Abstract
Low-temperature (1.4 K), single-molecule fluorescence-excitation spectra have been recorded for individual reaction center–light-harvesting 1 complexes from Rhodopseudomonas palustris and the PufX− strain of Rhodobacter sphaeroides. More than 80% of the complexes from Rb. sphaeroides show only broad absorption bands, whereas nearly all of the complexes from Rps. palustris also have a narrow line at the low-energy end of their spectrum. We describe how the presence of this narrow feature indicates the presence of a gap in the electronic structure of the light-harvesting 1 complex from Rps. palustris, which provides strong support for the physical gap that was previously modeled in its x-ray crystal structure.
Keywords: bacterial photosynthesis, light-harvesting complex, single-molecule spectroscopy
The primary reactions of purple bacterial photosynthesis take place within two well characterized pigment–protein complexes (1), the core reaction center (RC)-light-harvesting 1 (LH1) complex and the more peripheral light-harvesting 2 (LH2) complexes. Typically, light energy is absorbed by LH2 and is then transferred via LH1 to the RC. Here, it is used to drive a series of electron transfer reactions that result in the reduction of ubiquinone (UQ) (2). In the photosynthetic membrane, when UQ in the RC has been fully reduced to ubiquinol (UQH2), the quinol must leave the RC to transfer its reducing equivalents to the cytochrome b/c1 complex, as part of a rather simple cyclic electron transport pathway (3). Initial structural models of the RC–LH1 complex pictured the RC completely surrounded by a closed LH1 ring (see e.g., refs. 4 and 5). The basic structural unit of LH1 is an αβ-heterodimer, which binds two molecules of bacteriochlorophyll a (BChl a) and one or two molecules of carotenoid (1). These dimers then oligomerize around the RC to form the closed ring. The pairs of BChl a molecules from each dimer interact together to form a strongly coupled ring of pigments giving rise to the strong Qy absorption band in the 870- to 890-nm region. The spectroscopic properties of this ring reflect the strong excitonic coupling among these BChl a molecules (6). Based on these models an obvious question arises. How does UQH2 escape from such a core complex? There are two possible solutions: either the LH1 ring is not complete, i.e., there is a gap, or the LH1 structure is inherently flexible enough to allow UQH2 to diffuse through it (7–9).
It has been suggested that a protein found in both Rhodobacter sphaeroides and Rhodobacter capsulatus called PufX may provide an answer to this question (10). PufX appears to be a member of the LH1 ring, replacing one of the αβ dimers. This then introduces a gap through which it has been proposed that the UQH2 could pass. The real situation in Rb. sphaeroides, however, is more complicated. In WT membranes (PufX+) these core complexes are actually dimers (11). Deletion of PufX causes the core complexes to become monomeric and complete rings (12). Recently, a low-resolution (4.8 Å), x-ray crystal structure of the monomeric core complex from Rhodopseudomonas palustris has been described (13). Even though at this relatively low resolution the structure must be considered somewhat tentative, its main features seem quite clear. An elliptical LH1 complex surrounds the RC and adjacent to the RC UQ binding site, from where UQH2 must leave, there is a gap in the LH1 ring. This gap is associated with a protein called W, which replaces an LH1 αβ dimer and is believed to be an orthologue of PufX. The presence or absence of such a gap and its functional significance has become rather controversial (14, 15). The availability of monomeric core complexes with and without a physical gap in the LH1 ring provides an opportunity for an elegant use of low-temperature, single-molecule spectroscopy. Single-molecule spectroscopy can be used, as we describe in Fig. 1, to look for the spectroscopic signature of a gap in the electronic structure of LH1 BChl a molecules and thereby seek to test the crystallographic structure model.
