Abstract
Chromophore-assisted laser inactivation (CALI) is a light-mediated technique that offers precise spatiotemporal control of protein inactivation, enabling better understanding of the protein's role in cell function. EGFP has been used effectively as a CALI chromophore, and its cotranslational attachment to the target protein avoids having to use exogenously added labeling reagents. A potential drawback to EGFP-CALI is that the CALI phenotype can be obscured by the endogenous, unlabeled protein that is not susceptible to light inactivation. Performing EGFP-CALI experiments in deficient cells rescued with functional EGFP-fusion proteins permits more complete loss of function to be achieved. Here, we present a modified lentiviral system for rapid and efficient generation of knockdown cell lines complemented with physiological levels of EGFP-fusion proteins. We demonstrate that CALI of EGFP-CapZβ increases uncapped actin filaments, resulting in enhanced filament growth and the formation of numerous protrusive structures. We show that these effects are completely dependent upon knocking down the endogenous protein. We also demonstrate that CALI of EGFP-Mena in Mena/VASP-deficient cells stabilizes lamellipodial protrusions.
Keywords: capping protein, Mena/VASP, spatiotemporal, lentivirus
Analysis of loss-of-function phenotypes often leads to critical insights about protein function. However, long-term global protein inactivation via gene knockout or RNAi can lead to compensatory changes at the molecular level that are difficult to detect or control. Chromophore-assisted laser inactivation (CALI) circumvents these issues through instantaneous and spatially precise protein inactivation (1). With CALI, target proteins are inactivated by reactive photoproducts such as reactive oxygen species (ROS) that are generated by intense irradiation of photosensitizer chromophores (2). The short half-life of these ROS ensures that only proteins immediately adjacent to the chromophore are affected (3). CALI has been used to inactivate varied protein targets, but some skepticism remains about the specificity of this approach and its general applicability.
CALI specificity is achieved by selectively labeling the target protein with a chromophore. This targeting was originally done by microinjecting non-function-blocking antibodies labeled with malachite green or fluorescein into cells (4), a methodology that has proven to be technically difficult and has been used by only a limited number of laboratories. More recently, CALI strategies have been developed that use membrane-permeable chromophore components that recognize genetically encoded tags. These reagents include the biarsenical dyes FlAsH (5, 6) and ReAsh (7), which specifically recognize 12-aa tetracystine motifs, and a fluorescein-conjugated ligand (SLF′) whose intracellular target protein is FKBP12(F36V) (8). These membrane-permeable labeling strategies have increased the potential utility of CALI but still suffer from having to use exogenously added reagents, which introduces the risk of nonspecific labeling and collateral photodamage during irradiation (7, 9).
A third CALI approach is to use fluorescent proteins as photosensitizers. CALI using fluorescent protein fusions offers the unique advantage of having a CALI system in which all of the components are genetically encoded, thereby avoiding having to label the protein of interest with exogenous reagents. Cotranslational attachment of the fluorescent protein to its target provides the highest possible degree of specificity. EGFP is a widely used fluorescent protein that has been previously shown to be an effective CALI chromophore with both single photon (10) and multiphoton excitation (11). Whereas EGFP has many advantages for studies of protein localization, the drawback to using this fluorescent protein as a CALI chromophore is that it is a relatively poor photosensitizer. This deficit must be compensated for by using irradiation doses that are substantially higher than what is needed for inactivation by fluorescein, FlAsH, or ReAsH (12). Any changes in the experimental procedure of EGFP-CALI that improves its efficacy would allow more researchers to use this technique to locally and acutely inactivate proteins using CALI although retaining the benefits of EGFP as a localization tool.
One such improvement would be to perform CALI experiments in cells where the only functional version of the protein is EGFP-labeled. The presence of endogenous, unlabeled protein that is resistant to light inactivation has the potential to compensate for the loss of EGFP-tagged proteins, and can therefore mask the CALI effect. To address this problem one can use cells that are deficient for the endogenous protein and rescued with the EGFP-fusion complement. To facilitate production of complemented cell lines, we present a previously undescribed system where knockdown of endogenous protein and rescue with an EGFP-fusion protein can be accomplished in a single step. We demonstrate the CALI of EGFP-capping protein (CP) in rescued deficient cells and show that knockdown of the endogenous protein is essential for CP loss of function after laser irradiation. As a second example of this approach, we also demonstrate the CALI of EGFP-Mena in rescued fibroblasts derived from Mena−/−VASP−/− mice.
