Abstract
The IFN-induced resistance factor Mx1 is a critical component of innate immunity against influenza A viruses (FLUAV) in mice. Animals carrying a wild-type Mx1 gene (Mx1+/+) differ from regular laboratory mice (Mx1−/−) in that they are highly resistant to infection with standard FLUAV strains. We identified an extraordinary variant of the FLUAV strain, A/PR/8/34 (H1N1) (designated hvPR8), which is unusually virulent in Mx1+/+ mice. hvPR8 was well controlled in Mx1+/+ but not Mx1−/− mice provided that the animals were treated with IFN before infection, indicating that hvPR8 exhibits normal sensitivity to growth restriction by Mx1. hvPR8 multiplied much faster than standard PR8 early in infection because of highly efficient viral gene expression in infected cells. Studies with reassortant viruses containing defined genome segments of both hvPR8 and standard PR8 demonstrated that the HA, neuraminidase, and polymerase genes of hvPR8 all contributed to virulence, indicating that efficient host cell entry and early gene expression renders hvPR8 highly pathogenic. These results reveal a surprisingly simple concept of how influenza viruses may gain virulence and illustrate that high speed of virus growth can outcompete the antiviral response of the infected host.
Keywords: influenza virus reassortants, interferon, Mx GTPase, innate immunity, antiviral
Influenza A viruses (FLUAV) can cause severe disease in humans (1). Little is known about the viral factors and the molecular mechanisms that determine high pathogenicity of specific FLUAV strains such as the currently circulating Asian strains of H5N1 viruses or the H1N1 virus responsible for the 1918 influenza pandemic. Viral factors that play a key role in FLUAV virulence are the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA), the polymerase complex, and the nonstructural protein NS1 (2). HA mediates virus entry into the host cell and is a main determinant of organ tropism. NA supports virus entry and dissociation of progeny particles promoting viral spread. For the 1918 virus, it was shown that in addition to HA and NA, the gene segments encoding the viral polymerase complex contribute to high virulence and pathogenesis in infected mice (3). The viral NS1 protein is an antagonist of the type I IFN (IFN-α/β) system (4) that enhances virulence by suppressing IFN induction after virus infection. Furthermore, the NS1 of certain virus strains inhibits the establishment of an IFN-induced antiviral state (5).
Mice are frequently used as an animal model to study the pathogenesis of FLUAV. However, standard laboratory mice do not possess a complete antiviral defense system because they carry defective alleles of the Mx1 gene (6). The Mx1 gene is under tight transcriptional control of IFN-α/β and codes for a nuclear 72-kDa protein, which represents a decisive antiviral factor that controls FLUAV infections in mice (7, 8). Mx1 inhibits primary transcription of the FLUAV genome by a poorly defined mechanism (9). Because IFN-regulated Mx genes are also present in humans (10), Mx1+/+ mice, which carry functional Mx1 alleles, are believed to better mimic the innate immune system of humans than Mx1−/− mice.
Here we characterized an exceptional FLUAV strain with extraordinary high virulence for Mx1+/+ mice. We present evidence that high virulence resulted from extremely fast virus multiplication in the infected lungs. This property is mediated by a combination of viral genes encoding the viral surface proteins HA and NA and by the viral polymerase complex. We propose that the extremely high virus multiplication speed at the early stage of infection may efficiently outrun a timely antiviral response in the infected host.
Results
A Strain of Influenza A Virus with High Virulence for Mx1+/+ Mice.
By screening a library of laboratory viruses, we identified a strain of FLUAV with surprisingly high virulence for Mx1+/+ mice that usually survive challenges with high virus doses. Virulence of this virus strain increased further during passage in lungs of Mx1+/+ mice (11). Sequence analysis of the complete genome revealed that the highly virulent virus is closely related to but distinct from the Cambridge strain of A/PR/8/34 (12). The LD50 of this virus (designated hvPR8) was ≈10 pfu both in congenic Mx1+/+ B6.A2G-Mx1 and Mx1−/− C57BL/6 mice. hvPR8 thus behaved completely different from all other FLUAV strains that have been tested for virulence in Mx1+/+ mice, including another variant of A/PR/8/34 that is closely related to the Mount Sinai strain of A/PR/8/34 (13), which we frequently use in our laboratory (designated lvPR8). The LD50 of lvPR8 is ≈3 × 103 pfu in Mx1−/− and >106 pfu in Mx1+/+ mice (Table 1 and data not shown). hvPR8 and lvPR8 differ at one or more amino acid positions in each viral protein (Table 2). The largest number of differences are found in the HA and NA genes.
Table 1.