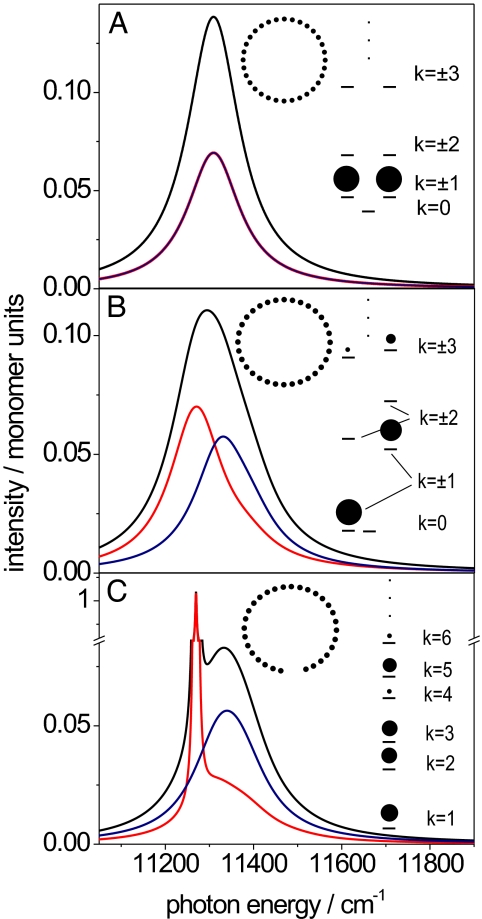
Fig. 1.
Comparison of different models for the geometrical arrangement of the BChl a molecules within an RC–LH1 complex and its influence on their resulting absorption spectra. The exciton energies are calculated in Heitler–London approximation (for details see ref. 16) taking only nearest-neighbor interactions into account. The BChl a molecules are represented by the dots, which are arranged as 32 molecules in a circle (A), 32 molecules in an ellipse (ellipticity 4%) (B), and 30 molecules in the same ellipse as in B but leaving a gap as indicated in the inserted structure (C). (Insets) The three modeled geometries, and the resulting energies of the exciton states, are shown. According to common practice, the exciton states are labeled for reasons of symmetry k = 0, ±1, ±15, k = 16 for the closed arrangements (A and B), and k = 1, 2, … 30 for the interrupted structure (C). The area of the spheres indicates the oscillator strength that is associated with the respective exciton state. For each model, two spectra with mutually orthogonal polarization (red and blue line) and the sum spectrum (black line) are displayed. The spectra have been calculated by using 11,850 cm−1 for the pigment site energies and 250 cm−1 for the interaction strengths. In each case for the corresponding spectra, the ultrafast relaxation between the higher exciton states is taken into account by dressing those transitions with a linewidth of 170 cm−1, whereas for the lowest exciton state, which decays on a slower time scale, a linewidth of 5 cm−1 was used. The vertical axis is given in units of monomer dipole strength, and the individual transitions have been normalized such that their area corresponds to the oscillator strength of the respective transition.
To illustrate the influence of symmetry (or indeed the lack of it) on the properties of the exciton states of the ring of BChl a molecules in RC–LH1 we have calculated the excited-state manifold for three different geometries: (i) a circular symmetric assembly of 32 BChl a molecules, (ii) an elliptical assembly of 32 BChl a molecules, and (iii) an overall elliptical assembly of 30 BChl a molecules that features a gap (as in the Rps. palustris structure) (Fig. 1). These calculations have been carried out as described (16). For the circular symmetric arrangement one obtains two nondegenerate and 15 pairwise degenerate exciton states (Fig. 1A). According to common practice we label the nondegenerate exciton states by the quantum numbers k = 0 and k = 16 and refer to the degenerate exciton states as k = ±1, ±2, ±15. Because of the high symmetry of the circular ring arrangement the oscillator strength is concentrated in the lowest degenerate pair of the exciton states, i.e., k = ±1. Introducing an elliptic distortion is equivalent to a reduction of the symmetry from circular to C2. The dominant effect of such a perturbation is that the pairwise degeneracies of the exciton states will be lifted and that oscillator strength from the k = ±1 states is redistributed to the k = ±3 states, leading to several spectral bands in the absorption spectrum. Because the relaxation of the higher exciton states occurs on an ultrafast time scale of ≈100 