Capping protein and Mena both localize to the barbed ends of actin filaments where they either negatively or positively regulate filament elongation; antagonism between the two factors governs the supramolecular organization of actin filaments in motile cells and has a profound effect on motility and migration (13–20). With EGFP-CALI, we recapitulate the loss-of-function phenotypes of Capping protein and Mena reported previously: CALI of capping protein creates a local increase in barbed ends and F-actin resulting in numerous protrusive structures (21), whereas CALI of Mena induces larger and more stable lamellipodial protrusions (17). CALI of CP and Mena cause distinct changes in local subcellular structure despite the overlapping localization of both proteins at the leading edge. In both cases, the phenotypic changes that occur upon CALI are positive (e.g., the generation of a new structure), strongly suggesting that CALI is leading to highly specific protein inactivation under these conditions.
Results
A Lentivirus System for Single-Step Generation of Deficient Cells Complemented with EGFP-Fusion Proteins.
To rapidly generate knockdown cells rescued with a functional EGFP-fusion protein, we modified a lentiviral vector to concomitantly express an short hairpin RNA (shRNA) from the PolIII U6 promoter and a functional EGFP-fusion protein from the 5′ LTR promoter of the murine stem cell virus (MSCV) (Fig. 1A). The LTR promoter drives modest expression levels (15), a detail that is important if a physiological level of the rescue protein is desired. This simultaneous knockdown/rescue system requires the complementing EGFP-fusion protein to be resistant to the RNAi because of mismatches in the RNAi target sequence. This system allows the production of deficient cells rescued with EGFP-fusions without having to generate a knockout mouse or having to perform multiple transfections with siRNA oligonucleotides and/or expression vectors. An additional advantage of simultaneous knockdown/rescue is that knockdown occurs in the presence of a functional copy of the knocked down gene, reducing the risk of coordinate regulatory and compensatory changes as the endogenous protein is lost.
Fig. 1.
A lentiviral system for simultaneous knockdown and rescue with a functional EGFP-fusion complement. (A) Schematic of the lentiviral expression vector used for simultaneous knockdown of rat CPβ (rCPβ) and rescue with EGFP-human CPβ (EGFP-hCPβ). Open triangles, LoxP sites. (B) DIC images of a WT Rat2 fibroblast (Top) and Rat2 fibroblasts infected with only the knockdown portion of the vector (Middle) or the complete knockdown/rescue construct (Bottom). (Scale bar: 20 μm.) (C) Western blot of WT Rat2 fibroblasts (left lane) and a clonally derived Rat2 cell line infected with the knockdown/rescue vector.
Capping protein (CapZ, abbreviated CP throughout) is an obligate heterodimer of alpha and beta subunits and is the predominant actin filament capping protein in most cells (22). To create a knockdown/rescue vector for CP, we used an shRNA that specifically targets rat CapZβ and an RNAi-resistant EGFP-human CapZβ (hereafter referred to as EGFP-CP) fusion. The phenotypic effects of CP knockdown using this shRNA sequence have been previously characterized and serve as a useful point of comparison for our studies (21). The loss of CP has dramatic effects on the actin cytoskeleton, most notably a substantial increase in filopodia. We confirmed this phenotype by infecting Rat2 fibroblasts with a version of the lentiviral vector that expresses only the knockdown shRNA, but not the rescue component (Fig. 1B Middle). In addition to increased filopodia, we also observed a large increase of ruffling in these cells (data not shown). Both of these effects are consistent with loss of CP activity and the subsequent increase in uncapped and rapidly growing actin filaments.
We infected Rat2 fibroblasts with the CP knockdown/rescue lentivirus and identified a clonal derivative with no detectable expression of endogenous CPβ that was complemented with a physiological level of EGFP-CP (Fig. 1C). This knockdown/rescue cell line was phenotypically indistinguishable from the control Rat2 cells (Fig. 1B Top and Bottom), indicating that the EGFP-CP functionally complemented the shRNA-induced phenotype. This result is consistent with data showing that Xenopus CPβ tagged with EGFP at the same terminus is functional and has similar on-rate and dissociation constants to actin barbed ends as unlabeled CP (23).