Virulence of parental and reassortant PR8 viruses in Mx1+/+ mice
| Virus* | LD50,† pfu |
|---|---|
| hvPR8 | <102 |
| lvPR8 | >106 |
| rhvPR8 (hv1,2,3,4,5,6,7,8) | <102 |
| rlvPR8 (lv1,2,3,4,5,6,7,8) | 6 × 105 |
| hvPR/lvPB2 (hv2,3,4,5,6,7,8lv1) | 3 × 102 |
| hvPR/lvPB1 (hv1,3,4,5,6,7,8lv2) | 1 × 102 |
| hvPR/lvPA (hv1,2,4,5,6,7,8lv3) | <102 |
| hvPR/lvHA (hv1,2,3,5,6,7,8lv4) | 103 |
| hvPR/lvNP (hv1,2,3,4,6,7,8lv5) | <102 |
| hvPR/lvNA (hv1,2,3,4,5,7,8lv6) | 6 × 102 |
| hvPR/lvM (hv1,2,3,4,5,6,8lv7) | <102 |
| hvPR/lvNS (hv1,2,3,4,5,6,7lv8) | <102 |
| hvPR/lvHN (hv1,2,3,5,7,8lv4,6) | >106 |
| lvPR/hvHA (hv4lv1,2,3,5,6,7,8) | 5 × 104 |
| lvPR/hvNA (hv6lv1,2,3,4,5,7,8) | >106 |
| lvPR/hvHN (hv4,6lv1,2,3,5,7,8) | 5 × 104 |
| lvPR/hvPPP (hv1,2,3lv4,5,6,7,8) | >106 |
| lvPR/hvPPPHN (hv1,2,3,4,6lv5,7,8) | <102 |
*Origin of gene segments in the various reassortant viruses is indicated.
†Groups of animals were intranasally infected with various doses of the indicated viruses. Animals were killed if they were severely ill or if weight loss exceeded 30%.
Table 2.
Comparison of amino acid sequences of lvPR8 and hvPR8
| Viral protein | Amino acid changes in hvPR8 |
|---|---|
| PB2 | I105M, N456D, I504V |
| PB1 | M174I, M205I, K208R, S216N, V587A |
| PB1-F2 | R59K, R60Q |
| PA | Q193H, I550L |
| HA | A65T, P86L, N144T, N146T, Δ147R,* E156A, P200S, E204D, N207K, T220S, I266V, M269R, Y309F, Y343S, H369Q, T398S, I405T |
| NP | A332G, L353V, N430T |
| NA | L15M, K128R, S131N, E314K, K371E, M403I, S451T |
| M1 | N82T |
| M2 | T27A, T39I |
| NS1 | D101E |
| NS2 | I89V |
*Δ indicates the absence of three nucleotides in the HA gene of lvPR8.
IFN Protects Against hvPR8 in Mx1+/+ but Not Mx1−/− Mice.
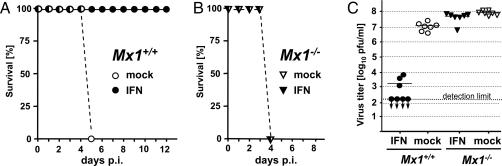
The exceptional virulence of hvPR8 in Mx1+/+ mice could reflect resistance to antiviral factor Mx1, ineffective induction of IFN, or other mechanisms. To test whether hvPR8 acquired resistance to Mx1, we set up an experiment in which Mx1 was induced by stimulation of mice with exogenous IFN before virus infection. Such treatment has previously been shown to be effective in Mx1+/+ mice against FLUAV (14). Groups of mice were treated intranasally with either buffer or hybrid IFN-αB/D, which is highly active in mouse cells (15), and 10 h later the animals were infected with ≈100 LD50 of hvPR8. We found that the IFN-treated Mx1+/+ mice were all protected from virus-induced disease, whereas the mock-treated controls all succumbed to the infection after ≈5 days (Fig. 1A). Interestingly, IFN treatment of Mx1−/− mice was virtually ineffective (Fig. 1B), demonstrating that the protective effects of IFN were mediated almost exclusively through the Mx system. The protective effect of IFN in Mx1+/+ mice resulted in at least 10,000-fold reduced virus titers in lung homogenates at 24 h after infection (Fig. 1C). In contrast, IFN did not significantly affect the multiplication of hvPR8 in Mx1−/− mice, as expected from the lack of protection in the survival experiments. These results demonstrated that the high virulence of hvPR8 is not due to mutations that confer resistance to Mx1.
Fig. 1.
Intranasal application of IFN protects Mx1+/+ but not Mx1−/− mice from lethal challenge with hvPR8. B6.A2G-Mx1 (Mx1+/+) (A) and C57BL/6 (Mx1−/−) (B) mice were treated with either buffer or 5 × 105 units of human IFN-αB/D (five animals per group). Ten hours later, the mice were infected with 1,000 pfu (equivalent to 100 LD50) of hvPR8, and their health status was recorded for up to 14 days. (C) Other groups of animals (n = 7) were treated as above but killed 24 h after infection. Virus titers in lung homogenates were determined. p.i., post infection.
hvPR8 and lvPR8 Are both Poor Inducers of Type I IFN.