fs (17) the absorption spectrum consists of either one or a few relatively broad spectral bands (Fig. 1 A and B, respectively). For both cases, i.e., circular and elliptical arrangement, the transitions from the k = ±1 exciton states are polarized perpendicular with respect to each other. Moreover, the lowest exciton state is optically forbidden, because a C2-type symmetry reduction alone does not give rise to oscillator strength in the k = 0 state. Very occasionally oscillator strength in the k = 0 state is introduced by random perturbations (16, 18), which have been omitted here for clarity. This low-probability event can be seen with some LH2 complexes from Rhodopseudomonas acidophila, which contain a complete nonameric circle of pairs of strongly interacting BChl a molecules (19, 20). Striking consequences for the absorption spectra are expected by introducing a symmetry-breaking gap in the BChl a assembly. The presence of such a gap in the LH1 ring is equivalent to the case of a linear excitonic system where because of the loss of symmetry the resulting exciton states are commonly referred to as k = 1, 2, … As has been well described for linear excitonic systems, such as J-aggregates (21), now the lowest exciton state, i.e., k = 1 in this case, gains considerable oscillator strength. Because the lifetime of this state is longer than a few hundred ps‖, the associated optical transition will appear as a relatively narrow feature in the low-energy wing of the absorption spectrum (see Fig. 1C). In contrast to the situation with LH2, in this case there is the expectation that this narrow feature will be present in most of the measured spectra from individual complexes.
Results and Discussion
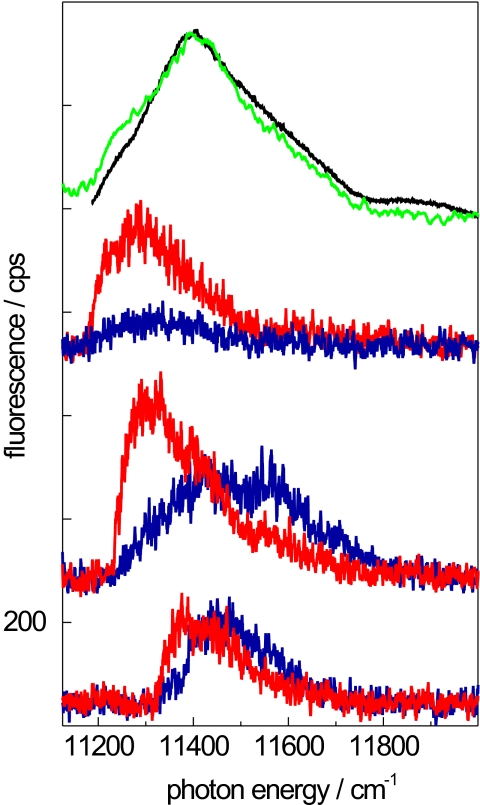
In the present study, we have compared individual core complexes from Rps. palustris (with an expected gap) and Rb. sphaeroides PufX−(closed structure) by using single-molecule spectroscopy. In Figs. 2 and 3 we compare several fluorescence-excitation spectra from RC–LH1 complexes from Rb. sphaeroides PufX− and Rps. palustris. In both figures the top traces show an ensemble spectrum (black line) together with the sum spectrum from 44 (or 41 for Rps. palustris) individual complexes (green line) from Rb. sphaeroides PufX−. In both cases the essential features of the ensemble spectra are reproduced by the sum spectra. For Rb. sphaeroides PufX− (Fig. 2), the ensemble spectrum shows a broad band centered at 11,409 cm−1 with a width of 329 cm−1. Fig. 2 Lower displays for each of three individual RC–LH1 complexes two fluorescence-excitation spectra that have been recorded with mutually orthogonal polarization of the excitation light. These spectra feature a few broad bands, which vary in width between 160 and 188 cm−1 (FWHM). In total we studied 44 individual RC–LH1 complexes from Rb. sphaeroides PufX−, and in only seven complexes were we able to observe a narrow spectral feature in the red wing that could be associated with the k = 0 transition. A similar low frequency of narrow lines was observed for individual LH2 complexes from Rps. acidophila, where a high-resolution x-ray crystal structure has shown that the complex contains a closed ring of BChl a molecules (20).
Fig. 2.