CALI of EGFP-CP Acutely Induces the Formation of Dorsal Protrusions in Deficient Cells, but Not in Cells Retaining Endogenous CP Expression.
We performed CALI of EGFP-CP in both knockdown/rescue (KDR) cells and cells which expressed EGFP-CP without knockdown of the endogenous gene. In both cell lines, EGFP-CP was expressed a comparable levels (data not shown). Cells were irradiated with light from the 488 nm line of an argon ion laser focused on the specimen plane with a beam diameter of ≈23 μm (≈1/4–1/6 of total cell area; Fig. 2A, white circle). After administration of a 100-ms dose of 1.5 mW/μm2 laser light, KDR cells exhibited dynamic changes on the dorsal surface of the cell, forming numerous protrusions [Fig. 2 A and B, supporting information (SI) Movie 1, and SI Movie 2]. However, in contrast to the filopodia phenotype that is seen with long-term CP depletion (Fig. 1B Middle) many of these structures most strongly resembled intrapodia, actin-based protrusions that occur on the dorsal surface of migrating growth cones (24, 25). The difference may represent the effects of long-term vs. instantaneous loss of CP. The formation and curvilinear extension of these protrusions was exclusive to the area that was irradiated (Fig. 2A and SI Movie 1). When these cells were irradiated with a smaller beam size (5 μm; 6.1 mW/μm2), no change in the dorsal surface was apparent (data not shown), despite the increase in photon flux. These data are qualitatively consistent with the rapid fluorescence recovery after photobleaching kinetics for EGFP-CP (Table 1: note that diffusional recovery would scale as bleached region diameter squared and substantial recovery would therefore occur only after ≈150 s, a time in agreement with when the phenotype begins to reverse as seen in Fig. 2D) and suggest that the CP CALI effects require inactivating a large enough fraction of the EGFP-CP pool to avoid rapid compensation from intact, nonirradiated EGFP-CP diffusing in from surrounding areas.
Fig. 2.
CALI of EGFP-CP induces the formation of dorsal ruffles and filopodia only in the knockdown/rescue background. (A) DIC images of Rat2 EGFP-CP knockdown/rescue (KDR) cell line before and 3 min after CALI of EGFP-CP (Left) and fluorescent images of EGFP-CP before and immediately after CALI (Right); area of irradiation is indicated with a black circle. (Scale bar: 20 μm.) (B) Time-lapse images of the irradiated regions of EGFP-CP KDR cells (Upper) and cells expressing EGFP-CP without knockdown of endogenous CP (Lower; EGFP-CP no KD) before and after CALI. (Scale bar: 10 μm.) (C) DIC images (Left) and Sobel filtered binary images (Right; for details see Methods) before (Upper) and 2 min after (Lower) CALI. The white circle represents the 12-μm-diameter center of the irradiated area that was used to measure the local edge detection index. Raw edge index values are shown next to their respective images. (D) Plot showing the normalized edge detection values in the irradiated region over time for EGFP-CP KDR cells (n = 20), EGFP-CP no KD cells (n = 8), and cells expressing EGFP (n = 12); the break in each line represents when the movies were paused to perform the laser irradiation. Error bars are SEM.
Table 1.
Cytoplasmic mobility of capping protein and Mena
| Protein | Cell line | Percent recovery | t1/2, s | n |
|---|---|---|---|---|
| EGFP-CP | Rat2 EGFP-CP KDR | 69.2 ± 1.9 | 6.6 ± 0.4 | 14 |
| EGFP-Mena | MVD7 | 50.3 ± 5.6 | 17.4 ± 3.9 | 10 |
Values are reported ± SEM.
When cells expressing EGFP-CP without knockdown of endogenous CPβ were irradiated with the full light dose, no detectable effect was observed (Fig. 2B and SI Movie 3), indicating that the unlabeled endogenous CP in these cells is resistant to CALI and completely masks the effect of EGFP-CP inactivation. As an additional control, we irradiated Rat2 cells expressing soluble EGFP to test for nonspecific laser-induced effects and again observed no morphological change in the irradiated area (SI Movie 4). To objectively quantify the magnitude of the morphological effect induced by CALI, we analyzed the differential interference contrast microscopy (DIC) time-lapse images with an edge detection method (see Methods for details). Application of a Sobel edge detection kernel to DIC images resulted in increased edge detection in regions that contained dorsal structures (Fig. 2C). A normalized index of these structures was generated and plotted against time to display the kinetics of this event (Fig. 2D). CALI of EGFP-CP in KDR cells resulted in an immediate increase in dorsal structures that peaked at ≈80 s after irradiation. This activity then gradually declined (Fig. 2D) until it eventually returned to pre-CALI levels (10–15 min after irradiation, data not shown). The same quantification scheme applied to cells expressing EGFP-CP (no knockdown) or EGFP alone showed no detectable increase in edge content in the irradiated area (Fig. 2D).