Because it was possible that the high virulence of hvPR8 in Mx1+/+ mice resulted from extraordinary suppression of virus-induced IFN synthesis, we determined the capacity of hvPR8 and lvPR8 to induce IFN-β in cell culture and lungs of infected mice. As positive control for these experiments, we used PR8delNS1, a NS1-deficient PR8 mutant virus which is a potent IFN inducer (16). To avoid complications resulting from differences in virus growth kinetics, the mice were infected with a high dose of virus (105 pfu) and killed 10 h later. As expected, we observed that infection with PR8delNS1 resulted in strong up-regulation of IFN-β synthesis (Fig. 2A), but the IFN-β levels in lungs of mice infected with either lvPR8 or hvPR8 were not above the background of mock-treated mice. These results demonstrated that both hvPR8 and lvPR8 are poor inducers of IFN. However, because of the limited sensitivity of the ELISA, it remained unclear whether hvPR8 inhibited the IFN response more strongly than lvPR8.
Fig. 2.
hvPR8 and lvPR8 are both poor inducers of IFN. (A) Groups of C57BL/6 mice (n = 5) were infected intranasally with 105 pfu of PR8delNS1, lvPR8, or hvPR8. Ten hours later, the animals were killed and IFN-β in lung extracts was determined by ELISA. (B) MDCK cells were infected with 0.5 pfu of PR8delNS1, lvPR8, or hvPR8 per cell. Thirteen hours later, RNA was isolated and transcript levels of IFN-β, viral NP, and β-actin were detected by RT-PCR.
In a second experiment, we therefore monitored virus-mediated IFN-β gene induction in Madin–Darby canine kidney (MDCK) cells by RT-PCR. PR8delNS1 strongly induced expression of IFN-β in cells infected with 0.5 pfu per cell for 13 h (Fig. 2B). hvPR8 and lvPR8 also induced the IFN-β gene, but to a substantially reduced extent. Importantly, we observed that the IFN-β transcript levels in cells infected with hvPR8 and lvPR8 did not differ notably (Fig. 2B). These results indicated that hvPR8 and lvPR8 are indistinguishable with regard to inhibition of IFN synthesis in infected cells, suggesting that the high virulence of hvPR8 is not due to enhanced suppression of the host IFN response.
hvPR8 Shows an Unusually Fast Growth Phenotype.
We next investigated in more detail the replication properties of hvPR8 in vitro and in vivo. hvPR8 showed a substantially higher rate of multiplication in MDCK cells than lvPR8 (Fig. 3A). Especially at early times after infection, virus titers in supernatants of MDCK cultures infected with hvPR8 were at least 20-fold higher than in supernatants of cells infected with the same dose of lvPR8. To determine the replication potencies of hvPR8 and lvPR8 in the mouse lungs in the absence of interference by the type I IFN system, we used Mx1−/− mice with defective type I IFN receptors (IFNAR10/0). Virus-induced IFN-α and -β is inactive in such mice so that possible differences between the two viruses in growth restrictions by the type I IFN system should not complicate the comparison. We infected the IFNAR10/0 mice by the intranasal route with 1,000 pfu of either hvPR8 or lvPR8 and determined the virus titers in lungs at 24 h after infection (Fig. 3B). Within this short period, hvPR8 grew to amazingly high titers of ≈5 × 107 pfu per lung, whereas lvPR8 grew to only ≈5 × 104 pfu. Interestingly, the growth rate difference between the two strains was much more pronounced when mouse lungs were analyzed instead of canine cell cultures (Fig. 3A), indicating that the high virulent phenotype of hvPR8 may be influenced by species-specific factors or by factors specifically expressed in lung cells. In any case, these data demonstrated that hvPR8 is able to multiply at much higher speed in mouse lungs than regular FLUAVs.
Fig. 3.
hvPR8 multiplies faster than lvPR8. (A) MDCK cells were infected with 0.001 pfu per cell of hvPR8 and lvPR8. Virus titers in supernatants were determined at the indicated times. (B) IFNAR10/0 mice were infected with 1,000 pfu of either lvPR8 or hvPR8. At 24 h after infection, the lungs were removed and virus titers were determined.