Fluorescence-excitation spectra of RC–LH1 complexes from Rb. sphaeroides PufX−. The top traces show the comparison between an ensemble spectrum (black line) and the sum of 44 spectra recorded from individual complexes (green line). The lower traces show spectra from single RC–LH1 complexes. For each individual complex two spectra, recorded with mutually orthogonal polarization of the excitation light, are displayed. All spectra were measured at 1.4 K at 10 W/cm2. The vertical axis is valid for the lowest traces, all other traces were offset for clarity. Supporting information (SI) Movie 1 shows a sequence of fluorescence-excitation spectra as a function of the polarization of the excitation light.
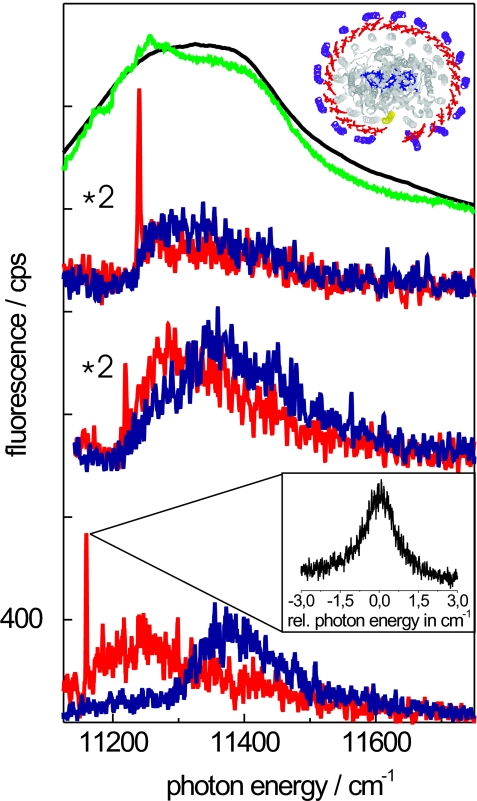
Fig. 3.
Fluorescence-excitation spectra of RC–LH1 complexes from Rps. palustris. The top traces show the comparison between an ensemble spectrum (black line) and the sum of 41 spectra recorded from individual complexes (green line). The lower traces show spectra from single RC–LH1 complexes. For each individual complex two spectra, recorded with mutually orthogonal polarization of the excitation light, are displayed. All spectra were measured at 1.4 K at 10 W/cm2. The vertical axis is valid for the lowest traces. The other traces from individual complexes were multiplied by a factor of two and offset for clarity. (Upper Inset) The x-ray crystal structure of Rps. palustris (13). (Lower Inset) The narrow spectral feature of the lowest spectrum on an expanded scale in relative energies. SI Movie 2 shows a sequence of fluorescence-excitation spectra as a function of the polarization of the excitation light.
In the case of the RC–LH1 complexes from Rps. palustris (Fig. 3), the ensemble spectrum shows a broad band centered at 11,322 cm−1 with a width of 378 cm−1 (FWHM) and two weak shoulders at 11,287 and 11,373 cm−1. Remarkable spectral details become visible upon removing ensemble averaging by recording spectra from individual complexes, as displayed in Fig. 3 Lower. The spectra from the individual RC–LH1 complexes show variations with respect to the spectral position of the bands, the width of the bands, and the mutual polarization of the bands. For almost all RC–LH1 complexes that could be studied (33 complexes corresponding to 80%) the fluorescence-excitation spectra feature a narrow spectral line at the low-energy side in addition to several broad bands at higher energy, similar to those seen in Fig. 2 with complexes from Rb. sphaeroides PufX−. For a few of these complexes the spectra (eight complexes corresponding to 20%) appeared to be red-shifted to such an extent that it prevented the recording of the full spectrum because the required spectral scan range of the excitation overlapped with the transmission window of the detection filter. In these cases, of course, the narrow spectral line could not be detected.