CALI of EGFP-CP Induces a Local Increase in Dorsal F-Actin and Barbed Ends.
The morphological changes induced by CALI in the KDR cells were consistent with loss of actin filament capping activity, but we sought to confirm the underlying mechanism of this effect using fluorescent assays of filament growth. To verify that the dorsal protrusive structures observed after CALI of EGFP-CP in KDR cells contained F-actin, cells were fixed and stained with fluorescent phalloidin after laser irradiation. There was perfect colocalization between the phalloidin signal and the dorsal protrusive structures observed with CALI treatment (Fig. 3A), showing that the dorsal protrusions induced by CALI were actin-based.
Fig. 3.
CALI of EGFP-CP in KDR cells generates a local increase in barbed ends of actin filaments and dorsal F-actin. (A) DIC image (Left), fluorescent image of Alexa Fluor 568-labeled phalloidin from a deconvolved three-dimensional stack (Center), and overlay (Right) from an EGFP-CP KDR cell after CALI. Area of irradiation is represented by the red circle. (Scale bar: 20 μm.) (Lower) Zoomed versions of Upper. (B) Fluorescent image of EGFP-CP immediately after laser irradiation (Left) and a pseudocolored fluorescent image of actin filament barbed-ends labeled with Alexa Fluor 568-conjugated G-actin at 3 min post-CALI. (C) Graph showing the relative barbed end increase in the irradiated regions of Rat2 fibroblasts expressing the following constructs: EGFP (n = 12), EGFP-CP KDR (n = 19), EGFP-CP no KD (n = 7), EGFP-Mena (n = 9), and Mena/VASP null MEFs (MVD7 cells) expressing EGFP-Mena (n = 5). Error bars are 95% confidence interval (C.I.). ∗, P < 0.01, compared with all other groups (Tukey's honestly significantly different (HSD) post hoc test; ANOVA P = 6.8 × 10−8).
Acute loss of CP activity should result in an increased number of barbed ends and enhanced filament growth. To test for increased numbers of barbed ends after CALI of EGFP-CP, we used a fluorescent assay that specifically detects them. This assay involves gently permeabilizing cells and adding fluorescently labeled actin monomers that polymerize onto free barbed ends in situ. After fixation, the fluorescent signal quantitatively reports the density of free or growing barbed ends in a particular cellular location (26). We compared the barbed end signal of the irradiated region with similar nonirradiated regions in the CALI-treated cells. CALI of EGFP-CP in KDR cells produced a two-fold average increase in barbed ends where the cells were irradiated (Fig. 3 B and C), whereas there was no detectable increase when we irradiated non-knockdown cells expressing EGFP-CP or soluble EGFP (Fig. 3C). As an additional control to ensure that the local barbed end increase was due to bona fide CP loss of function and not to nonspecific damage caused by irradiating EGFP at the ends of actin filaments, we also irradiated EGFP-Mena in Rat2 and in Mena/VASP null fibroblasts (MVD7 cells). In both cell lines, no significant barbed end increase was observed in the irradiated area (Fig. 3C).
CALI of EGFP-Mena in Rescued Deficient Cells Induces Larger, More Stable Lamellipodial Protrusions.