Increased multiplication speed might be caused by more efficient entry into host cells, more effective viral replication, or more efficient virus assembly and budding. To determine whether differences in early steps of the viral multiplication cycle were affected, we infected mouse L929 fibroblasts that do not support productive infection of A/PR/8/34 (data not shown). By using this cell line, we thus measured events in the viral multiplication cycle that occur before virus assembly and budding. When L929 cells were infected with hvPR8, viral NP was present at easily detectable concentrations in the cell lysates as early as 12 h after infection (Fig. 4A). In cells infected with lvPR8, the NP concentration was very low at 12 h after infection, and at 20 h after infection, the NP levels were still much lower than in cells infected with hvPR8 for only 12 h (Fig. 4A). Similar results were obtained when the activities of the two viruses were compared by using an FLUAV minigenome assay (Fig. 4B Left). In this system, a plasmid construct for an artificial viral RNA encoding chloramphenicol acetyltransferase (CAT) was transfected into L929 cells, and the cells were infected with the two viruses. In hvPR8-infected cells, CAT was clearly detectable by ELISA as early as 12 h after virus infection and the levels increased further over the next 8 h (Fig. 4B Left). By contrast, CAT expression in lvPR8-infected cells seemed to start later and remained at 3- to 5-fold lower levels.
Fig. 4.
Early steps of viral multiplication are highly efficient in hvPR8-infected cells. (A) L929 cells were infected with 0.5 pfu per cell of either hvPR8 or lvPR8 and lysed at the indicated times. Levels of viral NP and β-actin were detected by Western blotting. (B) L929 cells were transfected with a CAT-encoding viral minigenome construct. Twenty-four hours later, the cells were infected with 0.5 pfu per cell of either hvPR8 or lvPR8 (Left). Other cultures were transfected for 16 h with a mixture of the CAT-encoding viral minigenome construct and expression plasmids coding for the polymerase subunits PB1, PB2, PA, and NP of either hvPR8 or lvPR8 and Renilla luciferase, pRL-SV40 (Right). Cultures were harvested at the indicated times. CAT activity in the cell lysates was monitored by ELISA. For calculation of the viral polymerase activity, the CAT content was normalized to luciferase activity. The value obtained with the lvPR8 plasmids was set to 1. Mean values of three independent experiments are shown.
Although these data demonstrated that an early step of viral multiplication is highly efficient in hvPR8, it remained unclear whether the RNA polymerase of this virus was more active as compared with lvPR8. To determine viral polymerase activity, we again used the viral minigenome assay. But unlike in the previous experiment in which we had infected the cells with either hvPR8 or lvPR8, in the new experiments we reconstituted the viral RNA polymerase by transfecting expression plasmids encoding PB1, PB2, PA, and NP of either hvPR8 or lvPR8. We observed that the reconstituted polymerase complex of hvPR8 was ≈2-fold more active than the reconstituted complex of lvPR8 (Fig. 4B Right). Because the infection experiment (Fig. 4B Left) demonstrated that hvPR8 was 3- to 5-fold more active than lvPR8, enhanced viral polymerase activity alone is unlikely to account for the higher protein expression and replication activity of hvPR8 in the L929 cells. Thus, additional early steps of the viral replication cycle such as viral entry might also contribute substantially.
Virulence Factors of hvPR8.
To identify the genes of hvPR8 responsible for high virulence, we cloned all segments of hvPR8 and lvPR8 into ambisense expression plasmids and generated recombinant viruses carrying seven segments of hvPR8 and one segment of lvPR8. All segment exchanges were tolerated well, and the resulting viruses grew to high titers in embryonated chicken eggs. To minimize the risk that the recombinant viruses acquired spontaneous mutations that might complicate the picture, all reassortants were generated twice in two independent transfection experiments. To determine which segment exchanges might reduce virulence of hvPR8, we determined the LD50 of the various reassortants in Mx1+/+ mice. We found that viruses carrying either segment 1 (PB2), 2 (PB1), 4 (HA), or 6 (NA) of lvPR8 showed substantial increases in LD50 (Table 1). It should be noted that we did not observe a single case in which the two independently generated reassortants with identical gene constellation yielded discrepant results, indicating that the observed differences in virulence indeed resulted from the indicated segment exchanges and not from unwanted mutations elsewhere in the viral genome. Of the single segment exchange reassortants, virulence was reduced most strongly in hvPR/lvHA that carries HA of lvPR8. The LD50 value of this virus was ≈103 pfu, whereas the LD50 of recombinant hvPR8 (rhvPR8) was <102 pfu as with nonrecombinant parental hvPR8 (Table 1).