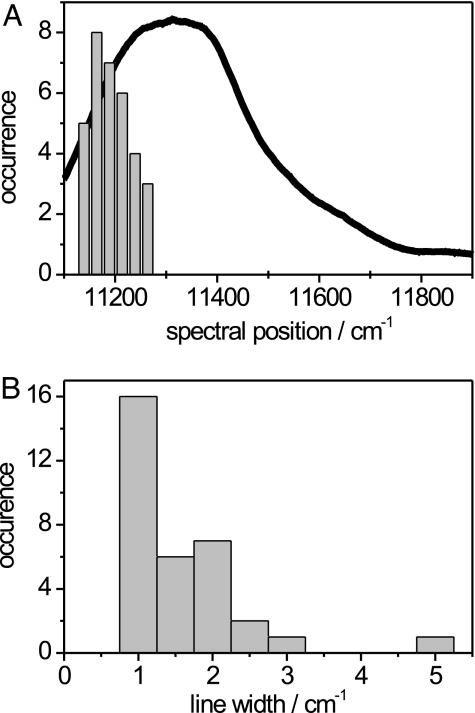
For the spectra from individual pigment–protein complexes to be compared, the incident polarization, for which the narrow feature yields the maximum intensity, was set arbitrarily to “horizontal” and provides the reference point. For the studied individual RC–LH1 complexes from Rps. palustris the distribution of the spectral positions of the narrow feature is shown in Fig. 4A together with an ensemble absorption spectrum. The histogram is centered at 11,195 cm−1 in the red wing of the ensemble spectrum and covers a spectral range of ≈125 cm−1, which illustrates the spectral heterogeneity (intercomplex disorder) of the ensemble of RC–LH1 complexes. These findings are consistent with hole-burning action spectra (22). The spectral width of the narrow absorption showed a distribution as well (Fig. 4B), which ranges from 1 to 5 cm−1 where most of the entries cover the range between 1 and 2 cm−1. This width would correspond to a lifetime of ≈33 ps, which is significantly shorter than the actual lifetime, indicating that these linewidths were determined by spectral diffusion rather than the lifetime limited value. This hypothesis has been verified by using a single-mode laser of <1-MHz spectral bandwidth that was scanned across the narrow feature at various scan speeds. It turned out that the observed width of the narrow feature increased for slower scan speeds, indicating spectral diffusion. The typical widths of the broad bands are similar to those observed for the Rb. sphaeroides PufX− complexes.
Fig. 4.
Statistics of the position and linewidths of the narrow spectral feature. (A) Distribution of the spectral positions of the narrow feature for 33 individual RC–LH1 complexes from Rps. palustris. The bin width is 25 cm−1, and the bold line corresponds to the ensemble absorption spectrum, which has been overlaid for illustration. (B) Distribution of the spectral bandwidth of the narrow feature for 33 individual RC–LH1 complexes from Rps. palustris. The bin width is 0.5 cm−1.
In principle, any deviation from circular symmetry will lift the degeneracies of the exciton states and redistribute the oscillator strength from the initially degenerate lowest pair of exciton states to adjacent states. However, in practice, random and elliptical deviations from circular symmetry can be distinguished (23) because random (energetic or structural) disorder will shift oscillator strength into the k = 0 circular exciton state as well, whereas a C2-type symmetry reduction, for example in an ellipse, will redistribute oscillator strength exclusively among the circular exciton states that differ in the quantum number k by Δk = 2, i.e., the k = 0 state is not coupled to the k = ±1 states. Based on these arguments we conclude that the RC–LH1 for Rb. sphaeroides PufX− features an overall elliptical shape, compatible with the model shown in Fig. 1B. For a few complexes, the presence of random disorder, which is unavoidable in such systems, leads to the observation of the k = 0 transition as well. Because the occurrence of k = 0 has also been observed previously with a similar probability for the circular LH2 complexes we consider effects like a spontaneous ring opening of the LH1–RC complexes or the presence of incomplete rings (24) as a less likely cause for the presence of the narrow spectral feature, even though we cannot completely dismiss them.
For the RC–LH1 complex from Rps. palustris we observe this narrow line for all complexes where we were able to record a full spectrum. Because, as in the case of the PufX− core complex from Rb. sphaeroides, it is also unlikely that the spectra from the Rps. palustris complex are dominated by random disorder, then how can we explain the presence of the narrow line at the red end of all of the fluorescence-excitation spectra? We propose that the ubiquitous presence of this narrow line can be explained by a gap in the electronic structure so that significant oscillator strength is shifted into the lowest exciton state. This important result is consistent with the model shown in Fig. 1C and in agreement with the x-ray crystal structure as shown in Fig. 3 Upper Inset (13).