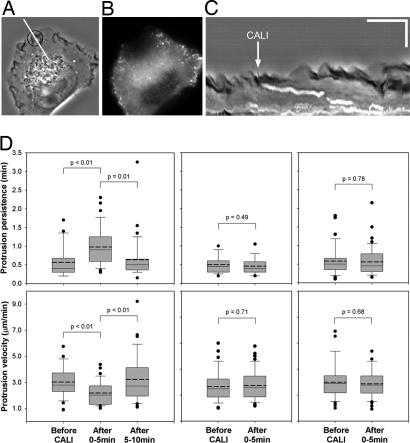
To verify the increased effectiveness of EGFP-CALI in the absence of endogenous proteins, we performed CALI of EGFP-Mena in the Mena/VASP null MVD7 cells. Loss of Mena/VASP at the leading edge of migrating cells results in altered lamellipodial dynamics: lamellipodia protrude more slowly and persist for longer periods, which correlates with faster whole cell migration speeds (15). MVD7 cells rescued with EGFP-Mena were irradiated for 300 ms with a laser power of 6.1 mW/μm2 distributed over a 5-μm-diameter beam. Irradiation parameters were changed from the CP experiments because fluorescence recovery after photobleaching (FRAP) analysis of EGFP-Mena revealed less total recovery and slower recovery kinetics than EGFP-CP (Table 1), indicating that CALI could be performed in smaller regions. Also MVD7 cells are ≈50% smaller than Rat2 fibroblasts. Lamellipodial protrusions were analyzed by using kymography, which characterizes the kinetic signature of lamellipodia undergoing cycles or protrusion and retraction (17, 27).
One of the hallmarks of using CALI to subcellularly inactivate target proteins is that the effect should be reversible providing that there is sufficient exchange of nonirradiated and irradiated protein: fluorescence recovery is indicative of nondamaged protein entering the effected region. FRAP analysis of EGFP-Mena revealed that there was a substantial local exchange of Mena (Table 1). (Note that, whereas lateral transport of “un-CALIed” protein is a requisite step in local functional exchange, additional reactions at barbed ends may delay the complete restoration of functionality.)
To test for reversibility of the CALI effects, protrusion parameters from these experiments were grouped into three categories: before CALI, 0–5 min after CALI, and 5–10 min after CALI. Irradiation of EGFP-Mena at the leading edge resulted in a transient change in lamellipodial dynamics: protrusion persistence increased whereas protrusion velocity decreased in the first 5 min after CALI (Fig. 4 C and D). These changes occurred in a manner that was consistent in direction and magnitude with previously published results (17). Both protrusion parameters returned to pre-CALI levels 5–10 min after irradiation (Fig. 4 C and D), showing the CALI effect was in fact reversible. Also, the effect was limited to the subcellular area that was irradiated as no effect on lamellipodial dynamics was observed in nonirradiated regions (Fig. 4D). To test for nonspecific effects of local fluorophore excitation, we irradiated MVD7 cells expressing soluble EGFP and observed no effect on lamellipodial dynamics (Fig. 4D).
Fig. 4.
CALI of EGFP-Mena in Mena/VASP-deficient cells induces more stable lamellipodial protrusions. (A–C) Phase contrast image of an MVD7 cell (A), fluorescent image of EGFP-Mena immediately after laser irradiation (B), and kymograph of a representative EGFP-Mena CALI experiment (C). (A) Area of irradiation is indicated with a black circle; the line used to generate the kymograph in is indicated by a white line. (In C, horizontal scale bar: 3 min; vertical scale bar: 5 μm.) (D) Box and whisker plots displaying results from kymograph analysis. Kymographs were drawn in the irradiated regions of cells expressing either EGFP-Mena (Left) (34 protrusions before CALI, 38 protrusions 0–5 min after CALI, 34 protrusions 5–10 min after CALI, n = 11 experiments), or EGFP (Center) (39 protrusions before CALI, 49 protrusions after CALI, n = 9 experiments), and also in the nonirradiated regions of cells expressing EGFP-Mena (Right) (37 protrusions before CALI, 53 protrusions 0–5 min after CALI, n = 11 experiments). The top whisker indicates the 90th percentile, the top line of the box is the 75th percentile, the middle line of the box is the 50th percentile, the bottom of the box is the 25th percentile, the bottom whisker indicates the 10th percentile, the mean is indicated with the dotted line, and outlying points are shown as closed black circles. P values for EGFP-Mena experiments are Tukey's HSD post hoc test (persistence ANOVA P = 0.01, velocity ANOVA P = 0.02). P values for all other experiments are from Student's t test.