It has previously been demonstrated that single exchanges of either segment 4 (encoding HA) or segment 6 (encoding NA) may impair the fitness of the resulting reassortant viruses because of a disturbance of functional interactions between the two envelope glycoproteins (17). We therefore generated reassortant hvPR/lvHN in which segment 4 and segment 6 were both derived from lvPR8 to guarantee optimal functional cooperativity between HA and NA. Nevertheless, we observed that hvPR/lvHN showed drastically reduced virulence (LD50 > 106 pfu) for Mx1+/+ mice which was comparable to the LD50 of rlvPR8 (Table 1). To further substantiate the view that the envelope proteins play a critical role for the virulence of hvPR8, we generated lvPR/hvHA that carries segment 4 of hvPR8 and the other seven segments of lvPR8. This virus was at least 12-fold more virulent in Mx1+/+ mice than rlvPR8 which carries all eight segments of lvPR8 (Table 1). No further virulence gain was observed in lvPR/hvHN that carries segments 4 and 6 of hvPR8. In agreement with this result, we found that lvPR/hvNA, which carries only segment 6 of hvPR8, was not more virulent for Mx1+/+ mice than rlvPR8 (Table 1), pointing toward a minor role of NA for virulence gain.
To determine whether the degree of virulence of the reassortant viruses correlated with their capacity to grow in lungs of infected animals, Mx1+/+ mice were infected and virus titers in the lungs were determined 72 h later (Fig. 5). Introduction of HA and NA from lvPR8 into the background of hvPR8 was detrimental. The resulting reassortant hvPR/lvHN grew more than 4,000-fold less well in lungs of Mx1+/+ mice than rhvPR8. Interestingly, hvPR/lvHN was even more attenuated than rlvPR8, indicating a suboptimal interplay between products of hvPR8 and lvPR8. In contrast, the reciprocal exchange of the envelope proteins had an enhancing effect. lvPR/hvHN grew to ≈50-fold higher titers in lungs of Mx1+/+ mice than recombinant lvPR8. These observations demonstrated that HA and NA are required but not sufficient for high virulence of hvPR8.
Fig. 5.
Growth behavior of recombinant viruses with swapped envelope proteins in Mx1+/+ mice. B6.A2G-Mx1 mice were infected with 1,000 pfu of rlvPR8 and rhvPR8 or reassortants lvPR/hvHN and hvPR/lvHN carrying the envelope proteins of the other strains, respectively. At 72 h after infection, the animals were killed and virus titers in lung homogenates were determined.
To assess the contribution of the viral polymerase complex, we first generated reassortant lvPR/hvPPP that contains the three polymerase subunits from hvPR8 and all other genes from lvPR8. lvPR/hvPPP was not pathogenic (LD50 > 106 pfu) in Mx1+/+ mice (Table 1), demonstrating that the polymerase complex of hvPR8 was unable to increase virulence in the presence of the surface glycoproteins of lvPR8. However, because other reassortants showed that HA and NA from hvPR8 play a dominant role in mediating virulence (Table 1), it was conceivable that the contribution of the viral polymerase complex could only be revealed in the context of the envelope glycoproteins of hvPR8. We therefore generated reassortant lvPR/hvPPPHN that contains the three polymerase subunits as well as HA and NA of hvPR8. As predicted, if the above assumption was correct, virulence of lvPR/hvPPPHN was at least 500-fold enhanced as compared with lvPR/hvHN (Table 1). These findings demonstrated that the viral polymerase contributes substantially to virulence of hvPR8. They further showed that the virulence-enhancing contribution of the polymerase complex is only evident if the virus is equipped with the glycoproteins of hvPR8.
Discussion
We identified an exceptional FLUAV strain that is highly pathogenic for mice carrying functional alleles of the influenza virus-resistance gene Mx1. This virus seems to represent a derivative of the Cambridge line of PR8 (12) that acquired virulence-enhancing mutations during passage in Mx1+/+ mice.
To explain the unusual phenotype of hvPR8 we first considered the possibility that this exceptional virus strain got inert to the antiviral action of the IFN-regulated resistance factor Mx1. We excluded this possibility by showing that Mx1+/+ mice, but not Mx1−/− mice, are highly resistant to hvPR8 if treated with IFN shortly before infection. We next considered the possibility that hvPR8 might carry mutations that lead to particularly strong suppression of the antiviral IFN response in infected cells. Enhanced virulence of human and avian FLUAV strains has been attributed to the NS1 gene that is known to suppress IFN synthesis in virus-infected cells (5, 16, 18, 19). We found that both hvPR8 and standard lvPR8 suppressed early IFN synthesis in lungs of infected mice very strongly, indicating that the NS1 proteins of both viruses act as functional IFN antagonists. Furthermore, no differences were noted between the two viruses with regard to their ability to induce IFN in infected MDCK cells, making it unlikely that enhanced ability to suppress the type I IFN system can account for the phenotype of hvPR8. In fact, the exchange of segment 8 that encodes NS1 did not detectably alter the properties of hvPR8 or lvPR8 (Table 1), supporting the view that NS1 does not determine the high-virulence phenotype of hvPR8. We favor the alternative possibility that hvPR8 escapes innate immune control of mice because of its ability to replicate much faster than standard virus strains. This particular feature of hvPR8 was observed in MDCK cells but was most impressive in mouse lungs. If equal doses of hvPR8 and lvPR8 were applied to mice, hvPR8 grew to ≈1,000-fold higher titers within the first 24 h after infection. Because the innate immune system is dormant in healthy animals and gets activated only after encountering pathogens (20–22), the success of pathogens like hvPR8 seems to be determined at least in part by the ability to replicate efficiently before the innate immune response has fully developed.