This work demonstrates that low-temperature, single-molecule spectroscopy is a powerful tool for revealing the effects of symmetry on the electronic structure of pigment–protein complexes. The interplay of the geometrical arrangement of the pigments and the transition probabilities of the various exciton states leads to key spectral features, such as narrow lines, that are clearly visible with single-molecule spectroscopy but are averaged out in conventional ensemble experiments. Such experiments provide valuable information for the theoretical modeling of energy-transfer processes within these systems and for a better understanding of their structure and function.
Methods
The RC–LH1 complexes from Rps. palustris and Rb. sphaeroides PufX− were isolated and purified as described (13, 25) and prepared for single-molecule spectroscopy as described (26). The samples were mounted in a cryostat, cooled to 1.4 K, and illuminated with a continuous wave Ti:Sapphire laser (spectral bandwidth 0.7 cm−1) through a home-built microscope. To obtain a well defined variation of the wavelength of the laser the intracavity birefringent filter was rotated with a motorized micrometer that ensured an accuracy and a reproducibility of 0.5 cm−1 for the laser frequency. The microscope could be operated either in wide-field mode in combination with a CCD camera for imaging purposes or in confocal mode in combination with an avalanche photodiode for spectroscopy. In both cases the detection bandwidth was 40 nm. The polarization of the incident radiation was controlled with a λ/2 plate in the excitation path that could be rotated in steps of 0.8°. For the experiments at enhanced spectral resolution a Ti:Sapphire single-mode laser (spectral bandwidth 0.5 MHz) was used. More experimental details can be found in ref. 27.
Supplementary Material
Acknowledgments
G.V. was supported by Ministero dell'Università e della Ricerca Grant PRIN2005 prot.2005027011. R.J.C. was supported by the Biotechnology and Biological Sciences Research Council.
Abbreviations
- BChl
bacteriochlorophyll
- LH1
light harvesting 1
- LH2
light harvesting 2
- RC
reaction center
- UQ
ubiquinone
- UQH2
ubiquinol.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611115104/DC1.
We have determined the room-temperature fluorescence lifetime of RC–LH1 complexes from Rps. palustris to be 200–300 ps (unpublished results).
References
- 1.Zuber H, Cogdell RJ. In: Anoxygenic Photosynthetic Bacteria. Blankenship RE, Madigan MT, Bauer CE, editors. Dordrecht, The Netherlands: Kluwer; 1995. pp. 315–348. [Google Scholar]
- 2.Feher G, Allen JP, Okamura MY, Rees DC. Nature. 1989;339:111–116. [Google Scholar]
- 3.Petty K, Jackson JB, Dutton PL. Biochim Biophys Acta. 1979;546:17–42. doi: 10.1016/0005-2728(79)90167-1. [DOI] [PubMed] [Google Scholar]
- 4.Karrasch S, Bullough PA, Ghosh R. EMBO J. 1995;14:631–638. doi: 10.1002/j.1460-2075.1995.tb07041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cogdell RJ, Gall A, Köhler J. Q Rev Biophys. 2006;39:227–324. doi: 10.1017/S0033583506004434. [DOI] [PubMed] [Google Scholar]
- 6.Robert B, Cogdell RJ, van Grondelle R. In: Light-Harvesting Antennas in Photosynthesis. Green BR, Parson WW, editors. Dordrecht, The Netherlands: Kluwer; 2003. pp. 169–194. [Google Scholar]