Discussion
Our results support a number of important conclusions about the use of EGFP as a CALI chromophore and more generally about the CALI technique. First, despite its low efficiency production of reactive photoproducts, our studies demonstrate that EGFP can be an effective CALI chromophore, confirming previous work (10, 11). Using two different proteins, we show that light-induced inactivation of functional EGFP-fusions lead to expected loss-of-function phenotypes. Second, EGFP-CALI can lead to highly selective protein inactivation without significant nonspecific photodamage. In the two examples of CALI reported here, the loss-of-function phenotypes induced by CALI led to positive changes in cellular structures or behaviors, suggesting that the effects are highly specific. Third, CALI of some proteins requires the elimination of unlabeled endogenous protein to be effective. Our knockdown/rescue lentivirus system will greatly facilitate the generation of cell lines meeting this criterion. Finally, different proteins will require empirically developed CALI regimes based on the rate of exchange between bound protein and the cytosolic pool as well as the diffusional mobility of the protein in the cytoplasm.
Is EGFP a suitable live-cell CALI chromophore? The answer is yes, but it must be recognized that EGFP is a low efficiency photosensitizer and requires excitation with intense light to achieve CALI. Neither a 100-W mercury arc lamp nor laser scanning confocal illumination produced CALI phenotypes in the EGFP-CP KDR cells (data not shown). At this juncture, suitable irradiation sources for EGFP-CALI include focused 488 nm laser light from high-powered Argon ion lasers or multiphoton excitation using a frequency doubled Ti-sapphire laser (11). However, the high-energy requirements of EGFP-CALI hold a hidden benefit: routine imaging and protein inactivation can be performed separately because very distinct excitation regimes are required for normal imaging and target inactivation. Newly developed fluorescent proteins such as Killer Red produce reactive photoproducts much more efficiently, and therefore illumination must be carefully controlled to avoid unintended photodamage during imaging (28). Moreover, because many functional EGFP fusions already exist, this technique can readily be applied to many proteins with minimal time spent generating new reagents.
Is protein inactivation by EGFP-CALI specific? Because most previous examples of CALI resulted in the loss of a cellular structure or disruption of a process, it was easy to dismiss them as resulting from nonspecific photodamage or phototoxicity rather than specific protein inactivation. In the case of Capping protein and Mena, these proteins restrain or inhibit cellular processes and inactivation by CALI leads to positive changes in cellular properties. It is highly unlikely that nonspecific photodamage could account for our observed positive cellular phenotypes. Because nonspecific damage is such an important concern with this technique, extensive controls were performed that ensured that laser irradiation alone had no detectable effects in any of our quantitative phenotypic assays. In the case of CP CALI, irradiation of EGFP-CP in cells without knockdown provided the best possible control for nonspecific CALI effects as it is the same functional protein with identical subcellular localization as the protein that induced the loss of function. Together, these results strongly argue against nonspecific effects due to phototoxicity and support the idea that EGFP-CALI can be used to specifically inactivate proteins with high spatial and temporal precision.
This study demonstrates that deficient cells complemented with tagged proteins are the ideal setting for a CALI experiment. We have identified a case with EGFP-CP where the presence of endogenous protein can completely mask CALI loss of function. Whereas EGFP-CALI has been shown to work in cells that retain expression of the endogenous protein [e.g., α-actinin (10)], this result may reflect a special case where the EGFP-tagged protein oligomerizes with the endogenous protein (29). In the CALI of EGFP-CP in KDR cells, we were able to use light fluxes that were 84% lower than the previously reported CALI of EGFP-α-actinin (10) (1.5 mW/μm2 vs. 10.5 mW/μm2, respectively) with identical irradiation times, thus showing that EGFP-CALI can be achieved with almost an order of magnitude less light delivery. The increased sensitivity of EGFP-CP to CALI is likely due to the fact that all of the CP is fused with EGFP, although other factors such as the intrinsic efficiency of protein inactivation due to the local protein environment of the EGFP tag may be involved.
In the case of CALI of CP, we found differences between the RNAi knockdown phenotype and the CALI phenotype: whereas long-term depletion of CP generated a mass increase in filopodia, instantaneous inactivation of CP produced a large number of intrapodia-like protrusions. Both of these phenotypes are consistent with actin polymerization (21, 24). One explanation of this difference is that in the case of CP RNAi, the cell has days to up-regulate pathways to deal with its lost ability to negatively regulate filament growth. For example, it has been recently shown that the actin bundling protein fascin is essential for filopodia formation (30) suggesting that fascin or other functional homologues may be up-regulated or activated when CP is knocked down. The dorsal protrusions induced by the instantaneous activation of CP with EGFP-CALI may reflect true function of CP on the actin network with minimal additional regulation. In this regard, it is of interest that intrapodia in growth cones can be strongly induced by washout of cytochalasin D (24), which depolymerizes actin filaments by capping them at their barbed ends (31). Thus the washout of the drug would result in rapid “uncapping” of actin filaments, a similar event to acute loss of CP through CALI. It will be of interest to compare the complementary effects of CALI and RNAi on other actin binding proteins.