The molecular basis of the speedy replication of hvPR8 is not completely understood. From experiments with L929 cells that do not support productive replication of FLUAV, it became clear that some early steps of the viral multiplication cycle occur with greatly enhanced efficacy in hvPR8 (Fig. 4). For highly pathogenic FLUAV isolates, a synergistic effect of the viral glycoproteins combined with enhanced polymerase activity has been demonstrated (3, 23–26). Using a viral minigenome assay, we found that the activity of the reconstituted polymerase complex of hvPR8 was ≈2-fold higher than the activity of the lvPR8-derived complex. This higher activity resulted in a 500-fold increase in virulence of reassortant lvPR/hvPPPHN virus in infected mice, as compared with lvPR/hvHN. Although considerable, the enhanced polymerase activity cannot sufficiently explain the high virulence phenotype of hvPR8. In addition, the viral envelope proteins HA and NA were strictly required for full virulence of hvPR8, indicating that early steps of the viral multiplication cycle including particle attachment to the host cell and membrane fusion also occur more efficiently.
An interesting aspect with implications for future drug treatment studies in mice was that exogenous IFN was virtually inactive against FLUAVs in standard mouse strains but highly efficient in mice with intact alleles of the Mx1 resistance gene. These results reinforce our previous conclusion that Mx1 is the decisive effector molecule that confers resistance toward FLUAV in mice (27). They further suggest that Mx1+/+ mice have to be used as an in vivo model system to evaluate the full potential of type I IFN in the defense against influenza viruses.
Altogether our work reveals a surprisingly simple and efficient viral strategy to successfully evade the IFN-induced innate immune system of the infected host. Unlike other recently identified evasion strategies which rely on the presence of nonstructural viral proteins which specifically target components of the innate immune response (5, 16, 18), the successful strategy used by hvPR8 is based on the concept that the host defense can be rendered ineffective if the pathogen multiplies extremely fast. The virus thus takes advantage of the fact that the antiviral defense system is not permanently active but rather gets induced after the first host cells encounter the pathogen. Because high speed of multiplication is also a hallmark of the recently resurrected 1918 pandemic influenza strain (3), it is tempting to speculate that the evasion strategy of hvPR8 that we identified here may also be used by other successful influenza viruses including the 1918 pandemic strain and the recent highly pathogenic Asian H5N1 viruses.
Materials and Methods
Viruses.
The FLUAV variant hvPR8 is closely related to the Cambridge strain of A/PR/8/34 (H1N1) (12), which is moderately pathogenic for Mx1+/+ mice. hvPR8 was generated by serial lung passages in Mx1+/+ mice (11). lvPR8 is closely related to the Mount Sinai strain of A/PR/8/34 (13), which is not pathogenic for Mx1+/+ mice even at high doses. It was generated by serial lung passages in Mx1−/− mice. Virus stocks were produced in embryonated chicken eggs. PR8delNS1 virus in a recombinant Mount Sinai strain of A/PR/8/34 was grown on Vero cells (16).
Generation of Recombinant Viruses.
RNA was isolated from virus-infected MDCK cells by using TriFast (PEQLab, Erlangen, Germany). Reverse transcription (RT) was performed by using random hexamer primers and MuLV reverse transcriptase. cDNAs were amplified with Pfx DNA polymerase (Invitrogen, Carlsbad, CA) by using primers corresponding to the 3′- and 5′-noncoding regions of the A/PR/8/34 segments. The PCR products were cloned into pDZ vector by using SapI. pDZ is an ambisense expression plasmid with a human RNA polymerase I promoter transcribing negative sense genomic RNA and a chicken β-actin promoter for expression of the recombinant gene products (28) (provided by P. Palese).
For the rescue of recombinant viruses, a mixture of eight pDZ plasmids (0.5 μg each) was transfected into 293T cells by using Lipofectamine 2000 (Invitrogen). To propagate the newly generated viruses, transfected 293T cells were cocultivated with MDCK cells for 48 h in DMEM containing 0.3% BSA, 10 mM Hepes (pH 7.5), and 1 μg/ml of l-1-tosylamido-2-phenylethyl chloromethyl ketone-Trypsin. Recombinant viruses in the supernatants of transfected cells were plaque-purified on MDCK cells. Virus stocks were prepared in embryonated chicken eggs and stored at −80°.
Mice.