- 7.Cogdell RJ, Fyfe PK, Barrett SJ, Prince SM, Freer AA, Isaacs NW, McGlynn P, Hunter CN. Photosynth Res. 1996;48:55–63. doi: 10.1007/BF00040996. [DOI] [PubMed] [Google Scholar]
- 8.Fotiadis D, Qian P, Philippsen A, Bullough PA, Engel A, Hunter CN. J Biol Chem. 2004;279:2063–2068. doi: 10.1074/jbc.M310382200. [DOI] [PubMed] [Google Scholar]
- 9.Aird A, Wrachtrup J, Schulten K, Tietz C. Biophys J. 2007;92:23–33. doi: 10.1529/biophysj.106.084715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barz WP, Vermeglio A, Francia F, Venturoli G, Melandri BA, Oesterhelt D. Biochemistry. 1995;34:15248–15258. doi: 10.1021/bi00046a033. [DOI] [PubMed] [Google Scholar]
- 11.Bahatyrova S, Frese RN, Siebert CA, Olsen JD, van der Werf KO, van Grondelle R, Niederman RA, Bullough PA, Otto C, Hunter CN, et al. Nature. 2004;430:1058–1062. doi: 10.1038/nature02823. [DOI] [PubMed] [Google Scholar]
- 12.Francia F, Wang J, Venturoli G, Melandri BA, Barz WP, Oesterhelt D. Biochemistry. 1999;38:6834–6845. doi: 10.1021/bi982891h. [DOI] [PubMed] [Google Scholar]
- 13.Roszak AW, Howard TD, Southall J, Gardiner AT, Law CJ, Isaacs NW, Cogdell RJ. Science. 2003;302:1969–1971. doi: 10.1126/science.1088892. [DOI] [PubMed] [Google Scholar]
- 14.Scheuring S, Goncalves RP, Prima V, Sturgis JN. J Mol Biol. 2006;358:83–96. doi: 10.1016/j.jmb.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 15.Qian P, Hunter CN, Bullough PA. J Mol Biol. 2005;349:948–960. doi: 10.1016/j.jmb.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 16.Matsushita M, Ketelaars M, van Oijen AM, Köhler J, Aartsma TJ, Schmidt J. Biophys J. 2001;80:1604–1614. doi: 10.1016/S0006-3495(01)76133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundström V, Pullerits T, van Grondelle R. J Phys Chem B. 1999;103:2327–2346. [Google Scholar]
- 18.Sener MK, Schulten K. Phys Rev E. 2002;65:031916. doi: 10.1103/PhysRevE.65.031916. [DOI] [PubMed] [Google Scholar]
- 19.van Oijen AM, Ketelaars M, Köhler J, Aartsma TJ, Schmidt J. Science. 1999;285:400–402. doi: 10.1126/science.285.5426.400. [DOI] [PubMed] [Google Scholar]
- 20.Ketelaars M, van Oijen AM, Matsushita M, Köhler J, Schmidt J, Aartsma TJ. Biophys J. 2001;80:1591–1603. doi: 10.1016/S0006-3495(01)76132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knoester J, Agranovich VM. In: Thin Films and Nanostructures. Agranovich VM, Bassani GF, editors. San Diego: Elsevier; 2003. pp. 1–96. [Google Scholar]
- 22.Wu H-M, Rätsep M, Jankowiak R, Cogdell RJ, Small GJ. J Phys Chem B. 1998;102:4023–4034. [Google Scholar]
- 23.Hu X, Ritz T, Damjanovic A, Schulten K. J Phys Chem B. 1997;101:3854–3871. [Google Scholar]
- 24.Pugh RJ, McGlynn P, Jones MR, Hunter CN. Biochim Biophys Acta. 1998;1366:301–316. doi: 10.1016/s0005-2728(98)00131-5. [DOI] [PubMed] [Google Scholar]
- 25.Francia F, Dezi M, Rebecchi A, Mallardi A, Palazzo G, Melandri BA, Venturoli G. Biochemistry. 2004;43:14199–14210. doi: 10.1021/bi048629s. [DOI] [PubMed] [Google Scholar]
- 26.van Oijen AM, Ketelaars M, Köhler J, Aartsma TJ, Schmidt J. Chem Phys. 1999;247:53–60. [Google Scholar]
- 27.Lang E, Baier J, Köhler J. J Microsc (Oxford) 2006;222:118–123. doi: 10.1111/j.1365-2818.2006.01579.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.