With the two examples presented in this study, the total number of proteins inactivated by EGFP-CALI now stands at five. Our results further validate the use of this technique and demonstrate that is a viable and efficient means of generating spatiotemporal protein inactivation. Our observation that endogenous, untagged protein can mask CALI effects is important for the design and interpretation of future CALI experiments. The knockdown/rescue lentiviral vector described in this work should greatly facilitate the application of CALI to other proteins. Because EGFP has many advantages for imaging and has been widely used to generate functional fusion proteins, EGFP-CALI using this approach will be a useful addition to the cell biologist's toolbox.
Methods
Generation of CP Single-Step Knockdown/Rescue Vector.
The human CPβ coding sequence was amplified by PCR from cDNA and cloned into the pMSCV vector downstream of EGFP. A 5′LTR-EGFP-CPβ PCR product was generated by splice-overlap extension PCR and cloned as a NotI/MfeI fragment into the pLenti-Lox 3.7 vector (32) to produce pLL5.0-EGFP-CPβ. The CPβ shRNA was generated by using the target sequence GCACGCTGAATGAGATCTA (referred to as T2 in ref. 21) and was cloned into pLL5.0-EGFP-CPβ to produce pLL5.1-shCP-EGFP-CPβ.
Cell Culture and Generation of CP Knockdown/Rescue Cells.
Rat2 and MVD7fibroblasts were cultured as described (15). pLL5.1-shCP-EGFP-CPβ was used generate lentivirus by standard protocols (32). Rat2 fibroblasts infected with this virus were cloned by fluorescence-activated cell-sorting and screened for CPβ expression by Western blot. Antibodies against CPβ (mAb 3F2.3) and α-Tubulin (E7) used for Western blotting were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa; the CPβ antibody recognizes both the rat and human form of the protein.
CALI Protocol.
EGFP-CP CALI experiments were performed by using a Spectra Physics Stabilite 2017 (Spectra Physics Laser Incorporated) argon ion laser (488 nm line, 2 W of beam power at the laser head) focused onto a 23.4 μm diameter spot (1/e2 diameter) through a ×60 1.45 N.A. PlanApo TIRF objective (Olympus). Irradiation was controlled with a fast Uniblitz (Vincent Associates) shutter. Laser power at the specimen plane dropped to 625 mW because of optical losses and was measured by placing the sensor of a laser power meter (Model FM with LS 10 head, Coherent Inc.) directly above the objective. Irradiation time was 100 ms, resulting in a 62.5-mJ dose of energy for the CALI experiment. Laser light was focused onto the specimen plane through a 488-nm laser filter set (Z488/10, HQ525/50, Z488RDC, Chroma Technology Corporation), which had significantly higher light yield than the filter set described above used to visualize EGFP. EGFP-Mena CALI was performed essentially as above with the following modifications: laser light was focused on the specimen plane through a ×100 PlanApo 1.3 Phase objective to produce a spot with a 5.0-μm diameter. Power (525 mW) at the laser head resulted in 120 mW of irradiation power at the specimen plane after optical losses. Irradiation time was 300 ms, resulting in a total energy dose of 36 mJ for the CALI experiment.
For more information on imaging and data analysis, see SI Methods.
Supplementary Material
Acknowledgments
We thank T. Marshall and L. Cai (University of North Carolina, Chapel Hill) for reagents and assistance with the barbed end assay, Z. Rajfur for laser and microscope assistance, and Y. Chen for help with FRAP analysis. Funding for this work was provided by the Sontag Foundation, by Carolina Center of Cancer Nanotechnology Excellence/National Cancer Institute Grant 1U54CA119343 (to J.E.B.), and by National Institutes of Health Cell Migration Consortium Grant GM064346 (to K.J.).
Abbreviations
- CALI
chromophore-assisted laser inactivation
- CP
CapZ capping protein
- EGFP-CP
EGFP-human CapZβ
- shRNA
short hairpin RNA
- KDR
knockdown/rescue
- DIC
differential interference contrast microscopy
- FRAP
fluorescence recovery after photobleaching.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701801104/DC1.