Standard C57BL/6 mice with defective Mx1 alleles were purchased from Harlan (Horst, The Netherlands). Congenic B6.A2G-Mx1 mice (29) carrying intact Mx1 alleles were bred locally. IFNAR10/0 mice on the C57BL/6 background were provided by Ulrich Kalinke (Paul-Ehrlich-Institut, Langen, Germany). Six- to 8-week-old animals were used for the challenge experiments, which were performed in accordance with the local Animal Care Committee. Animals were killed if severe symptoms developed. LD50 values were calculated as described (30).
Infection of Mice.
Animals were anesthetized by i.p. injection of a mixture of ketamine (100 μg per gram body weight) and xylazine (5 μg gram per body weight) and infected intranasally with the indicated doses of virus in 50 μl of PBS containing 0.3% BSA.
IFN Treatment of Mice.
Anesthetized mice were intranasally treated with 5 × 105 units of human hybrid IFN-αB/D (15) (provided by H. Hochkeppel, Novartis, Basel, Switzerland).
Determining Virus Titers in Lungs.
Lung homogenates were prepared by grinding the tissue by using a mortar and sterile quartz sand. Homogenates were suspended in 1 ml of PBS, and tissue debris was removed by low speed centrifugation. Virus titers in supernatants were determined on MDCK cells by serial 10-fold dilutions in PBS containing 0.3% BSA. The number of fluorescent cell foci detected after staining with a rabbit antiserum specific for NP was counted. Virus titers are expressed as fluorescent pfu.
Titration of IFN-β in Mouse Lungs.
Supernatants of lung homogenates were assayed for IFN-β content by ELISA (PBL Biomedical Laboratories, New Brunswick, NJ).
Virus Growth Kinetics in MDCK Cells.
Confluent MDCK cells were infected with 0.001 pfu per cell for 1 h. The cells were then washed three times with PBS and incubated in DMEM containing 0.3% BSA, 10 mM Hepes, and 1 μg/ml of l-1-tosylamido-2-phenylethyl chloromethyl ketone-Trypsin. Culture supernatants were collected at different time points and progeny viruses were determined.
Analysis of IFN-β Gene Induction by RT-PCR.
MDCK cells were infected with 0.5 pfu per cell for 13 h, and total RNA was isolated by using TriFast and subjected to reverse transcription. PCR (30 cycles) was performed by using primers specific for canine IFN-β (GenBank accession no. XM_538679, primers from nucleotide positions 1–81 and 594–575), NP of A/PR/8/34 (GenBank accession no. NC_004522, primers from nucleotide positions 580–600 and 1120–1104), and canine β-actin (GenBank accession no. XM_536230, primers from nucleotide positions 378–401 and 805–781).
Minireplicon Assay.
Murine L929 cells were transfected with a plasmid encoding CAT in negative-sense orientation on an RNA minigenome flanked by modified noncoding regions of FLUAV segment 4 derived from plasmid pHL1104 (31) (provided by G. Hobom, Freiburg, Germany). In this construct, expression of the minigenome is under control of the murine RNA polymerase I promoter and terminator. At 24 h after transfection, the cells were infected with 0.5 pfu per cell of the various viruses and harvested at the indicated times. For reconstitution of the viral polymerase, L929 cells were cotransfected with the plasmid encoding the minigenome together with expression plasmids coding for PB2, PB1, PA, and NP and incubated for 16 h. In addition, the cells were cotransfected with an expression plasmid coding for Renilla luciferase under the control of the SV40 promoter, pRL-SV40. The CAT protein content in cell lysates was determined by ELISA (Roche, Indianapolis, IN) and normalized to the luciferase activity.
Western Blot Analysis.
L929 cells were infected with virus at 0.5 pfu per cell for the indicated times and lysed in buffer containing 50 mM Tris (pH 7.5), 280 mM NaCl, 0.5% Nonidet P-40, 2 mM EDTA, and 0.5% Triton X-100. Lysates were subjected to low speed centrifugation and supernatants were treated with SDS and 2-mercaptoethanol. Proteins were separated by SDS polyacrylamide gel electrophoresis and transferred onto polyvinyliden-fluoride membranes. The blots were probed with rabbit antiserum specific for NP and monoclonal mouse antibody against β-actin (Sigma, St. Louis, MO). Horseradish peroxidase-labeled secondary antibodies and the ECL detection system (Roche) were used to detect primary antibodies.
Acknowledgments
We thank Dr. Gerd Hobom for providing CAT minigenome plasmids, Drs. Dimitriy Zamarin (Mount Sinai School of Medicine, New York, NY) and Peter Palese for providing vector pDZ, Dr. Heinz Hochkeppel for providing recombinant IFN-αB/D, Dr. Ulrich Kalinke for the IFNAR10/0 mice, and Simone Gruber for excellent technical assistance. Parts of this work were supported by a grant to G.K. from the Müller-Fahnenberg-Stiftung (University of Freiburg), and by the National Institutes of Health-supported Center to Investigate Viral Interferon Antagonism (U19AI62623) and National Institutes of Health Grants R01AI46954 and P01AI58113 (to A.G.-S.).
