eker F, Griebenow K, Cao X, Nafie L. J Am Chem Soc. 2004;126:2768–2776. doi: 10.1021/ja039452c. [DOI] [PubMed] [Google Scholar]
- 38.Marqusee S, Robbins VH, Baldwin RL. Proc Natl Acad Sci USA. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schweitzer-Stenner R. J Phys Chem B. 2004;108:16965–16975. [Google Scholar]
- 40.Ham S, Cho M. J Chem Phys. 2003;118:6915–6922. [Google Scholar]
- 41.Gorbunov RD, Kosov DS, Stock G. J Chem Phys. 2005;122:224904–224915. doi: 10.1063/1.1898215. [DOI] [PubMed] [Google Scholar]
- 42.Torii H, Tasumi M. J Raman Spectrosc. 1998;29:81–86. [Google Scholar]
- 43.Krimm S, Bandekar J. Adv Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 44.Long DA. The Raman Effect: A Unified Treatment of the Theory of Raman Scattering by Molecules. New York: Wiley; 2002. [Google Scholar]
- 45.Schweitzer-Stenner R. Vib Spec. 2006;42:98–117. [Google Scholar]
- 46.Eker F, Griebenow K, Schweitzer-Stenner R. Biochemistry. 2004;43:6893–6898. doi: 10.1021/bi049542+. [DOI] [PubMed] [Google Scholar]
- 47.Schweitzer-Stenner R, Measey T, Hagarman A, Eker F, Griebenow K. Biochemistry. 2006;45:2810–2819. doi: 10.1021/bi052282r. [DOI] [PubMed] [Google Scholar]
- 48.Measey T, Schweitzer-Stenner R. J Raman Spectrosc. 2006;37:248–254. [Google Scholar]
- 49.Schweitzer-Stenner R, Measey T, Kakalis L, Jordan F, Pizzanelli S, Forte C, Griebenow K. Biochemistry. 2007;46:1587–1596. doi: 10.1021/bi062224l. [DOI] [PubMed] [Google Scholar]
- 50.Measey T, Hagarman A, Eker F, Griebenow K, Schweitzer-Stenner R. J Phys Chem B. 2005;109:8195–8205. doi: 10.1021/jp045762l. [DOI] [PubMed] [Google Scholar]
- 51.Vila JA, Baldoni HA, Ripoli DR, Gosh A, Scheraga H. Biophys J. 2004;86:731–742. doi: 10.1016/S0006-3495(04)74151-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamm P, Manho L, Hochstrasser RM. J Phys Chem B. 1998;102:6123–6138. [Google Scholar]
- 53.Choi J, Cho M. J Chem Phys. 2004;120:4383–4392. doi: 10.1063/1.1644100. [DOI] [PubMed] [Google Scholar]
- 54.Lednev IK, Karnoup AS, Sparrow MC, Asher AS. J Am Chem Soc. 1999;121:8074–8086. doi: 10.1021/ja003381p. [DOI] [PubMed] [Google Scholar]
- 55.Gnanakaran S, Garcia AE. J Phys Chem B. 2003;107:12555–12557. [Google Scholar]
- 56.Measey T, Schweitzer-Stenner R. Chem Phys Lett. 2005;408:123–127. [Google Scholar]
- 57.Wright PE, Dyson HJ, Lerner RA. Biochemistry. 1988;27:7167–7175. doi: 10.1021/bi00419a001. [DOI] [PubMed] [Google Scholar]
- 58.Hagarman A, Measey T, Doddasomayajula R, Dragomir I, Eker F, Griebenow K, Schweitzer-Stenner R. J Phys Chem B. 2006;110:6979–6986. doi: 10.1021/jp0561625. [DOI] [PubMed] [Google Scholar]