References
- 1.Jay DG. Proc Natl Acad Sci USA. 1988;85:5454–5458. doi: 10.1073/pnas.85.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao JC, Roider J, Jay DG. Proc Natl Acad Sci USA. 1994;91:2659–2663. doi: 10.1073/pnas.91.7.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linden KG, Liao JC, Jay DG. Biophys J. 1992;61:956–962. doi: 10.1016/S0006-3495(92)81902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beermann AE, Jay DG. Methods Cell Biol. 1994;44:715–732. doi: 10.1016/s0091-679x(08)60940-1. [DOI] [PubMed] [Google Scholar]
- 5.Marek KW, Davis GW. Neuron. 2002;36:805–813. doi: 10.1016/s0896-6273(02)01068-1. [DOI] [PubMed] [Google Scholar]
- 6.Poskanzer KE, Marek KW, Sweeney ST, Davis GW. Nature. 2003;426:559–563. doi: 10.1038/nature02184. [DOI] [PubMed] [Google Scholar]
- 7.Tour O, Meijer RM, Zacharias DA, Adams SR, Tsien RY. Nat Biotechnol. 2003;21:1505–1508. doi: 10.1038/nbt914. [DOI] [PubMed] [Google Scholar]
- 8.Marks KM, Braun PD, Nolan GP. Proc Natl Acad Sci USA. 2004;101:9982–9987. doi: 10.1073/pnas.0401609101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo J, Chen H, Puhl H. L., 3rd, Ikeda SR. J Physiol. 2006;576:477–492. doi: 10.1113/jphysiol.2006.113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajfur Z, Roy P, Otey C, Romer L, Jacobson K. Nat Cell Biol. 2002;4:286–293. doi: 10.1038/ncb772. [DOI] [PubMed] [Google Scholar]
- 11.Tanabe T, Oyamada M, Fujita K, Dai P, Tanaka H, Takamatsu T. Nat Methods. 2005;2:503–505. doi: 10.1038/nmeth770. [DOI] [PubMed] [Google Scholar]
- 12.Tour O. Nat Methods. 2005;2:491–492. doi: 10.1038/nmeth0705-491. [DOI] [PubMed] [Google Scholar]
- 13.Schafer DA, Cooper JA. Annu Rev Cell Dev Biol. 1995;11:497–518. doi: 10.1146/annurev.cb.11.110195.002433. [DOI] [PubMed] [Google Scholar]
- 14.Schafer DA, Welch MD, Machesky LM, Bridgman PC, Meyer SM, Cooper JA. J Cell Biol. 1998;143:1919–1930. doi: 10.1083/jcb.143.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- 16.Cooper JA, Schafer DA. Curr Opin Cell Biol. 2000;12:97–103. doi: 10.1016/s0955-0674(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 17.Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 18.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 19.Pollard TD, Borisy GG. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 20.Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. J Biol Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 22.Schafer DA, Korshunova YO, Schroer TA, Cooper JA. J Cell Biol. 1994;127:453–465. doi: 10.1083/jcb.127.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, Scita G, Watanabe N. J Cell Biol. 2006;175:947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rochlin MW, Dailey ME, Bridgman PC. Mol Biol Cell. 1999;10:2309–2327. doi: 10.1091/mbc.10.7.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaefer AW, Kabir N, Forscher P. J Cell Biol. 2002;158:139–152. doi: 10.1083/jcb.200203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan AY, Raft S, Bailly M, Wyckoff JB, Segall JE, Condeelis JS. J Cell Sci. 1998;111:199–211. doi: 10.1242/jcs.111.2.199. [DOI] [PubMed] [Google Scholar]
- 27.Surrey T, Elowitz MB, Wolf PE, Yang F, Nedelec F, Shokat K, Leibler S. Proc Natl Acad Sci USA. 1998;95:4293–4298. doi: 10.1073/pnas.95.8.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. Nat Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 29.Edlund M, Lotano MA, Otey CA. Cell Motil Cytoskeleton. 2001;48:190–200. doi: 10.1002/1097-0169(200103)48:3<190::AID-CM1008>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 30.Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. J Cell Biol. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown SS, Spudich JA. J Cell Biol. 1981;88:487–491. doi: 10.1083/jcb.88.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, et al. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.