Abbreviations
- CAT
chloramphenicol acetyltransferase
- FLUAV
influenza A virus
- hvPR8
highly virulent influenza A/PR/8/34 virus
- NA
neuraminidase
- NS1
nonstructural protein-1
- MDCK
Madin–Darby canine kidney.
Footnotes
References
- 1.Palese P. Nat Med. 2004;10:S82–S87. doi: 10.1038/nm1141. [DOI] [PubMed] [Google Scholar]
- 2.Neumann G, Kawaoka Y. Emerg Infect Dis. 2006;12:881–886. doi: 10.3201/eid1206.051336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Sastre A. Virology. 2001;279:375–384. doi: 10.1006/viro.2000.0756. [DOI] [PubMed] [Google Scholar]
- 5.Seo SH, Hoffmann E, Webster RG. Nat Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- 6.Staeheli P, Grob R, Meier E, Sutcliffe JG, Haller O. Mol Cell Biol. 1988;8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Cell. 1986;44:147–158. doi: 10.1016/0092-8674(86)90493-9. [DOI] [PubMed] [Google Scholar]
- 8.Haller O, Frese M, Kochs G. Rev Sci Tech. 1998;17:220–230. doi: 10.20506/rst.17.1.1084. [DOI] [PubMed] [Google Scholar]
- 9.Pavlovic J, Haller O, Staeheli P. J Virol. 1992;66:2564–2569. doi: 10.1128/jvi.66.4.2564-2569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haller O, Kochs G. Traffic. 2002;3:710–717. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- 11.Haller O. In: Current Topics in Microbiology and Immunology. Henle W, et al., editors. Vol 92. Heidelberg: Springer; 1981. pp. 25–52. [DOI] [PubMed] [Google Scholar]
- 12.Winter G, Fields S, Brownlee GG. Nature. 1981;292:72–75. doi: 10.1038/292072a0. [DOI] [PubMed] [Google Scholar]
- 13.Schickli JH, Flandorfer A, Nakaya T, Martinez-Sobrido L, Garcia-Sastre A, Palese P. Philos Trans R Soc London B. 2001;356:1965–1973. doi: 10.1098/rstb.2001.0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haller O, Arnheiter H, Gresser I, Lindenmann J. J Exp Med. 1981;154:199–203. doi: 10.1084/jem.154.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangemi JD, Lazdins J, Dietrich FM, Matter A, Poncioni B, Hochkeppel HK. J Interferon Res. 1989;9:227–237. doi: 10.1089/jir.1989.9.227. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 17.Wagner R, Matrosovich M, Klenk HD. Rev Med Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Jiang Y, Jiao P, Wang A, Zhao F, Tian G, Wang X, Yu K, Bu Z, Chen H. J Virol. 2006;80:11115–11123. doi: 10.1128/JVI.00993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kochs G, Koerner I, Thiel L, Kothlow S, Kaspers B, Noicolas R, Summerfield A, Pavlovic J, Stech J, Staeheli P. J Gen Virol. 2007 doi: 10.1099/vir.0.82764-0. in press. [DOI] [PubMed] [Google Scholar]
- 20.Fujita T. Science. 2006;314:935–936. doi: 10.1126/science.1135756. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Sastre A, Biron CA. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 22.Haller O, Kochs G, Weber F. Virology. 2006;344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatta M, Gao P, Halfmann P, Kawaoka Y. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 24.Kobasa D, Takada A, Shinya K, Hatta M, Halfmann P, Theriault S, Suzuki H, Nishimura H, Mitamura K, Sugaya N, et al. Nature. 2004;431:703–707. doi: 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
- 25.Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. Proc Natl Acad Sci USA. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salomon R, Franks J, Govorkova EA, Ilyushina NA, Yen HL, Hulse-Post DJ, Humberd J, Trichet M, Rehg JE, Webby RJ, et al. J Exp Med. 2006;203:689–697. doi: 10.1084/jem.20051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staeheli P, Haller O. Interferon. 1987;8:1–23. [PubMed] [Google Scholar]
- 28.Quinlivan M, Zamarin D, Garcia-Sastre A, Cullinane A, Chambers T, Palese P. J Virol. 2005;79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staeheli P, Dreiding P, Haller O, Lindenmann J. J Biol Chem. 1985;260:1821–1825. [PubMed] [Google Scholar]
- 30.Reed LJ, Muench H. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 31.Neumann G, Hobom G. J Gen Virol. 1995;76:1709–1717. doi: 10.1099/0022-1317-76-7-1709. [DOI] [PubMed] [Google Scholar]